INTRODUCTION
Metabolic associated fatty liver disease (MAFLD), previously recognized as nonalcoholic fatty liver disease (NAFLD), is a prevalent condition affecting an estimated 25% of the global population, making it the most common liver disorder[1]. The global prevalence of MAFLD has significantly increased in recent years, reaching up to 23.4% in 2019, compared to 17.6% in 1990[2]. This alarming increase in prevalence has led to a substantial socioeconomic burden, with MAFLD becoming a major cause of liver-related morbidity and mortality. In 2019, global deaths due to MAFLD nearly doubled from 93.7 thousand in 1990 to 169.0 thousand, and disability-adjusted life years increased from 2.7 million to 4.4 million. The economic impact is also significant, with healthcare costs for MAFLD patients in the United States averaging 19908 dollars per year, and those requiring liver transplantation facing costs up to 129276 dollars[3]. The etiology of MAFLD is intricate, stemming from a complex interplay of numerous contributing factors. Clinical manifestations of the disease span a spectrum from benign fatty infiltration to more severe forms such as metabolic dysfunction-associated steatohepatitis, progressing to fibrosis, cirrhosis, and hepatocellular carcinoma. Current treatment options for MAFLD primarily include lifestyle modifications, pharmacological interventions targeting insulin resistance and lipid metabolism, and emerging immunomodulatory therapies. However, the growing understanding of the role of immune cells in MAFLD pathogenesis highlights the potential for novel therapeutic strategies targeting immune pathways to improve disease outcomes[1].
Immune cells and MAFLD represent a burgeoning area of research at the intersection of immunology and hepatology[4,5]. Research into the relationship between immune cells and MAFLD has unveiled a complex scenario where immune responses contribute to the initiation and progression of liver steatosis, inflammation and fibrosis[6,7]. Immune cells infiltrate the liver and modulate the local immune microenvironment, thereby influencing disease severity[8-10]. Understanding the precise role of these immune cells in MAFLD is crucial for developing targeted therapies[11,12]. For instance, macrophage polarization into proinflammatory (M1) or anti-inflammatory (M2) phenotypes has been implicated in the regulation of inflammation and fibrosis in the liver[13,14]. Following liver injury, Kupffer cells release inflammatory mediators and chemokines to recruit circulating monocytes. These monocytes differentiate into M1 macrophages, which promote the activation of hepatic stellate cells and subsequent fibrosis through transforming growth factor-β secretion. Conversely, M2 macrophages, which are more abundant in chronic liver injury, contribute to fibrosis by enhancing extracellular matrix deposition. However, recent studies also suggest that M2 macrophages can mediate fibrosis regression under certain conditions, highlighting their dual role in liver disease progression[15,16]. Similarly, the activation state and subset distribution of T cells are considered potential targets for intervention to modulate immune responses and ameliorate disease outcomes[17].
Bibliometric analysis is a critical tool for unearthing the underlying patterns, trends and gaps in the literature[18]. The application of bibliometric analysis to uncover trends in immunology and hepatology represents a novel and valuable approach, offering unique insights into the evolving landscape of research in these fields. This methodological rigor facilitates a comprehensive understanding of the thematic evolution and the interdisciplinary connections to shape the current narrative on immune cells and MAFLD.
MATERIALS AND METHODS
Data collection
To ensure the rigor and relevance of our analysis, we have explicitly defined the inclusion criteria for studies. We chose to restrict our search to the Web of Science Core Collection (WoSCC) database. The WoSCC is renowned for its high-quality indexing and citation tracking, providing a comprehensive and reliable source of bibliometric data.
To enhance the representativeness and retrievability of the data, we extracted records from the WoSCC, using the following search terms regardless of language or document type: Topic = (“immune cells” OR “T cells” OR “B cells” OR “Natural Killer cells” OR “NK cells” OR “Dendritic cells” OR “Macrophages” OR “Neutrophils” OR “Eosinophils” OR “Basophils” OR “Mast cells” OR “Monocytes” OR “Tregs” OR “Plasma cells” OR “Lymphocytes” OR “Myeloid cells”) and translational science = (MAFLD OR NAFLD OR NASH OR “Metabolic associated fatty liver disease” OR “Non-alcoholic fatty liver disease” OR “Non alcoholic fatty liver disease” OR “Metabolic steatohepatitis” OR “Non-alcoholic steatohepatitis” OR “Non alcoholic steatohepatitis”). The search period was restricted from January 1, 2004 to May 20, 2024. To further enhance the quality of the studies included, three researchers from different backgrounds conducted a screening process, excluding communications, brief reports, book reviews, and editorials. Ultimately, a total of 1936 articles were included for analysis. These data were utilized for the visualization analysis of countries/regions, institutions, authors, journals, fields, co-cited references, keywords, genes, and diseases. Gene and disease data were sourced from the Citexs Research Assistant (https://www.citexs.com) for the visualization analysis of genes and diseases.
Statistical analysis
The co-occurrence of countries/regions, institutions, authors, fields, keywords, genes, and diseases was analyzed using VOSviewer 1.6.18 (Centre for Science and Technology Studies, Leiden University, The Netherlands), Pajek 64 5.16 (University of Ljubljana, Slovenia), and Scimago Graphica 1.0.35 (United States). VOSviewer was chosen for its robust capabilities in mapping co-occurrence networks, providing an intuitive visualization of research landscapes. It is particularly effective for identifying clusters and patterns within large datasets. In the visualizations generated by these software tools, nodes are represented by spheres and text labels, with node size indicative of the sphere’s volume; different colors denote distinct clusters; lines between nodes signify co-occurrence relationships; and the thickness of these lines correspond to the strength of co-occurrence.
