Published online Aug 27, 2010. doi: 10.4254/wjh.v2.i8.295
Revised: July 7, 2010
Accepted: July 14, 2010
Published online: August 27, 2010
This editorial reviews the recent evidence showing that Mallory-Denk bodies (MDBs) form in hepatocytes as the result of a drug-induced shift from the 26s proteasome formation to the immunoproteasome formation. The shift is the result of changes in gene expression induced in promoter activation, which is induced by the IFNγ and TNFα signaling pathway. This activates TLR 2 and 4 receptors. The TLR signaling pathway stimulates both the induction of a cytokine proinflammatory response and an up regulation of growth factors. The MDB- forming hepatocytes proliferate as a result of the increase in growth factor expression by the MDB- forming cells, which selectively proliferate in response to drug toxicity. All of these mechanisms are induced by drug toxicity, and are prevented by feeding the methyl donors SAMe and betaine, supporting the epigenetic response of MDB formation.
- Citation: French SW, Bardag-Gorce F, Li J, French BA, Oliva J. Mallory-Denk body pathogenesis revisited. World J Hepatol 2010; 2(8): 295-301
- URL: https://www.wjgnet.com/1948-5182/full/v2/i8/295.htm
- DOI: https://dx.doi.org/10.4254/wjh.v2.i8.295
A great deal of new information on the subject has been published since the last review on Mallory-Denk body (MDB) pathogenesis published in 2007[1]. In that review it was emphasized that MDBs contain keratins K18 and 8, ubiquitin and p62. The relevant proteins and cellular processes that contribute to MDB formation include chronic stress-induced protein misfolding and consequential proteasome overload, a K8-greater-than-K18 ratio, and transamidation of K8 and other proteins. However, the mechanisms involved in the formation of the MDB aggresome have remained elusive. Three new mechanisms of MDB formation have recently been explored. The first is epigenetic mechanisms. The second is the shift from the 26s proteasome to the immunoproteasome. The third is the chronic activation of the Toll-like signaling pathways which stimulate proinflammatory and cell growth pathways. The 3 mechanisms combine to form MDBs.
The first indication that MDB formation is the result of epigenetic changes in gene expression came about when it was demonstrated that feeding S-adenosyl-methionine (SAMe) prevented MDB formation when diethyl-1,4-dihydro-2,4,6-trimethyl-3,5-pyridine decarboxylate (DDC) was re-fed to drug-primed mice[2]. Microarray analysis of the livers from these mice showed that the drug treatment phenotype was remembered by the liver cells at 9 wk, 11 wk and 4 mo after withdrawal of the drug, suggesting that the epigenetic changes were heritable. More significant was that this memory was completely prevented by feeding SAMe with DDC because it meant that SAMe, a methyl donor, had silenced the changes in gene expression that had induced MDB formation. Methylation of H3K9 of histones and DNA leads to gene silencing. Consequently, no induction of MDBs resulted when SAMe was fed. Data mining of the microarray changes in gene expression by the MDB forming liver cells showed that SAMe feeding prevented the changes induced in gene expression caused by drug re-feeding associated with MDB formation. Most notably, SAMe prevented the up regulation of HSP70, caspase 3, Map3K14, glutathione synthase, sequestosome 1 (p62), HDAC 9, alpha fetal proteins (Afp), Kruppel-like factor 6 (KLF-6), Egr2, glutathione S transferase mu2 (Gstm2), ubiquitin D (FAT10), gamma-glutamyl transferase 1 and glutathione peroxidase 2. These changes in gene expression stimulate changes in growth factors, apoptosis, chaperones, antioxidants and preneoplasia.
To further substantiate the changes in gene expression, qPCR was done. FAT10 and KLF6 were markedly up regulated by DDC, and this was prevented by feeding SAMe. The up regulation of FAT10, KLF6, Afp and Gstm2 was observed at all 3 time intervals (9 wk, 11 wk and 4 mo) and SAMe prevented these changes in gene expression at every time interval[2]. Feeding SAMe also affected the expression of acetylation and methylation enzymes (Dnmt3A, HDC9) induced by DDC re-feeding. Parameters involving oxidative stress (GSH levels, 4HNE, and carbonyl protein levels) were not changed during MDB formation, and were unaffected by SAMe feeding. MDB formation by drug-primed liver cells in primary cultures was totally prevented by SAMe in vitro. These results clearly established the role of epigenetic changes in gene expression during MDB formation and prevention by the methyl donor SAMe[2].
