Published online Jan 27, 2025. doi: 10.4254/wjh.v17.i1.100377
Revised: October 26, 2024
Accepted: November 19, 2024
Published online: January 27, 2025
Processing time: 144 Days and 3 Hours
The gut microbiome is associated with hepatic encephalopathy (HE), but research results on the gut microbiome characteristics of patients with liver cirrhosis with and without HE are inconsistent.
To study the gut microbiota characteristics of patients with liver cirrhosis with and without HE.
We searched the PubMed, Web of Science, EMBASE, and Cochrane databases using two keywords, HE, and gut microbiome. According to the inclusion and exclusion criteria, suitable literature was screened to extract data on the diversity and composition of the fecal microbiota in patients with liver cirrhosis with and without HE. The data were analyzed using RevMan and STATA.
Seventeen studies were included: (1) A meta-analysis of 7 studies revealed that the Shannon index in liver cirrhosis patients with HE was significantly lower than that in patients without HE [-0.20, 95% confidence interval (CI): -0.28 to -0.13, I2 = 20%]; (2) The relative abundances of Lachnospiraceae (-2.73, 95%CI: -4.58 to -0.87, I2 = 38%) and Ruminococcaceae
The gut microbiomes of patients with liver cirrhosis with and without HE differ. Some gut microbiomes may distinguish liver cirrhosis patients with or without HE and determine patient prognosis.
Core Tip: The gut microbiome is associated with hepatic encephalopathy (HE), but research results on the gut microbiome characteristics of patients with liver cirrhosis with and without HE are inconsistent. We aimed to conduct a meta-analysis of the gut microbiota characteristics of patients with liver cirrhosis with and without HE, to provide a reference for the targeted regulation of the gut microbiota and reduction in HE incidence.
- Citation: Xu XT, Jiang MJ, Fu YL, Xie F, Li JJ, Meng QH. Gut microbiome composition in patients with liver cirrhosis with and without hepatic encephalopathy: A systematic review and meta-analysis. World J Hepatol 2025; 17(1): 100377
- URL: https://www.wjgnet.com/1948-5182/full/v17/i1/100377.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i1.100377
Hepatic encephalopathy (HE) is one of the most common complications of liver cirrhosis and a leading cause of death in patients with liver disease, with an increasing incidence rate[1,2]. In recent years, increasing research has focused on the relationships between the gut microbiome and the occurrence and development of diseases[3]. A difference in the gut microbiota has been reported between patients with cirrhosis and normal patients[4], which is also associated with complications in patients with cirrhosis[5]. Cirrhosis and portal hypertension are related to systemic inflammation and portal vein blood stasis. These conditions further damage intestinal barrier function, alter intestinal permeability, and lead to bacterial translocation[6]. Ultimately, changes in the composition and function of the intestinal microbiota are involved in the development of HE[7]. The gut microbiome may serve as a reasonable biomarker for liver cirrhosis diagnosis and prognosis[5]. However, in existing reports, there was inconsistency in the differences in the gut microbiota among cirrhosis patients with and without HE. Therefore, this study conducted a meta-analysis of the gut microbiota characteristics of patients with liver cirrhosis with and without HE, to provide a reference for targeted regulation of the gut microbiota to reduce the incidence of HE in patients.
The PubMed, Web of Science, EMBASE, and Cochrane databases were searched. MeSH terms and free words were used to search for literature related to gastrointestinal microbiomes and HE. The search period ended on June 1, 2024. At the same time, a search was conducted for references. The specific retrieval strategy is shown in Supplementary Table 1. This meta-analysis was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines and was registered in PROSPERO (CRD42024569064).
The inclusion criterion was as follows: Patients with liver cirrhosis with or without HE who underwent fecal gut microbiota testing. The gut microbiota diversity index and gut microbiota composition were measured. The exclusion criterion was patients with liver cancer. Patients without cirrhosis. Duplicate studies, case reports, reviews, meta-ana
Main research findings: Differences in gut microbiota diversity and composition between HE and non-HE patients with liver cirrhosis. The alpha diversity of the gut microbiota included the Shannon, Simpson, and Chao 1 indices. Secondary research findings: Effects of drug therapy on the gut microbiota. Relationship between the gut microbiota and metabolism. The value of the gut microbiota in distinguishing patients with liver cirrhosis with and without HE and predicting patient prognosis.