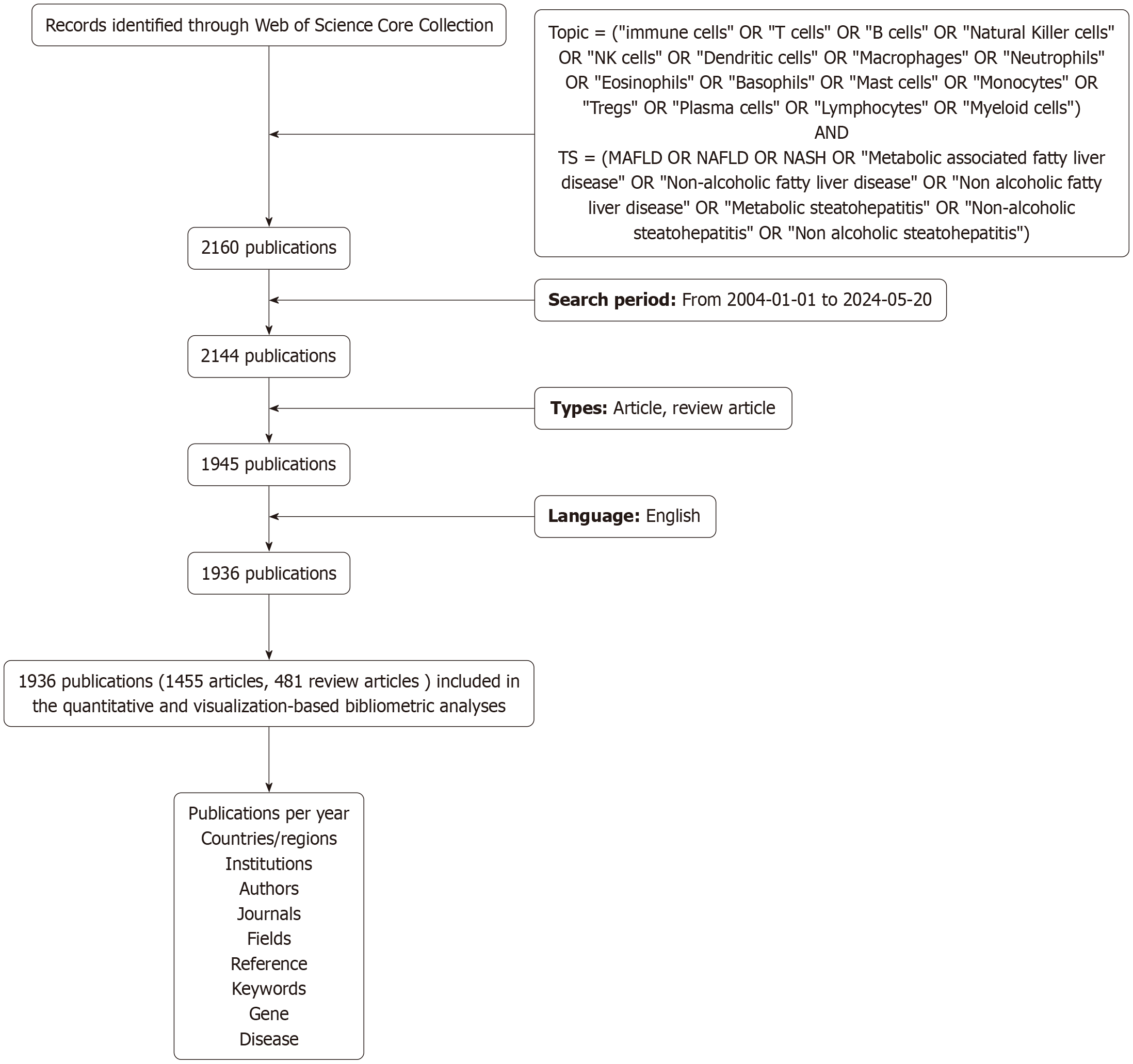
The software Citespace 6.3.R1 (Chen CM, China) was used for visualization analysis of journals, co-cited references, and keywords, with the generation of relevant visual maps. CiteSpace was selected for its specialized functions in analyzing co-citation networks and identifying emerging research hotspots through timeline analysis. It allows for the detection of key references and emerging trends in the literature. For the co-citation cluster analysis map in CiteSpace, the parameters were set as follows: Time slicing from 2004 to 2024, one year per slice, and selection criterion (k = 4). Distinct spheres represent unique co-cited references, with sphere size proportional to the number of citations the publication has received. Lines between spheres indicate co-citation relationships. The superimposed rings within each sphere, varying in size and color, represent the quantity of cited references and their respective periods. For the keyword frequency cluster timeline analysis in CiteSpace, the parameters were set as time slicing from 2004 to 2024, one year per slice, and selection criterion (k = 10). The Clusterprofiler, enrichplot and ggplot2 R packages were utilized for the visualization of Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes enrichment analysis of the extracted genes. For the construction and visualization of protein-protein interaction (PPI) networks extracted from the proteins, the STRING online platform (http://string-db.org) and Cytoscape 3.8.2 (Cytoscape Consortium, United States) were used. Figure 1 shows the flowchart for the search strategy and selection process in this study.
Figure 1 Flowchart of search strategy and selection process.
MAFLD: Metabolic associated fatty liver disease.
RESULTS
Publication trends, collaboration, and authorship contributions
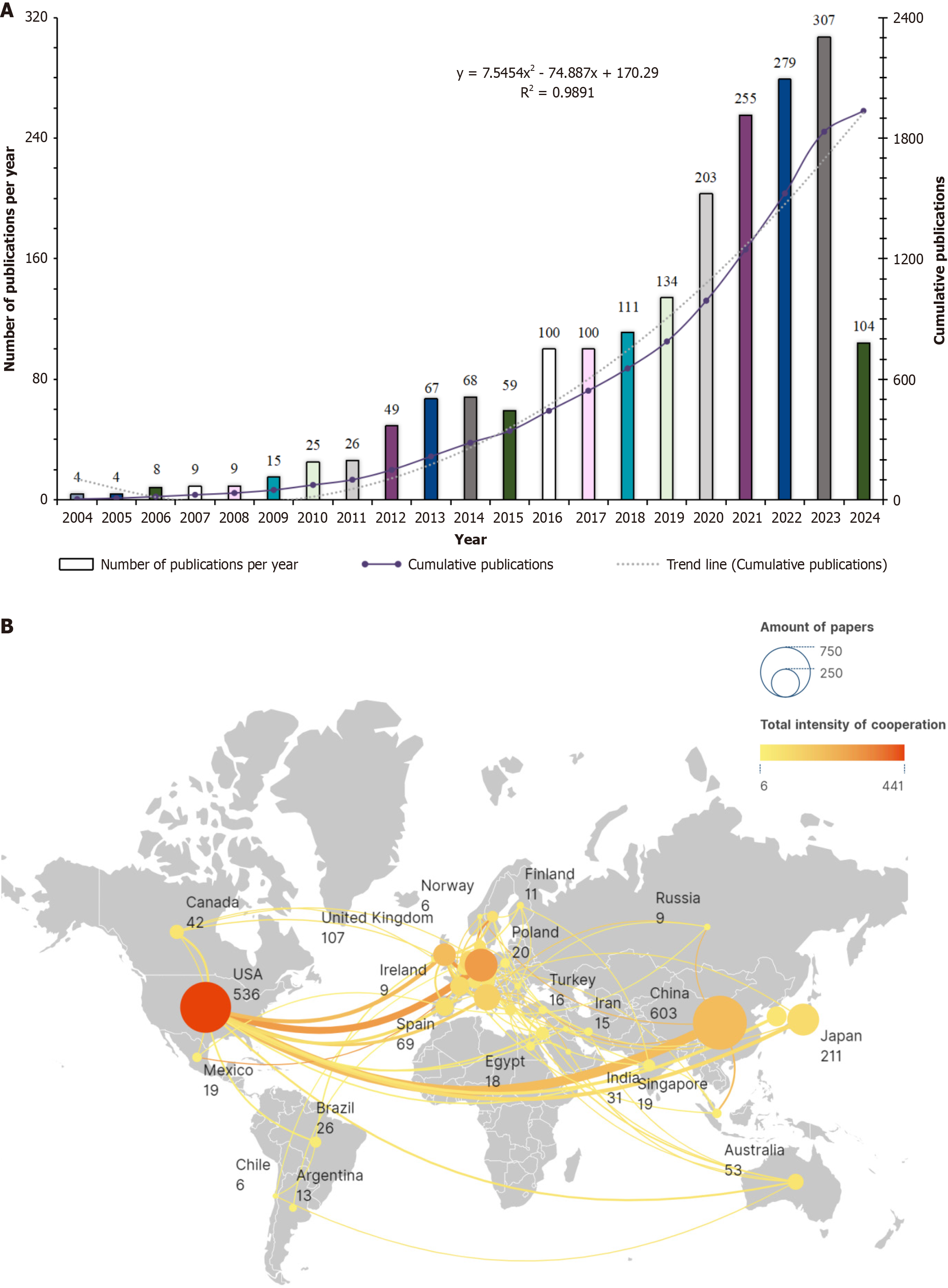
From January 1, 2004 to May 20, 2024, the literature on immune cells and MAFLD showed a significant increase, with 1936 articles. The average annual publication rate is 92.19 articles, with a consistent rise in both annual and cumulative volumes, except for a minor dip in 2015. The most notable growth was in 2006 with a 100% increase, followed by 2012 and 2016, with increases of 88.46% and 69.49%, respectively. This surge in publication volume indicated heightened research interest in immune cells and MAFLD during these years. A polynomial function, y = 7.5454x2 - 74.887x + 170.29 (R2 = 0.9891), where x is the year and y is the cumulative publication volume, was used to model the growth trend, showing a strong fit and suggesting an annual increase in research focus (Figure 2). Geographically, the research was conducted across 65 countries, with the United States showing the most collaboration, particularly with China, Germany and the UK. China led in publication volume with 603 articles, significantly ahead of Italy (143 articles) and the United States (536 articles). Switzerland stood out with the highest average citation rate per article at 134.53, followed by Australia with 99.92 (Figure 2B).