To further document the role of epigenetics in the formation of MDBs, FAT10 (UbD) was used as an immunofluorescent marker of the MDB- forming liver cell phenotype[3]. The FAT10 positive cells that formed MDBs proliferated over 7 d of DDC re-feeding to drug-primed mice. The FAT10 liver cell phenotype showed a growth advantage over the intervening normal FAT10 negative hepatocytes in the livers of the mice re-fed DDC[3]. The proliferative response was completely prevented by feeding SAMe. FAT10 protein was markedly increased by Western blot analysis, as well as by immunohistochemistry. Morphologically, the FAT10-positive MDB-forming hepatocytes are often in mitosis, and the nuclei stain positive for cyclin D1 and proliferation cell nuclear antigen, indicating that they were proliferating. The individual FAT10-positive liver cells persisted in the liver among normal hepatocytes for 9 mo after drug withdrawal, at which time FAT10-positive tumors formed. This correlated with a decrease in 8-oxyguanine DNA-glycosylase (OGG1), which would favor the failure of DNA damage repair. Nuclear extracts from drug re-fed mice showed a decrease in Dnmt-3B protein, as was predicted by the microarray results. Tissue cultures of liver cells from drug-primed mice formed MDBs spontaneously, and TSA (a deacetylase inhibitor) prevented MDB formation, whereas 5-aza-cytidine, a transmethylase inhibitor, did not[3].
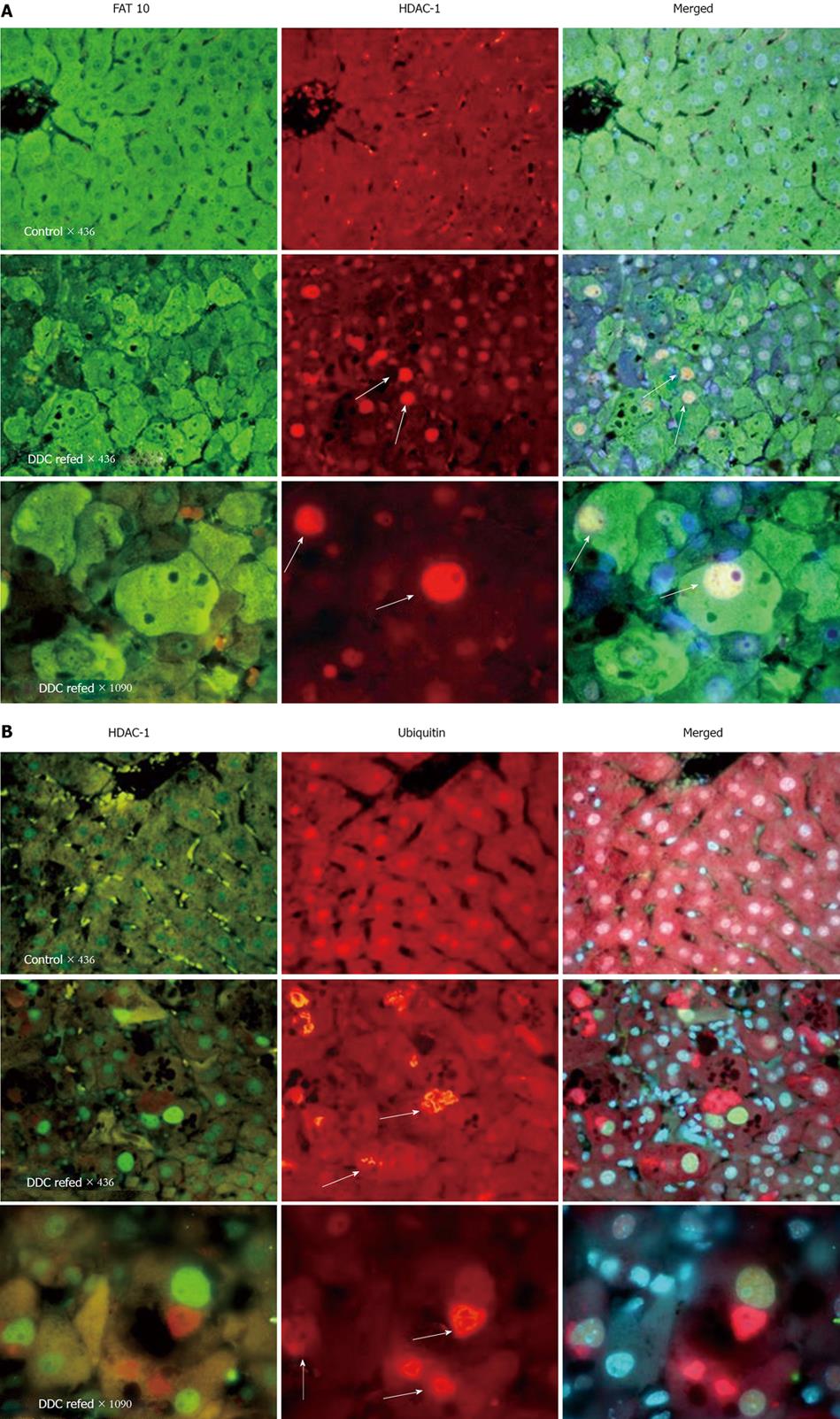
When microarray analysis was carried out on the livers of mice fed DDC for 10 wk and re-fed DDC for 7 d after 1 mo of drug withdrawal, the number of changes in gene expression in the liver was remarkable (3343 genes). The change in expression was completely reversible after drug withdrawal for 1 mo[4]. Almost all of the KEGG functional pathways were up-regulated, especially cell adhesion molecules, actin cytoskeleton, the Toll-like receptor signaling pathway, cytokine-cytokine interaction, and the NFκB signaling pathway. The largest increases in expression were in FAT10 (119 fold), alpha fetal proteins (68 fold) and two growth factors, Ctgf and Gadd 45 g. Western blots confirmed the increased expression of Ctgf and Gadd 45 g. H1F-1 alpha, measured by means of Western blot of nuclear extracts, showed a