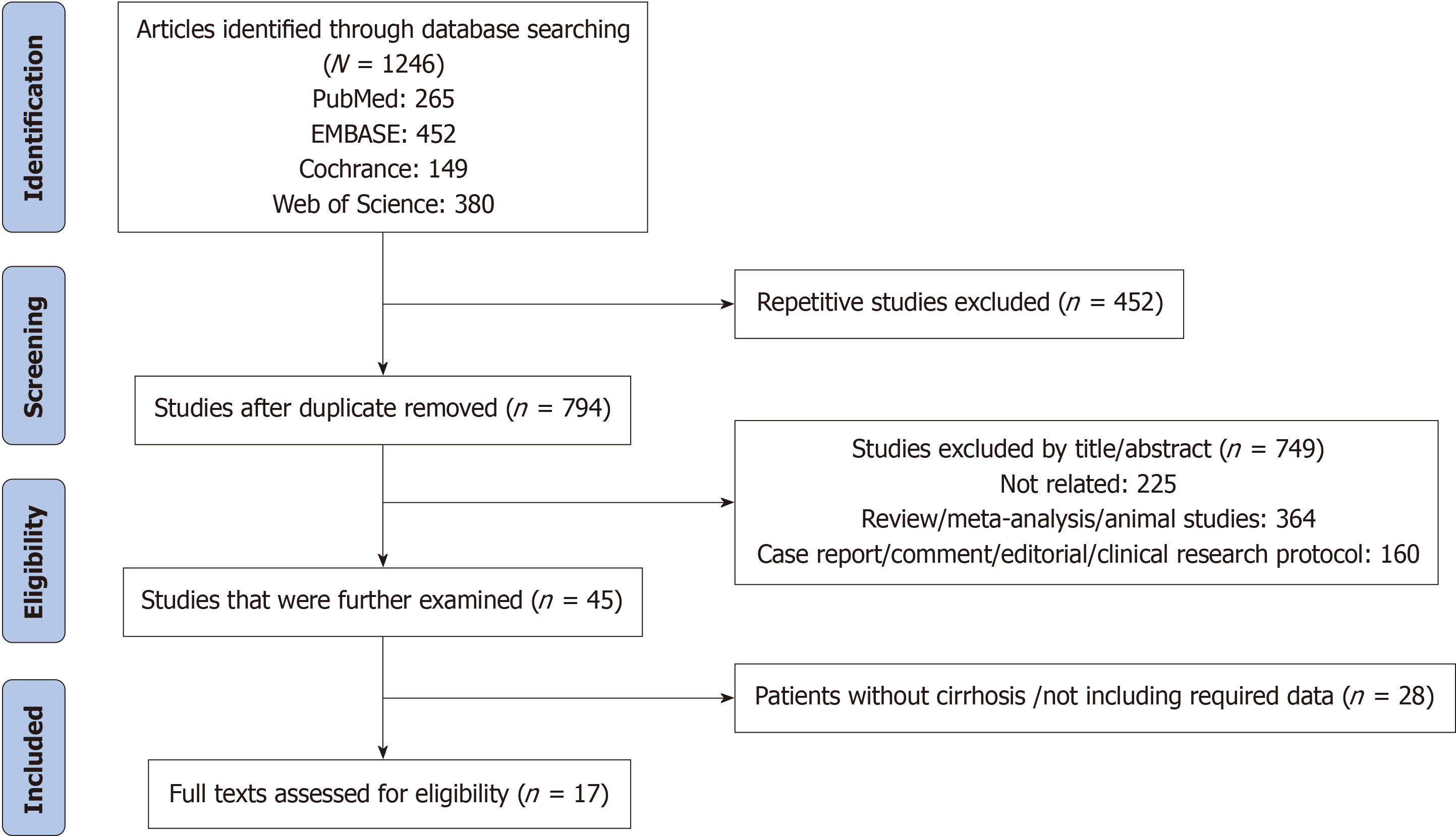
Two independent evaluators (Xiao-Tong Xu and Min-Jie Jiang) selected studies that met the criteria by screening titles and abstracts. The quality evaluation of the literature was based on the quality evaluation table, as shown in Table 1. For ambiguous parts, a third evaluator was required to participate in the discussion and make the final decision. The filtering process is shown in Figure 1.