Figure 2 Publication trends, collaboration and authorship contributions.
A: Publication trend analysis chart; B: Study the national and regional documents and cooperation relationship map. USA: United State.
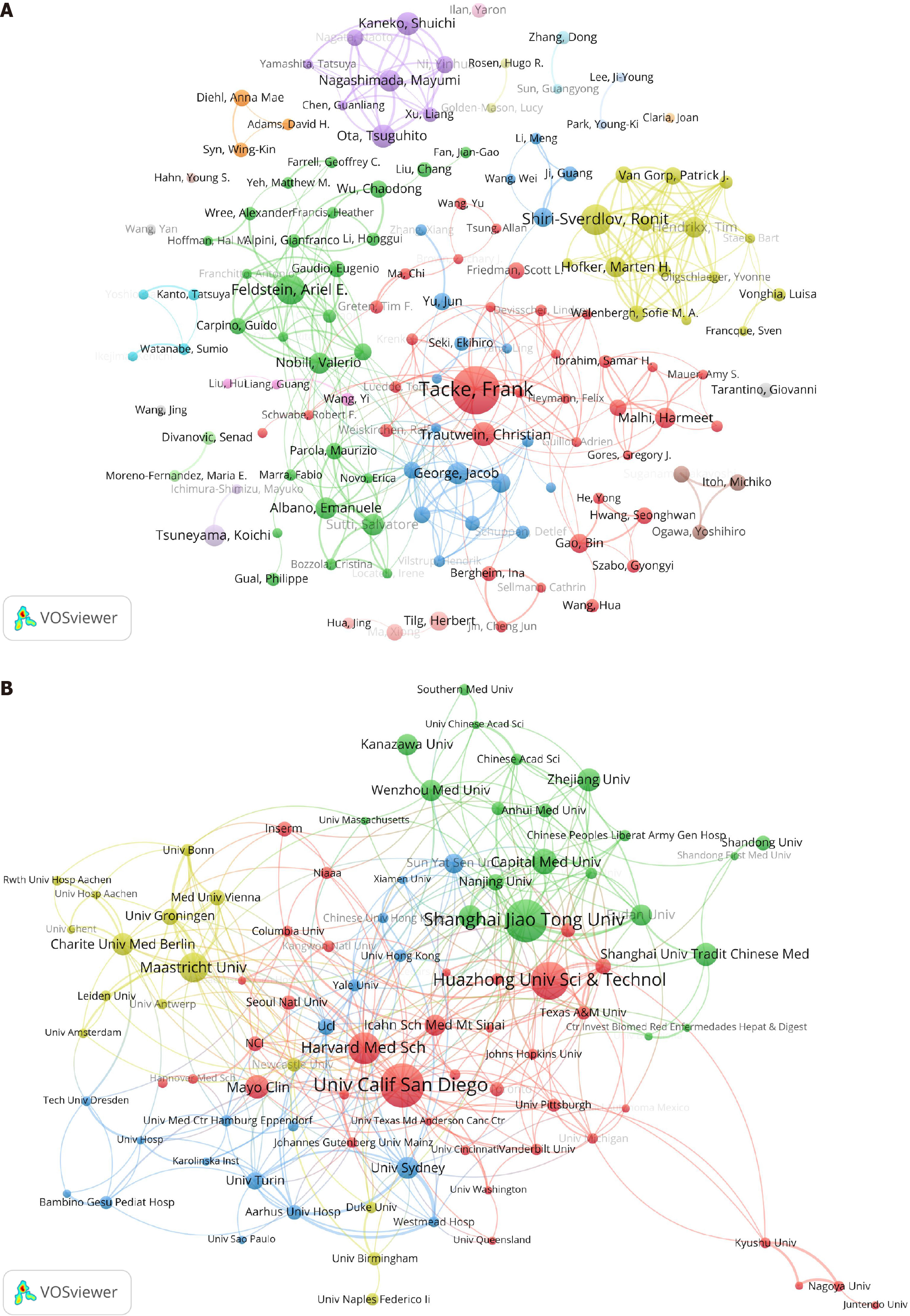
In terms of author contributions, 11994 authors contributed to the 1936 articles on immune cells and MAFLD. By setting a minimum publication threshold of six articles per author, the author publication and collaboration network map was created. R. Shiri-Sverdlov ranked first in total link strength, indicating the strongest willingness to collaborate, with M.H. Hofker being the most frequent collaborator. E. Albano and S. Sutti showing the highest collaboration intensity, indicating the closest cooperative relationship. F. Tacke led in publication count with 35 articles, followed by R. Shiri-Sverdlov, A.E. Feldstein, C. Trautwein and T. Ota, with 21 articles, 20 articles, 16 articles and 15 articles respectively (Figure 3A). The publication contributions of institutions were also analyzed, with 2409 institutions publishing 1936 articles on immune cells and MAFLD. By setting a minimum publication threshold of 10 articles per institution, a visualization map of institutional publications and collaboration relationships was created. Harvard Medical School had the highest total link strength value, indicating the strongest willingness to collaborate, with the Massachusetts General Hospital and Harvard Medical School having the closest collaboration. The University of Sydney and Westmead Hospital had the highest collaboration intensity, indicating the closest cooperative relationship. The University of California San Diego topped the publication count with 50 articles, followed by Shanghai Jiao Tong University and Huazhong University of Science and Technology, with 48 articles and 43 articles, respectively (Figure 3B). This analysis highlighted the global interest and interconnectedness in the field of immune cells and MAFLD research, with significant contributions from various authors and institutions worldwide. The United States and China stood out as major players in terms of collaboration and publication volume, while Switzerland and Australia were noted for their high citation rates per article.