decrease in HIF-1 alpha both in the DDC- fed for 10 wk and in the DDC-re-fed. Gene mining showed that Sirt 3, (a deacetylase) expression, was down regulated by DDC re-feeding. By Western blot, the expression of HDAC was reduced by DDC feeding. HDAC-1 was increased in the nuclei of FAT10-positive/MDB-forming hepatocytes induced by DDC re-feeding, as indicated by immunofluorescent antibody staining (Figure 1).
H3K9ac was increased by DDC feeding and re-feeding. SAMe did not affect the H3K9ac response to DDC refeeding[5]. The same was true for the decrease in H3K18ac. SAMe also did not prevent the down regulation of Sirt-3 caused by DDC feeding. The histone acetyltransferase (GCN5) levels were increased after DDC refeeding but SAMe did not prevent this. On the other hand, feeding SAMe prevented the decrease in H3K9me3 and H3K4m3 caused by DDC refeeding. H3K9me3 causes gene silencing and H3K4me3 causes gene up regulation globally. SAMe prevented the down regulation of the histone methyltransferase SUV39H1 and the up regulation of the histone methyltransferase SET 7/9. The former methyl transferase methylates H3K9 and the latter methylates H3K4. DDC refeeding increased the expression of the histone demethylase LSD1 but SAMe did not prevent this. SAMe prevented the up regulation of H2A ubiquitination[5]. DDC refeeding altered the expression of enzymes involved in the metabolism of methionine and these changes were prevented by SAMe[5]. Refeeding DDC, up regulated MAT2a, AMD, and Mthfr and down regulated Ahcy and Gnmt. Gnmt demethylates SAMe. Mthfr is a potent inhibitor of Gnmt.
Betaine, another methyl donor, was also protective, like SAMe, when fed to drug-primed mice refed DDC[6]. This was true as assessed by microarray analysis and KEGG functional pathways, although the effect of betaine feeding on these parameters was less dramatic. The same was true where betaine feeding prevented MDB formation induced by DDC refeeding. Betaine feeding also reduced the proliferation of FAT10 positive cells induced by DDC refeeding. This was correlated with the prevention of the up regulation of FAT10 mRNA expression induced by DDC. Betaine feeding also markedly reduced the replication of FAT10 positive MDB forming hepatocytes when DDC was refed[6].