| Ref. | Representativeness of the exposed cohort | Selection of the nonexposed cohort | Ascertainment of exposure | Demonstration that outcome of interest was not present at the start of the study | Comparability between exposed and nonexposed cohort | Assessment of outcome | Follow-up was long enough for outcomes to occur | Adequacy of follow-up of cohorts | Total score |
| Wang et al[9], 2023, China | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 9 |
| Bajaj et al[12], 2012, United States | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 8 |
| Bajaj et al[13], 2015, United States | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Bajaj et al[14], 2019, United States | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 8 |
| Bajaj et al[55], 2018, United States | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Sung et al[15], 2019, China | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 9 |
| Bloom et al[8], 2021, Japan | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 8 |
| Saboo et al[56], 2022, United States | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 7 |
| Yukawa-Muto et al[10], 2022, Japan | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Li et al[57], 2022, China | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 8 |
| Hua et al[58], 2022, China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 7 |
| Bajaj et al[59], 2022, United States | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Yu et al[11], 2021, China | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 9 |
| Baja et al[60], 2020, United States | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 9 |
| Ahluwalia et al[16], 2016, United States | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 7 |
| Zhang et al[17], 2013, China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 |
| Bajaj et al[61], 2012, United States | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 8 |
Author, country, year, age, gender, etiology, liver function score, gut microbiota detection method, sequencing region of liver cirrhosis patients with or without HE. Diversity of gut microbiota alpha diversity (Chao 1, Simpson, and Shannon indices) and the composition of the gut microbiota.
A single group rate analysis was conducted for studies that reported only the composition proportion of the fecal gut microbiota. We extracted mean values and standard deviations from studies with quantitative data on the fecal gut microbiota for analysis. The collected data were analyzed using RevMan and STATA with random effects or fixed effects.
A total of 1246 articles were initially searched in the database, and after screening, 17 articles were finally included, including 782 cases of liver cirrhosis with HE and 839 cases of liver cirrhosis without HE. The literature screening process is shown in Figure 1. Two studies were conducted in Japan, 6 in China, and 9 in the United States, and relevant in