Figure 3 Distribution of authors and research institutions.
As shown in the figure, the nodes are composed of circles and text labels, with different colors representing distinct clusters. The thickness of the lines between circles indicates the strength of collaboration between authors/institutions. A: A visual map for VOSviewer network among authors; B: A visual map for VOSviewer network among institutions.
Distribution of disciplines and journals
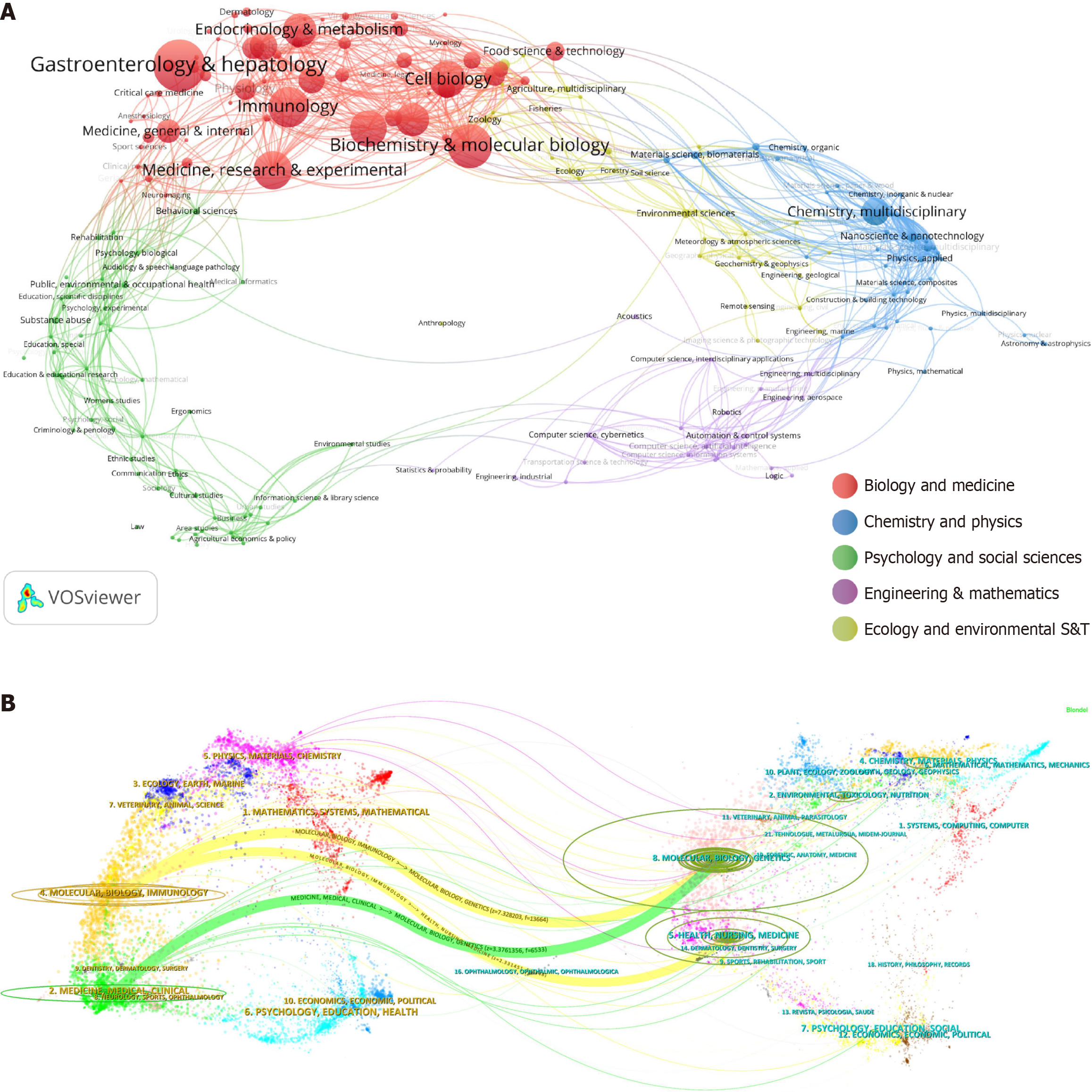
Statistical analysis from WoSCC categorized 227 subject areas, with VOSviewer visualizing1936 immune cell and MAFLD articles into five major fields, mainly biology and medicine. Subfields like gastroenterology and hepatology, biochemistry and molecular biology, and immunology were prominent (Figure 4A). The dual map shows journal positions relative to disciplines, with immune cell and MAFLD research in molecular, biology, immunology, medicine, and clinical journals, citing molecular, biology, genetics, health, nursing, and medicine. The map’s points and curves represent journals and citation links, revealing inter disciplinary citation flows (Figure 4B).
Figure 4 Distribution of disciplines and journals.
A: Field analysis chart. The VOSviewer software was used for visualization analysis, clustering the 1936 articles into five major fields; B: Dual-map overlay of journals. The figure is divided into two parts: The left side represents the citing journals, and the right side represents the cited journals. The more papers a journal publishes, the longer the vertical axis of the ellipse; the more authors involved, the longer the horizontal axis of the ellipse.