Betaine feeding also prevented the effect of DDC refeeding on the metabolism of methionine[6]. For instance, betaine partially prevented the increase in the expression of Mhtfr induced by DDC refeeding. Mhtfr elevation inhibits Gnmt activity[7]. Betaine plus DDC refeeding increased the expression of MAT1a[6]. It prevented the decrease in expression of Gnmt, Ahcy and Bhmt caused by DDC refeeding. Bhmt converts homocysteine to methionine and reduces the levels of homocysteine. It partially prevented the decrease of AMD-1 caused by DDC refeeding[6]. Betaine feeding also prevented the decrease in SAH levels caused by DDC refeeding[6]. SAH inhibits the methylating activity of Gnmt. It is likely that betaine prevents MDBs by preventing the changes in methionine metabolism, and consequently the reduced methylation caused by DDC refeeding.
When DDC is refed, the activity of the 26s proteasome is decreased by the shift of the expression of the proteasome catalytic subunits from the 26s proteasome proteins to that of the immunoproteasome[8]. The consequence of the loss of 26S proteasome activity is aggresome formation (MDBs) where the lack of protein turnover leads to accumulation of altered and ubiquitinated proteins[9,10]. Microarray analysis of the livers from DDC fed mice showed the up regulation of FAT10 and the catalytic subunits of the immunoproteasome (MECL-1, LMP2 and 7) as well as the immunoproteasome regulatory subunit PA28 alpha. The up regulation was limited to FAT10 over expressing liver cells, which formed MDBs as seen by immunofluorescence microscopy[2,3]. The FAT10 stained positive hepatocytes also over expressed LMP2 and 7 and MECL-1 when viewed by confocal microscopy[8]. Thus only the MDB forming cells and not the intervening hepatocytes had shifted from the 26s proteasome to the immunoproteasome. When liver homogenates from control mice, DDC fed, DDC withdrawn 1 mo and DDC refed 7 d were assayed by Western blot there was an increase in the expression of LMP2 and 7 and MECL-1 only in the group fed DDC for 10 wk and in the group fed DDC for 10 wk, withdrawn from DDC 1 mo and then refed DDC for 7 d. This contrasted with the Western blot results for the 26s proteasome catalytic subunit B5. B5 protein (chymotrypsin-like subunit) was decreased when DDC was fed or refed. The effect of DDC feeding and refeeding on the 26s proteasome chymotrypsin-like catalytic activity was tested[8]. DDC feeding for 10 wk caused a loss of activity which returned to control levels after 1 mo withdrawal. The activity was again reduced when DDC was refed 7 d. SAMe fed with DDC refeeding prevented the loss of the 26s proteasome chymotrypsin-like activity[8]. The accumulation of polyubiquitinated proteins occurred 10 wk after DDC feeding and also after 7 d of DDC refeeding[8]. Again, SAMe fed with DDC refeeding prevented the accumulation of polyubiquitinated proteins caused by DDC. The results clearly indicated that DDC feeding or refeeding caused a switch from the 26s proteasome, which was down regulated, to the immunoproteasome which was up regulated. The immunoproteasome increased at the expense of the 26s proteasome in the MDB forming hepatocytes. The switch was completely prevented when SAMe was fed with DDC. This result was true for the catalytic subunits LMP2 and 7 and MECL-1. When qRT-PCR was done to assay the gene expression of LMP2 and 7 and MECL-1, the increases in the expression of these genes was also prevented by SAMe feeding[8].
The question was then addressed, what was the mechanism that drives the switch in the proteasome expression? The fact that SAMe prevented the switch indicated that an epigenetic mechanism was responsible. However, it remained to be determined as to what changes in gene expression induced the switch. An increase in tumor necrosis factor α (TNFα) and interferon γ (IFNγ) expression stimulates the expression of LMP2 and FAT10[11]. Because of this, the gene expression of the TNFα and IFNγ receptors (IFNγR) was measured by quantitive polymerase chain reaction[8]. The expression of IFNγR1 and IFNγR2, and TNFR2 and 21a were markedly increased by DDC refeeding. SAMe fed with DDC refeeding completely prevented these changes. The expression of TNFα was markedly increased by DDC refeeding and SAMe prevented the increase when fed with DDC refeeding[8]. IFNγ added to primary hepatocyte cultures from DDC-withdrawn mice induced a 4-fold increase in the number of hepatocytes that formed MDBs in vitro[8].