| Ref. | Amplified region/reference databases | Characteristics of gut microbiota/alpha diversity indices | |||||
| Age | Gender (male/female) | MELD | Child-Pugh score/class | Etiology | PPI | ||
| Wang et al[9], 2023, China | N/A, N/A | Cirrhosis with HE (n = 30): (1) Higher of Burkholderiaceae, Ralstonia, Parabacteroides-distasonis; (2) Phylum: Firmicutes (58.08%), Proteobacteria (18.83%), Bacteroidetes (18.19%), and Actinobacteria (2.5%); (3) Genera: Bacteroides (10.59%), Enterococcus (22.98%), Prevotella (2.21%), Escherichia-Shigella (11.9%), Ruminococcus-gnavus (2.13%), and Streptococcus (5.9%); and (4) Shannon (3.84 ± 2.01), Simpson (0.78 ± 0.19), and Chao 1(231.26 ± 66.41) | |||||
| 56.13 ± 13.20 | 19/11 | 19.67 ± 5.95 | Child A: 0, Child B: 9, Child C: 21 | HBV (9)/HCV (1)/alcohol (12)/HBV + alcoholic (1)/HCV + alcohol (1)/others (6) | N/A | ||
| Cirrhosis without HE (n = 30): (1) Higher of Aegypius-monachus, Erysipelatoclostridiaceae, and Erysipelotrichales; (2) Phylum: Firmicutes (58.42%), Proteobacteria (15.08%), Bacteroidetes (20.14%), and Actinobacteria (1.6%); (3) Genera: Bacteroides (14.80%), Enterococcus (16.36%), Prevotella (2.55%), Escherichia-Shigella (2.55%), Ruminococcus-gnavus (5.35%), and Streptococcus (3.82%); and (4) Shannon (4.11 ± 1.61), Simpson (0.81 ± 0.18), and Chao 1 (211.16 ± 56.35) | |||||||
| 52.23 ± 9.00 | 20/10 | 13.90 ± 6.51 | Child A: 9, Child B: 7, Child C: 14 | HBV (14)/HCV (1)/alcohol (8)/HBV + alcoholic (3)/HCV + alcohol (1)/others (3) | N/A | ||
| Bajaj et al[12], 2012, United State | N/A, RDP | Cirrhosis with HE (n = 17): Leuconostocaceae (2.19 ± 1.08), Clostridiales-Incertae Sedis XIV (0.99 ± 0.21), Ruminococcaceae (5.68 ± 1.42), Lachnospiraceae (9.54 ± 3.72), Enterobacteriaceae (10.02 ± 4.13), Streptococcaceae (4.05 ± 1.98), Fusobacteriaceae (1.36 ± 0.95), Alcaligenaceae (2.61 ± 0.74); N/A | |||||
| 56 ± 3 | 16/1 | MELD17 ± 6 | N/A | Alcohol (10)/others (7) | 16 | ||
| Cirrhosis without HE (n = 8): Leuconostocacea (1.69 ± 0.95), Clostridiales_Incertae Sedis XIV (1.29 ± 0.35), Fusobacteriaceae (2.75 ± 2.75), Lachnospiraceae (12.08 ± 2.47), and Ruminococcaceae (9.04 ± 2.62); N/A | |||||||
| 55 ± 5 | 7/1 | MELD 12 ± 5 | N/A | Alcohol (3)/others (5) | 7 | ||
| Bajaj et al[13], 2015, United State | N/A | Cirrhosis with HE (n = 43): Clostridiales XIV (4.5%), Lachnospiraceae (16%), Ruminococcaceae (7.4%), Fusobacteriaceae (1.0%), Prevotellaceae (5.0%), Enterococcaceae (1.0%), Enterobacteriaceae (1.0%), Erysipelotrichaceae (0.6%), Bacteroidaceae (24.9%), and Streptococcacae (1.9%); N/A | |||||
| 56 ± 16 | 34/9 | 17.2 ± 7.2 | 9.7 ± 4.4 | HCV (20)/alcohol (6)/HCV + alcohol (9)/NASH (7)/others (1) | 19 | ||