Co-cited references and burst detection
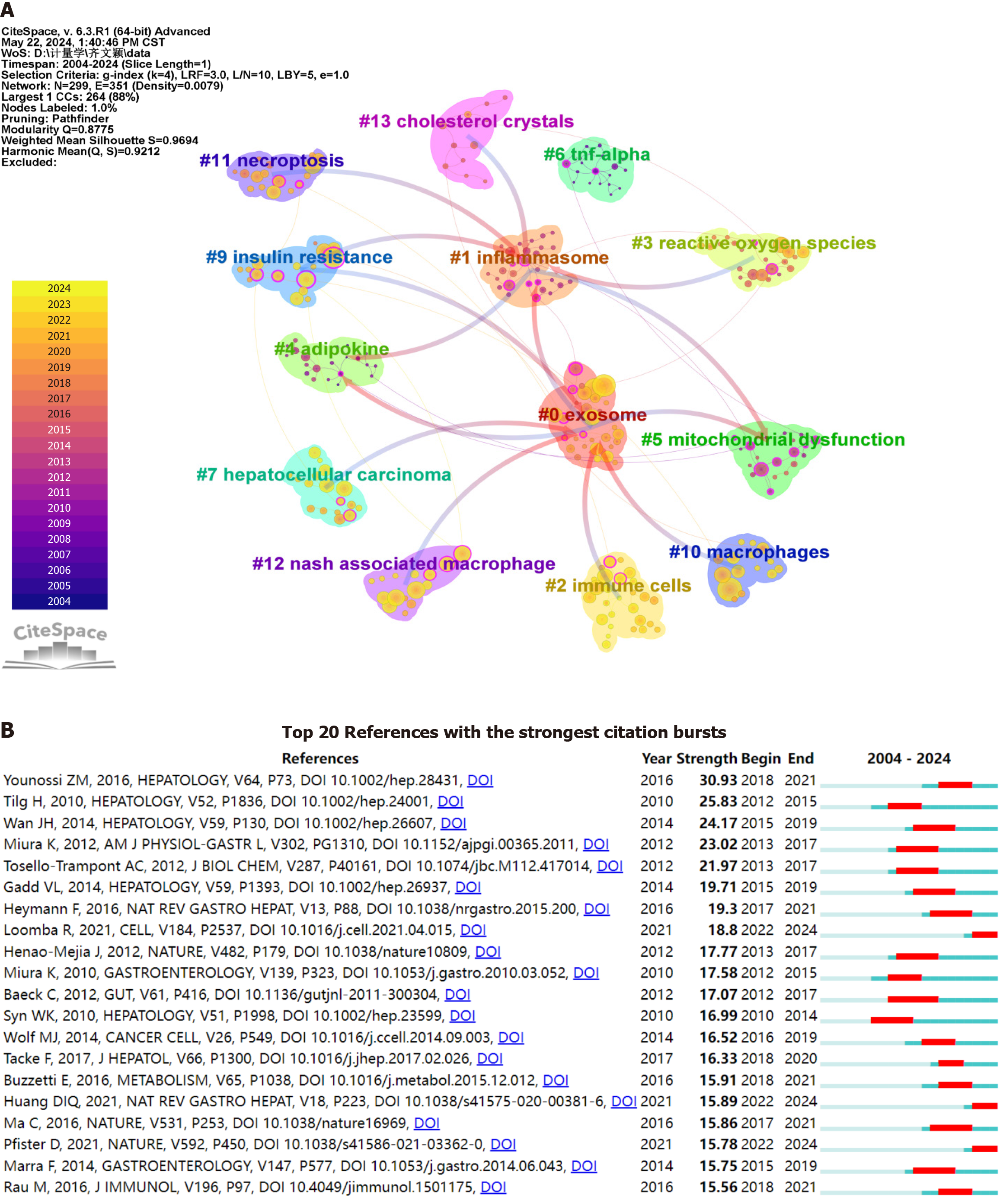
Using CiteSpace, a bibliometric analysis was performed on literature co-citation trends related to immune cells and MAFLD from 2004 to 2024. The analysis utilized time slicing and a selection criterion of k = 4, with circle sizes indicating co-citation frequency and colors denoting citation timeliness. Notably, the modularity and silhouette values of 0.8775 and 0.9694, respectively, confirmed significant and convincing clustering. The literature was categorized into 13 distinct clusters, highlighting key research areas in the field (Figure 5A).
Figure 5 Co-citation clustering and citation burst analysis.
A: Cluster analysis map of co-cited references. The parameters in CiteSpace were set as follows: Time slicing (2004-2024), one year per slice, and selection criterion (k = 4). The size of the superimposed rings (i.e., the cumulative size of the rings on the timeline) is proportional to the number of co-citations. Purple indicates relatively early citation years, while yellow indicates more recent citation years. Overlapping colors suggest that the article was cited in each corresponding year. Lines between circles represent co-citation relationships. Nodes marked in magenta are key nodes with centrality greater than 0.1; B: Citation burst analysis map. A citation burst refers to a significant increase in the number of citations a particular article receives within a specific time frame. The red areas in the graph indicate the periods during which the citations of each article surged.
Bibliometric analysis was conducted using CiteSpace to identify the top 20 most cited documents related to immune cells and MAFLD, with a focus on citation bursts between January 1, 2004 and May 20, 2024. A citation burst refers to a significant increase in the number of citations a particular article receives within a specific time frame, with the red areas in the graph indicating the periods of such surges in citations. This suggests that these recent publications have had a notable impact on the current research landscape in the area of immune cells and MAFLD (Figure 5B).
Research hotspots and frontier analysis
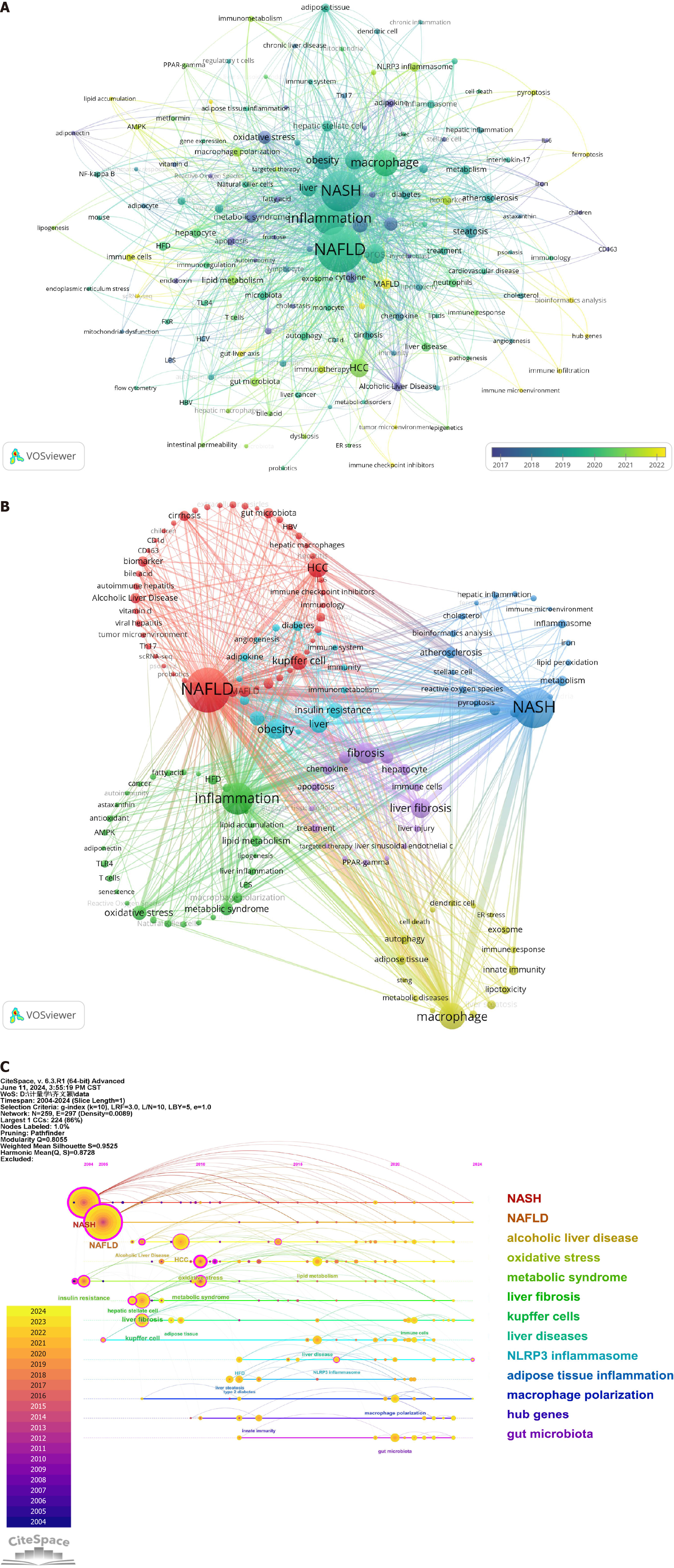
Employing VOSviewer, a co-occurrence cluster analysis of article keywords was conducted, with a minimum occurrence threshold of six times. From an initial 3085 keywords, 145 were selected post-deduplication to create a visual map. The map’s nodes, composed of circles and labels, reflect keyword frequency and average year of appearance, with color gradients indicating temporal trends (Figure 6A). Notable early keywords included “alcoholic liver disease” and “oxidative stress,” while “MAFLD” and “bioinformatics analysis” emerged later, with “hub genes” appearing most recently, suggesting emerging research interests.