Further in vitro studies on the mouse Hepa1-6 cell line were done to test whether TNFα and IFNγ would induce the expression of the immunoproteasome catalytic subunits and FAT10[12]. It was found that IFNγ but not TNFα alone induced the expression of FAT10, LMP2 and 7 and MECL-1. However, there was a further synergistic increase in the expressions of FAT10, LMP2 and 7 and MECL-1 when both IFNγ and TNFα were added to the culture. The same response was found when downstream phosphorylation of STAT3 and pSTAT1 was measured by Western blot. TNFα, IFNγ and the combination of TNFα and IFNγ activated D1 on the promoter of FAT10. The ISRE sequence (interferon sequence responsive element) was involved in the increased expression of FAT10. In the absence of the D1 ISRE sequence, the treatment with the TNFα-IFNγ co-treatment could not induce the activity of the promoters D2 and D3. The Hepa1-6 cells were subjected to long term treatment with TNFα and IFNγ to see if MDBs would form in vitro in response to these cytokines. MDB-like aggresomes formed, beginning at 21 d of culture. The aggresomes stained positive for FAT10, ubiquitin and CK8, as is characteristic for MDBs[12]. In summary: the data supports the hypothesis that proinflammatory cytokines are responsible for up regulation of the immunoproteasome and consequently, down regulation of the 26s proteasome, which causes the accumulation of undigested proteins and MDB formation in FAT10 positive hepatocytes.
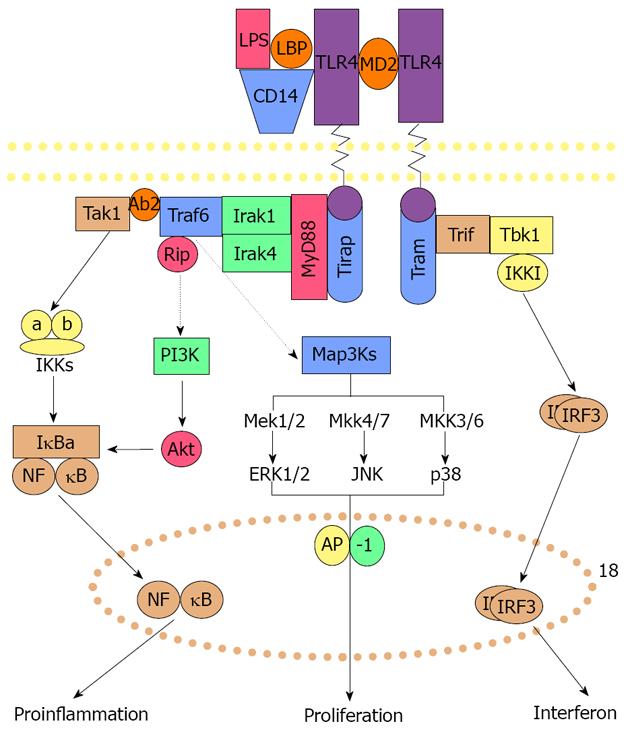
An explanation for the increase in TNFα and IFNγ stimulation of the proinflammatory and cell growth response that leads to MDB formation is needed. TNFα and IFNγ activate the Toll-like receptor (TLR) signaling pathway[13-15]. We therefore investigated this possibility in our model. Mice refed DDC developed an up regulation of the expression of both TLR 2 and 4 as indicated by qPCR[16]. SAMe feeding prevented the up regulation of both TLRs by DDC refeeding. Western blot confirmed the increase in TLR 4 and 2 induced by DDC refeeding and the prevention by SAMe. Downstream components of the TLR signaling pathway were also up regulated, including MyD88 and TRAF-6. SAMe feeding prevented both of these up regulations[16]. To test if the proinflammatory response was activated by DDC refeeding, interleukin (IL)-1 beta was measured by Western blot. DDC markedly increased the expression of IL-1 beta and, conversely, feeding SAMe prevented this[16]. CD-14, which binds lipopolysaccharide (LPS) at the TLR 4 receptor, was also up regulated by DDC refeeding and this was prevented by SAMe feeding. These increased gene expressions would increase the response of the TLR signaling pathway to LPS in the DDC-fed mice.
How would the increase in the proinflammatory signaling pathway and growth of the FAT 10 positive hepatocytes cause the formation of MDBs? The TLR pathway activity stimulates both activation of NFκB mediated proinflammatory response and the AP-1 mediated growth response, as illustrated in Figure 2.