| Cirrhosis without HE (n = 59): ClostridialesXIV (6.6%), Lachnospiraceae (21.3%), Ruminococcaceae (8.7%), Fusobacteriaceae (0%), Prevotellaceae (5.3%), Enterococcaceae (0%), Enterobacteriaceae (3%), Erysipelotrichaceae (1.9%), Bacteroidaceae (24.5%), and Streptococcacae (4.4%); N/A | |||||||
| 54 ± 13 | 50/9 | 8.6 ± 2.5 | 6.2 ± 5.1 | HCV (27)/alcohol (14)/HCV + alcohol (7)/NASH (7)/others (4) | 21 | ||
| Bajaj et al[14], 2019, United State | N/A, N/A | Cirrhosis with MHE (n = 122): (1) PHES: Higher of Lactobacillales, Bacilli, and Micrococcaceae; (2) ICT: Higher of Lactobacillales, Enterobacteriales, Bacilli, Enterobacteriaceae, Protebacteria, Gammaprotebacteria, Micrococcaceae, Eubacteriaceae, Streptococcaceae, Actinomycetales; (3) Stroop: Higher of Lactobacillales, Lactocillaceae, Actinobacteria; and (4) Shannon MHE on PHES (1.9 ± 0.7), MHE on ICT (2.0 ± 0.7), and MHE on Stroop 1.9 ± 0.6 | |||||
| 60.1 ± 7.0 | 122/0 | 11.3 ± 4.7 | N/A | N/A | 77 | ||
| Cirrhosis without HE (n = 145): (1) PHES: Higher of Clostridiales, Incertae Sedis XI and XIII, Pasteurellales, Pasteurellaceae, Lachnospiraceae, and Clostridia; (2) ICT: Higher of Bacteroidaceae, Bacteroidales, Bacteroidia, Bacteroidetes; (3) Stroop: Higher of Eubacteriaceae, GP1, Telmatobacter, Breoghania, Caulobacterales, Telmatobacter, Acidobacteria, proteobacteria-incertaesedis, Hyphomonadaceae, and Listeriaceae; and (4) Shannon non-MHE on PHES (2.2 ± 0.6), non-MHE on ICT (2.1 ± 0.6), and non-MHE on Stroop (2.1 ± 0.6) | |||||||
| 59.4 ± 9.1 | 85/60 | 14.3 ± 7.1 | N/A | N/A | 54 | ||
| Bajaj et al[55], 2018, United State | V1-2/RDP | Cirrhosis with prior HE (n = 10): Higher abundance of Erysipelothriceaea, Erysipelotrichia, and Erysipelotrichales; N/A | |||||
| N/A | N/A | 13.2 ± 3.1 | N/A | N/A | N/A | ||
| Cirrhosis without prior HE (n = 14): Higher abundance of Selomonadales and Negativicutes; N/A | |||||||
| N/A | N/A | 7.8 ± 1.7 | N/A | N/A | N/A | ||
| Sung et al[15], 2019, China | V3-4, N/A | Cirrhosis with HE (n = 62): (1) Firmicutes (41.4%), Bacteroidetes (36.5%), Proteobacteria (15.8%) and Actinobacteria (3.4%); (2) Schlegelella (1.172 ± 2.924), Megasphaera (1.105 ± 1.518), Veillonella (11.007 ± 12.473), Enterococcus (1.369 ± 5.038), Lactobacillus (1.086 ± 3.184), XI (1.603 ± 7.644), Prevotella (1.254 ± 4.101), Sedis (0.401 ± 0.923), Bacteroides (1.780 ± 4.062), and Parabacteroides (0.788 ± 2.946); and (3) Shannon (2.6 ± 0.59) and Chao 1 (146.83 ± 47.35) | |||||
| 56.7 ± 12.2 | 49/13 | 18.5 ± 6 | Child A: 0; Child B: 22; Child C: 40 | HBV (18)/HCV (19)/alcoholic (36)/others (4) | 22 | ||
| Cirrhosis without HE (n = 15): (1) Firmicutes (on average 332%), Bacteroidetes (47.2%), Proteobacteria (15.5%), and Actinobacteria (1.4%); (2) Schlegelella (0.029 ± 0.085), Megasphaera (1.031 ± 1.861), Veillonella (6.015 ± 7.01073), Enterococcus (0.02 ± 0.031), Lactobacillus (0.011 ± 0.017), XI (0.039 ± 0.081), Prevotella (0.022 ± 0.084), Sedis (1.788 ± 2.968), Bacteroides (1.607 ± 2.290), and Parabacteroides (1.334 ± 2.269); and (3) Shannon (2.64 ± 0.45) and Chao 1 (194.53 ± 52.93) | |||||||