Figure 6 Research hotspots and frontier analysis.
A: Keyword timeline map; B: Keyword frequency cluster analysis chart; C: Hotspot keyword frequency cluster timeline analysis chart. NASH: Negatively regulates nonalcoholic steatohepatitis; NAFLD: Nonalcoholic fatty liver disease.
Further analysis categorized keywords into distinct clusters, each representing different research directions. The red cluster focuses on MAFLD with keywords such as “NAFLD”, “HCC”, and “Kupffer cell”, while the blue cluster centers on metabolic dysfunction-associated steatohepatitis with terms such as “NASH”, “atherosclerosis”, and “NLRP3 inflammasome”. The green cluster is associated with inflammation, featuring keywords such as “inflammation” and “oxidative stress”, and the yellow cluster pertains to macrophages with terms such as “macrophage” and “adipose tissue”. The purple cluster is linked to fibrosis with keywords including “fibrosis” and “liver fibrosis”, and the light blue cluster is related to obesity with terms such as “obesity” and “insulin resistance” (Figure 6B).
Figure 6C presents a timeline analysis, illustrating the evolution of keyword co-occurrences. Central nodes in magenta indicate high centrality, while the timeline’s horizontal alignment of keywords within clusters reflects their interconnectivity and field significance. The progression from top to bottom and left to right delineates the emergence and development of research topics, offering a macroscopic view of knowledge and research trends in the immune cells and MAFLD domain. This visualization is instrumental for researchers to grasp overarching trends and identify pivotal areas of study, such as the continuous development of clusters from 0 NASH to 12 gut microbiota, highlighting the dynamic nature of research in this field.
Cluster analysis of associated diseases
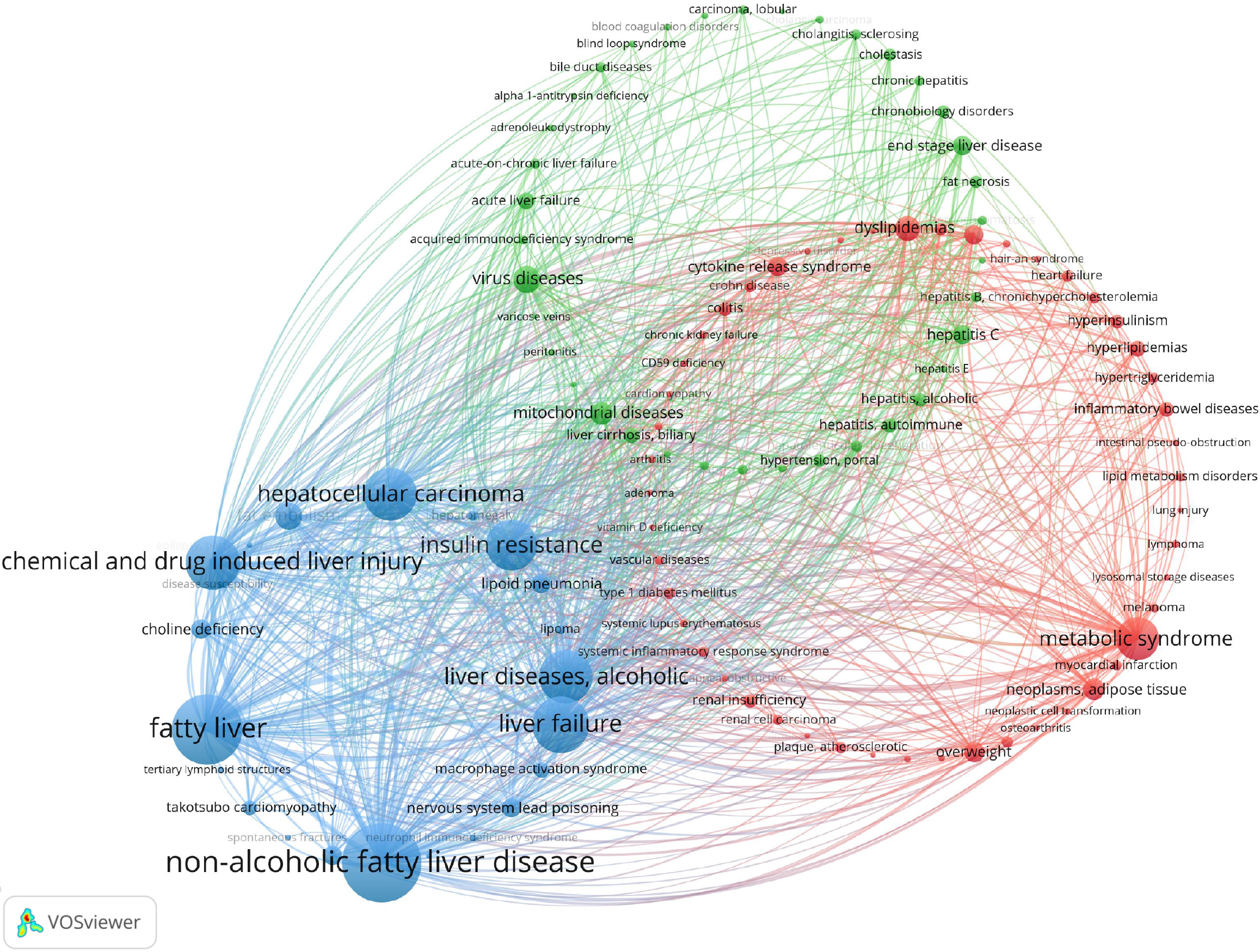
Using VOSviewer, a co-occurrence analysis of 1421 immune cell and MAFLD-related diseases from 1936 articles was conducted, with a 20-occurrence threshold. The resulting map visualizes disease frequency and relationships, with three main clusters identified. The blue cluster focuses on NAFLD and related liver conditions. The red cluster centers on metabolic syndrome and its complications. The green cluster addresses viral diseases, including hepatitis and liver cirrhosis, highlighting key research directions in the field (Figure 7).