NFκB activation is increased in response to DDC feeding both in vivo and in primary hepatocyte cultures when MDBs form[17-20]. Activation of p38, pERK and JNK pathways have been shown to be up regulated in vitro when MDBs were formed by DDC-withdrawn hepatocytes in primary culture[20-21]. JNK, p38 and ERK activate AP-1 (Figure 2) which leads to cell proliferation. It has been shown that DDC feeding also causes AP-1 activation[19].
Thus both the proinflammatory pathways and growth pathways are activated as a consequence of TLR signaling, which is enhanced by TNFα and IFNγ generated by NFκB activation. Growth of FAT10 positive cells results when AP-1 is activated. MDBs are formed in FAT10 positive hepatocytes as a result of the IFNγ stimulated switch in the expression of the catalytic subunits from the 26s proteasome to the immunoproteasome.
There are two major components involved in the mechanism of MDB formation. The first component is the switch of the metabolism of methionine away from the S-adenosylmethionine-methyltransferase activity pathway to the decarboxylated S-adenosylmethionine pathway[10] and by down regulating Gnmt expression which catalyzes SAMe utilization in the methyltransferase methylation of histones H3K4 and H3K9[5] and DNA[3]. Gnmt activity was also inhibited by the upregulation of Mthfr which is a potent inhibitor of Gnmt[6]. SAMe[5] and betaine[6] both prevented this shift in methionine metabolism, as well as the epigenetic changes in histone and DNA methylating enzymes induced by DDC refeeding. This argues strongly in favor of the epigenetic changes playing a major role in the mechanism of MDB formation.
The second component involves the methyl donors S-adenosylmethionine and betaine. Both prevented the shift from the 26s proteasome to the formation of the immunoproteasome[8], and prevent the up regulation of the TLR 2/4 signaling pathways[16]. Consequently MDB formation was prevented[2,6]. Blocking the up regulation of the TNFα and IFNγ receptor up regulation[16] and TLR2/4 signaling prevented the activation of the NFκB and API up regulation of growth and proinflammation genes.
In conclusion, since both the methyl donors SAMe and betaine were proven to be effective in blocking the two mechanisms involved in MDB formation and the associated growth and proinflammatory gene expression, both donors should be effective in preventing MDB formation in liver in tumor formation in patients if used in clinical trials in the prevention and treatment of alcoholic liver disease.
Peer reviewers: Can-Hua Huang, PhD, Oncoproteomics group, The State Key Laboratory of Biotherapy, Sichuan University, Gaopeng ST, High-Tech Zone, Chengdu 610041, Sichuan Province, China; Nattiya Hirankarn, MD, Associated Professor, Immunology Unit, Department of Microbiology, Faculty of Medicine, Chulalongkorn University, Rama 4 road, Bangkok 10330, Thailand
| 1. | Zatloukal K, French SW, Stumptner C, Strnad P, Harada M, Toivola DM, Cadrin M, Omary MB. From Mallory to Mallory-Denk bodies: what, how and why? Exp Cell Res. 2007;313:2033-2049. |
| 2. | Li J, Bardag-Gorce F, Dedes J, French BA, Amidi F, Oliva J, French SW. S-adenosylmethionine prevents Mallory Denk body formation in drug-primed mice by inhibiting the epigenetic memory. Hepatology. 2008;47:613-624. |
| 3. | Oliva J, Bardag-Gorce F, French BA, Li J, McPhaul L, Amidi F, Dedes J, Habibi A, Nguyen S, French SW. Fat10 is an epigenetic marker for liver preneoplasia in a drug-primed mouse model of tumorigenesis. Exp Mol Pathol. 2008;84:102-112. |
| 4. | Bardag-Gorce F, Dedes J, French BA, Oliva JV, Li J, French SW. Mallory body formation is associated with epigenetic phenotypic change in hepatocytes in vivo. Exp Mol Pathol. 2007;83:160-168. |