| 58.1 ± 12.3 | 12/3 | 16.8 ± 5.58 | Child A: 0; Child B: 8; Child C: 7 | HBV (6)/HCV (2)/alcohol (8)/others (0) | 4 | ||
| Bloom et al[8], 2021, Japan | N/A, NCBI | Cirrhosis with HE (prior or current) (n = 33): (1) Higher of Bifidobacteriaceae, Enterococcaceae, Strptoccaceae, Lactobacteriaceae, and Enterobacteriaceae; and (2) Simpson (4.2 ± 2.64) | |||||
| 57 ± 10 | 19/14 | 21 ± 8 | N/A | Alcohol (16)/NASH (10)/viral and alcohol (4)/viral (1)/others (2) | 28 | ||
| Cirrhosis without HE (n = 16): (1) Higher of Lachnospiraceae, Bacteroidaceae, Ruminococcaceae, Veillonellaceae, AKKermansiaceae, and Eubacteriaceae; and (2) Simpson (10.88 ± 9.46) | |||||||
| 59 ± 10 | 11/5 | 16 ± 10 | N/A | Alcohol (6)/NASH (2)/viral and alcohol (3)/vral (4)/others (1) | 11 | ||
| Saboo et al[56], 2022, United State | V1-2/RDP | Cirrhosis with HE (n = 57): Lower of Lachnospriaceae, Bacteroidaceae, and Ruminococcaceae; N/A | |||||
| 59.6 ± 6.4 | 46/11 | 15.1 ± 7.2 | N/A | Alcohol (20)/HCV (22)/NASH (12)/others (3) | 28 | ||
| Cirrhosis without HE (n = 125): Higher of Lachnospriaceae, Bacteroidaceae, and Ruminococcaceae; N/A | |||||||
| 60.8 ± 7 | 97/28 | 9.6 ± 3.8 | N/A | Alcohol (54)/HCV (45)/NASH (21)/others (5) | 50 | ||
| Yukawa-Muto et al[10], 2022, Japan | N/A | Decompensated cirrhosis with HE (n = 26): (1) Higher of Streptococcus salivarius, Lactobacillus spp., Bifidobacterium dentium, Lactobacillus mucosae, Lactobacillus fermentum, Veillonella atypica, Veillonella parula, and Lactobacillus rhamnosus; and (2) Shannon (6.47 ± 0.38) | |||||
| 68.4 (52-85) | 15/11 | 11.7 (6-19) | N/A | HBV (3)/HCV (9)/NASH (7)/alcohol (3)/PBC (3)/AIH (1) | N/A | ||
| Decompensated cirrhosis without HE (n = 27): (1) Lower of Streptococcus salivarius, Lactobacillus spp., Bifidobacterium dentium, Lactobacillus mucosae, Lactobacillus fermentum, Veillonella atypica, Veillonella parula, and Lactobacillus rhamnosus; and (2) Shannon (6.90 ± 0.5) | |||||||
| 67.6 (54-81) | 16/11 | 8.2 (6-12) | N/A | HBV (0)/HCV (26)/NASH (1)/alcohol (0)/PBC (0)/AIH (0) | N/A | ||
| Li et al[57], 2022, China | V3-4/RDP | Cirrhosis with HE (n = 33): (1) Higher of Ruminococaceae, Lachnospiraceae, Ruminococcus, Rumi_uncultured, Roseburia, Dialister, Dialister, Coprococcus, Blautia, and Anaerostipes; and (2) Shannon (2.84 ± 0.45) and Chao 1 (221.39 ± 39.36) | |||||
| 51 (44-58) | 19/14 | 16.84 (13.6, 19.27) | 8 (7-11); Child A: 3; Child B: 21; Child C: 9 | HBV + HCV (25)/others (8) | N/A | ||
| Cirrhosis without HE (n = 73): (1) Lower of Ruminococaceae, Lachnospiraceae, Ruminococcus, Rumi_uncultured, Roseburia, Dialister, Dialister, Coprococcus, Blautia, and Anaerostipes; and (2) Shannon (3.04 ± 0.39) and Chao 1 (244.24 ± 58.72) | |||||||
| 52 (45-58) | 46/27 | 14.34 (12.15, 16.47) | 7 (6-8); Child A: 34; Child B: 35; Child C: 4 | HBV + HCV (50)/others (23) | N/A | ||