Figure 7
Disease association cluster analysis map.
Cluster analysis of associated genes
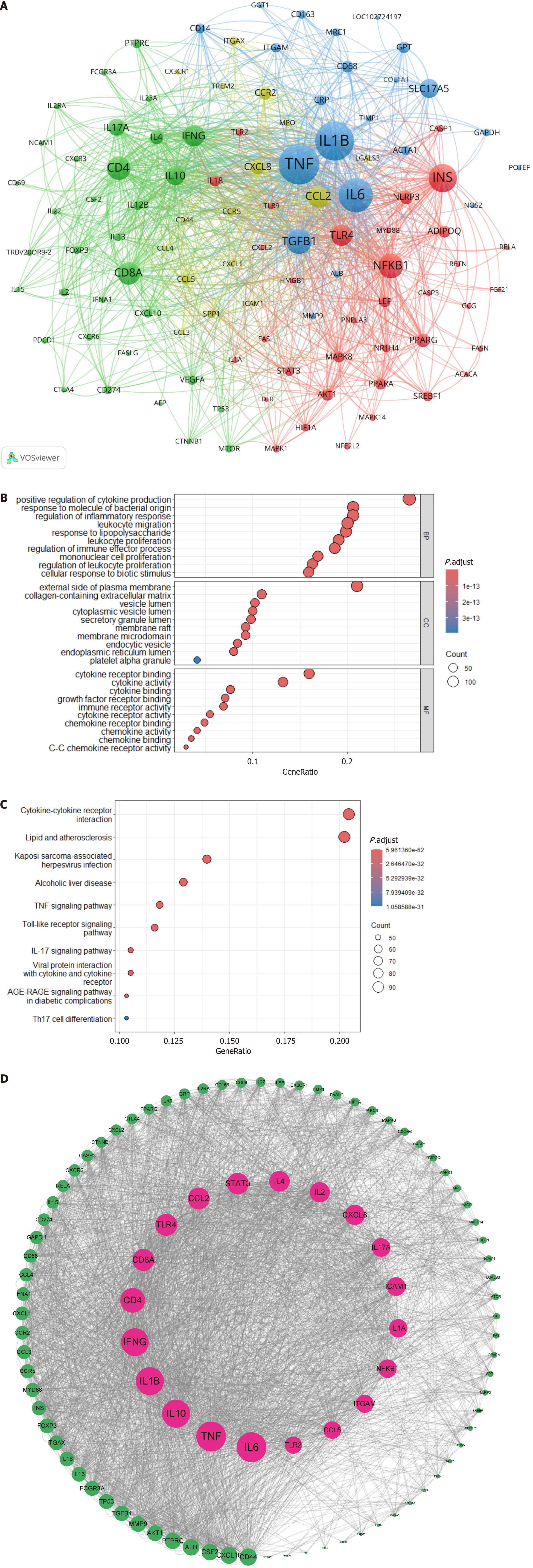
Using VOSviewer software, a co-occurrence cluster analysis was conducted on genes related to immune cells and MAFLD research. The Citexs platform extracted a total of 3254 genes from 1936 articles, with a minimum occurrence threshold of 40 times foreach gene to be included in the visualization map. In the map, nodes composed of circles and labels represent genes, with the size of the circle being positively correlated to the frequency of gene occurrence and the thickness of the lines between circles indicating the strength of the relationships between genes. Different colored nodes form different clusters, each representing gene clusters in different fields. Notably, the red cluster with the highest heat is for insulin (INS); the yellow cluster for chemokine CC ligand (CCL)2; the green cluster for cluster of differentiation (CD)4; and the blue cluster for tumor necrosis factor (TNF) (Figure 8A).
Figure 8 Cluster analysis of associated genes.
A: Gene association cluster analysis map. This map visualizes the co-occurrence of genes related to immune cells and metabolic associated fatty liver disease. Different types of genes were classified by clustering algorithms and represented by four distinct colors: Inflammation-related genes are indicated in blue; chemokine-related genes in yellow; immune-related genes in green; and lipid metabolism-related genes in red; B: Gene Ontology enrichment analysis bubble chart. Bubble size denotes gene count, and color depth reflects enrichment level; C: Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis bubble chart. The x-axis shows the number of enriched genes, and the y-axis lists pathways; D: Protein-protein interaction network construction analysis map. Nodes represent proteins, and edges indicate interactions.
The GO bubble chart illustrates GO Terms with bubble size denoting gene count and color depth reflecting enrichment. The Gene Ratio of the x axis shows the proportion of GO-associated genes in the genome. Higher values suggest significant biological roles. The y axis categorizes terms into biological processes, molecular functions and cellular components. Enriched terms in biological processes included cytokine regulation and inflammatory response, cellular components included plasma membrane and extracellular matrix, and molecular functions included cytokine binding and activity (Figure 8B).
The Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis selected the top 10 signaling pathways and plotted a bubble chart, with the x axis representing the number of significantly enriched genes in each pathway and the y axis representing different signaling pathways. The Gene Ratio refers to the proportion of genes associated with the GO term in the background gene set (the entire genome of the human species under study); red represents significant pathways after enrichment, and blue represents pathways with low significance. The immune cells and MAFLD research field was significantly related to signaling pathways such as cytokine-cytokine receptor interaction and lipid and atherosclerosis (Figure 8C).
Through the Citexs Research Assistant (https://www.citexs.com), big data platform, the top 100 proteins mentioned in the articles (with a minimum occurrence of 42 times) were imported into the STRING platform, with Homo sapiens as the species and a high confidence level (0.700) for PPI network information. The network information obtained was imported into the Cytoscape software to calculate the node degree value.
After importing the selected genes into the STRING database, a PPI network was obtained. The network contained 100 nodes and 1474 edges, with an average node degree value of 29.5. Using Cytoscape software, the node degree value was calculated, and based on the degree value, a core protein PPI network map was constructed from smallest to largest degree. The top 10 targets were interleukin (IL)-6, TNF, IL-10, IL-1b, interferon g, CD4, CD8A, toll-like receptor 4, CCL2, and signal transducer and activator of transcription (STAT) 3, which may be core proteins in the field of immune cells and MAFLD (Figure 8D).