| 5. | Bardag-Gorce F, Oliva J, Villegas J, Fraley S, Amidi F, Li J, Dedes J, French B, French SW. Epigenetic mechanisms regulate Mallory Denk body formation in the livers of drug-primed mice. Exp Mol Pathol. 2008;84:113-121. |
| 6. | Oliva J, Bardag-Gorce F, Li J, French BA, Nguyen SK, Lu SC, French SW. Betaine prevents Mallory-Denk body formation in drug-primed mice by epigenetic mechanisms. Exp Mol Pathol. 2009;86:77-86. |
| 7. | Purohit V, Abdelmalek MF, Barve S, Benevenga NJ, Halsted CH, Kaplowitz N, Kharbanda KK, Liu QY, Lu SC, McClain CJ. Role of S-adenosylmethionine, folate, and betaine in the treatment of alcoholic liver disease: summary of a symposium. Am J Clin Nutr. 2007;86:14-24. |
| 8. | Bardag-Gorce F, Oliva J, Li J, French BA, French SW. SAMe prevents the induction of the immunoproteasome and preserves the 26S proteasome in the DDC-induced MDB mouse model. Exp Mol Pathol. 2010;88:353-362. |
| 9. | Bardag-Gorce F, Vu J, Nan L, Riley N, Li J, French SW. Proteasome inhibition induces cytokeratin accumulation in vivo. Exp Mol Pathol. 2004;76:83-89. |
| 10. | Bardag-Gorce F, Riley NE, Nan L, Montgomery RO, Li J, French BA, Lue YH, French SW. The proteasome inhibitor, PS-341, causes cytokeratin aggresome formation. Exp Mol Pathol. 2004;76:9-16. |
| 11. | Lukasiak S, Schiller C, Oehlschlaeger P, Schmidtke G, Krause P, Legler DF, Autschbach F, Schirmacher P, Breuhahn K, Groettrup M. Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene. 2008;27:6068-6074. |
| 12. | Oliva J, Bardag-Gorce F, Lin A, French BA, French SW. The role of cytokines in UbD promoter regulation and Mallory-Denk body-like aggresomes. Exp Mol Pathol. 2010;89:1-8. |
| 13. | Winder AA, Wohlford-Lenane C, Scheetz TE, Nardy BN, Manzel LJ, Look DC, McCray PB Jr. Differential effects of cytokines and corticosteroids on toll-like receptor 2 expression and activity in human airway epithelia. Respir Res. 2009;10:96. |
| 14. | Romieu-Mourez R, François M, Boivin MN, Bouchentouf M, Spaner DE, Galipeau J. Cytokine modulation of TLR expression and activation in mesenchymal stromal cells leads to a proinflammatory phenotype. J Immunol. 2009;182:7963-7973. |
| 15. | Franklin BS, Parroche P, Ataíde MA, Lauw F, Ropert C, de Oliveira RB, Pereira D, Tada MS, Nogueira P, da Silva LH. Malaria primes the innate immune response due to interferon-gamma induced enhancement of toll-like receptor expression and function. Proc Natl Acad Sci USA. 2009;106:5789-5794. |
| 16. | Bardag-Gorce F, Oliva J, Lin A, Li J, French BA, French SW. SAMe prevents the up regulation of toll-like receptor signaling in Mallory-Denk body forming hepatocytes. Exp Mol Pathol. 2010;88:376-379. |
| 17. | Yuan QX, Nagao Y, French BA, Wan YJ, French SW. Dexamethasone enhances mallory body formation in drug-primed mouse liver. Exp Mol Pathol. 2000;69:202-210. |
| 18. | Nan L, Wu Y, Bardag-Gorce F, Li J, French BA, Wilson LT, French SW. The p105/50 NF-kappaB pathway is essential for Mallory body formation. Exp Mol Pathol. 2005;78:198-206. |
| 19. | Nagao Y, Yuan Q-X, Wan Y-J Y, French BA, French SW. Pathogenesis of Mallory body formation: Studies using the drug-primed mouse model. Hepatol Res. 1998;13:42-64. |
| 20. | Nan L, Dedes J, French BA, Bardag-Gorce F, Li J, Wu Y, French SW. Mallory body (cytokeratin aggresomes) formation is prevented in vitro by p38 inhibitor. Exp Mol Pathol. 2006;80:228-240. |
| 21. | Wu Y, Nan L, Bardag-Gorce F, Li J, French BA, Wilson LT, Dedes J, French SW. The role of laminin-integrin signaling in triggering MB formation. An in vivo and in vitro study. Exp Mol Pathol. 2005;79:1-8. |