DISCUSSION
General information
This bibliometric analysis reveals a significant surge in research interest in immune cells and MAFLD since 2006, marked by a polynomial growth trend in publications. The United States and China have emerged as dominant forces in this field, fostering robust global collaboration networks and propelling scientific advancements. Their leadership in interdisciplinary research and capacity-building has accelerated discoveries and informed global health policies. While smaller nations face challenges in competing for resources and recognition, international cooperation and knowledge transfer can bolster global research capacity and promote a more equitable scientific landscape[19].
The study also highlights the importance of interdisciplinary approaches, particularly in gastroenterology, biochemistry, and immunology, with journal distribution and co-citation analysis identifying influential works and emerging research focal points. Immunology has illuminated the roles of macrophage polarization (M1/M2 phenotypes) and natural killer cells in MAFLD, showing their impact on inflammation, tissue repair, and disease progression via the Janus kinase/STAT pathway[16,20,21]. These insights pave the way for targeted therapies modulating the hepatic immune microenvironment. Gastroenterology has emphasized the gut-liver axis, revealing that gut microbiota composition correlates with MAFLD severity and that modulating it can improve liver inflammation and metabolic function. Increased gut permeability is also linked to systemic inflammation and type 2 diabetes[22]. These findings not only elucidate the pathophysiology of MAFLD but also provide a basis for developing therapeutic strategies targeting the gut microbiota.
Key research hotspots in immune cells and MAFLD include oxidative stress and Kupffer cells[23,24]. Oxidative stress induces mitochondrial dysfunction and activates inflammatory pathways like nuclear factor kappa B, with antioxidants showing promise in preclinical models[25]. Kupffer cells drive liver inflammation by releasing cytokines (e.g., TNF-α) and chemokines. Modulating their function reduces inflammation and fibrosis[26,27]. These hotspots highlight the importance of integrating immunological and metabolic insights to develop novel therapies for MAFLD. The above-mentioned methods have made significant contributions to the integration of immunology and bioinformatics, particularly in identifying clinically reliable therapeutic targets through bioinformatics analysis[24,28].
Key signaling pathways and core proteins
The cluster analysis using Citexs and VOSviewer revealed the genetic and pathological intricacies of MAFLD. Identified clusters highlighted the disease complexity, with key signaling pathways such as cytokine-cytokine receptor interaction and lipid and atherosclerosis playing crucial roles. Additionally, core proteins INS, CCL2, CD4, and TNF were identified as key players in MAFLD pathogenesis. Activation of INS via insulin receptor controls glucose uptake and storage processes, indicating therapeutic potential for MAFLD[29]. CCL2 is a chemokine that plays a key role in recruiting monocytes and T cells to sites of inflammation, such as atherosclerotic plaques and areas within the liver during MAFLD. Elevated levels of CCL2 are associated with increased macrophage infiltration in the liver, contributing to the progression of steatohepatitis and fibrosis[30-32]. In MAFLD, CD4+ T cells may influence the disease by modulating the immune response and the balance between proinflammatory and anti-inflammatory cytokines. These cells can also affect the activation of other immune cells, such as macrophages, which play a significant role in the pathogenesis of liver disease[33]. In MAFLD, TNF is involved in the regulation of inflammation and INS resistance. It contributes to the development of steatohepatitis and fibrosis by inducing the expression of other inflammatory molecules and promoting the activation and recruitment of immune cells to the liver[34]. In summary, INS, CCL2, CD4 and TNF are integral to the pathophysiological mechanisms of immune cell interactions and MAFLD, highlighting their potential as therapeutic targets and diagnostic markers in the context of this metabolic disorder.
Important disease targets
The identification of IL-6, TNF, IL-10, IL-1b, interferon g, CD4, CD8, toll-like receptor 4, CCL2 and STAT3 as core proteins in the realm of immune cells and MAFLD is of significant interest due to their pivotal roles in the immune response and inflammation; both of which are integral to the pathophysiology of MAFLD[35-37]. In particular, the identification of IL-6, TNF, and STAT3 as key proteins has garnered significant attention due to their potential roles in modulating inflammatory responses and metabolic pathways in MAFLD. IL-6 is a pleiotropic cytokine involved in the regulation of immune responses, inflammation, and metabolic processes. Elevated levels of IL-6 are associated with the progression of liver diseases, including MAFLD, and it is thought to contribute to the inflammatory cascade and INS resistance observed in these conditions. Clinically, targeting IL-6 signaling pathways has shown promise in reducing inflammation and improving metabolic function in patients with chronic inflammatory diseases[38,39]. TNF is a potent pro-inflammatory cytokine that plays a central role in the regulation of inflammation and INS resistance in MAFLD. It contributes to the development of steatohepatitis and fibrosis by inducing the expression of other inflammatory molecules and promoting the activation and recruitment of immune cells to the liver. In the context of MAFLD, targeting the TNF signaling pathway could reduce hepatic inflammation and improve INS sensitivity[34,40]. STAT3 is a transcription factor that mediates cellular responses to cytokines and growth factors. STAT3 is involved in various cellular processes, including cell growth, apoptosis, and immune responses. Its activation is associated with the development of inflammation and cancer, and it may represent a convergence point for multiple signaling pathways in MAFLD. Clinically, STAT3 inhibitors have shown potential in preclinical studies for treating various cancers and inflammatory diseases. Targeting STAT3 in MAFLD could provide a novel therapeutic approach to modulate inflammation and prevent disease progression[41].
Limitations
First, the data were sourced from the WoSCC, which may introduce regional biases as it predominantly includes publications from Western countries and may under represent research from other regions. Additionally, the included articles were in English, which could introduce language bias. Future studies may consider incorporating additional databases and conducting multilingual searches to mitigate these biases. Unlike meta-analysis, which can provide more precise estimates of effect sizes or outcomes[19,42], bibliometric analysis focuses on identifying research trends, hotspots, and potential therapeutic targets through bibliometric and visual analysis methods. However, the analytical tools employed in this study also have limitations that warrant acknowledgment. For instance, VOSviewer relies heavily on user-defined thresholds, which may influence the granularity of the results. Additionally, the clustering algorithms used in CiteSpace may introduce biases in the interpretation of co-citation clusters. To mitigate these limitations, we employed multiple validation steps and cross-referenced our results with those from other analytical methods. We hope that in the future, more analytical software can be developed to provide a more scientific approach to analyzing such data.
CONCLUSION
This study mapped the growth of immune cell and MAFLD research since 2006, led by the United States and China. It highlighted oxidative stress, metabolic syndrome, liver fibrosis and Kupffer cells as key areas. Bioinformatics and gene analysis, particularly of IL-6, TNF and STAT3, have advanced the field, signaling a trend towards collaborative, specialized research. The identification of research hotspots and the visualization of knowledge domains offer a roadmap for future research endeavors. Despite the progress, there remains a need for further investigation into the mechanisms linking immune cells to MAFLD, the development of novel therapeutics, and the optimization of treatment strategies. The insights gained from this bibliometric study can inform research priorities, guide funding decisions, and stimulate innovative approaches to address the complex challenges posed by MAFLD.