| Hua et al[58], 2022, China | N/A | Cirrhosis with HE (n = 5): (1) Higher of Pasteurellales, Pasteurellaceae, Haemophilus, and Selenomonas; (2) Higher of Veillonella (4.12 ± 6.09), Pasteurellales (2.39 ± 4.04), Pasteurellaceae (2.39 ± 4.04), Haemophilus (2.34 ± 3.96), Selenomonas (0.016 ± 0.04), Lachnospiraceae (11.48 ± 6.53), Coprococcus (0.311 ± 0.292), and Dorea (0.855 ± 0.716); and (3) N/A | |||||
| 60.11 ± 9.54 | 4/1 | N/A | N/A | HBV (5) | N/A | ||
| Cirrhosis without HE (n = 15): (1) Higher of Veillonella; (2) Higher of Veillonella (4.40 ± 6.13), Blautia (1.36 ± 1.88), Turicibacterales (0.012 ± 0.039), Turicibacter (0.012 ± 0.039), Turicibacteraceae (0.012 ± 0.039), Adlercreutzia (0.053 ± 0.014), Holdemania (0.009 ± 0.016), and Coprococcus (0.305 ± 0.43); (3) Lachnospiraceae (9.08 ± 6.38); and (4) N/A | |||||||
| 61.73 ± 13.55 | 11/4 | N/A | N/A | HBV (15) | N/A | ||
| Bajaj et al[59], 2022, United State | V1-2/RDP | Cirrhosis with MHE (n = 180): Higher of Pediococcus, Lacticaseibacillus, Pseudomonas, Doudenibacillus, Lactobacillus, Enterococcus, Escherichia-Shigella, Limosilactobacillus, Klebsiella, Sutterella, Megamonas, Veillonella, Absiella, and Prevotellamassila; N/A | |||||
| N/A | N/A | N/A | N/A | N/A | N/A | ||
| Cirrhosis without MHE (n = 141): Higher of Enterobacter, Weissella, Anaerobutyrium, Romboutsia, and Citrobacter; N/A | |||||||
| N/A | N/A | N/A | N/A | N/A | N/A | ||
| Yu et al[11], 2021, China | N/A, NCBI | Cirrhosis with HE (n = 5): (1) Higher of Bacteroidetes and Proteobacteria; (2) Lower of Firmicutes, Candidatus Saccharibacteria, and the Verrucomicrobia phyla; and (3) Shannon (1.78 ± 0.52), Simpson (0.71 ± 0.15), and Chao 1 (48.42 ± 17.78) | |||||
| 58.2 ± 7.04 | 3/2 | N/A | N/A | N/A | N/A | ||
| Cirrhosis without HE (n = 16): (1) Lower of Bacteroidetes and Proteobacteria; (2) Higher of Firmicutes, Candidatus Saccharibacteria, and the Verrucomicrobia phyla; and (3) Shannon (2.23 ± 0.4), Simpson (0.82 ± 0.07), and Chao 1 (68.14 ± 20.05) | |||||||
| 55.33 ± 5.86 | 10/6 | N/A | N/A | N/A | N/A | ||
| Bajaj et al[60], 2020, United State | N/A, MYSQL | Cirrhosis with HE 16S rRNA (n = 29): (1) Bacteroidaceae (35%), Prevotellaceae (0%), Lactobacillaceae (1%), Lachnospiraceae (11.3%), Ruminococcaceae (4.4%), Streptococcaceae (1%), Veillonellaceae (0%), and Enterobacteriaceae (1%); and (2) Shannon (1.49 ± 0.37) | |||||
| 58.8 ± 11.7 | 20/9 | 14.8 ± 7 | N/A | N/A | 28% | ||
| Cirrhosis without HE (n = 51): (1) Bacteroidaceae (33%), Prevotellaceae (0%), Lactobacillaceae (1%), Lachnospiraceae (26%), Ruminococcaceae (10%), Streptococcaceae (0.1%), Veillonellaceae (0%), and Enterobacteriaceae (0%); and (2) Shannon (1.6 ± 0.4) | |||||||
| 57.9 ± 7.4 | 36/15 | 9.5 ± 3.4 | N/A | N/A | 18% | ||
| Ahluwalia et al[16], 2016, United State | N/A | Cirrhosis with prior HE (n = 85): (1) Phylum: Firmicutes (24 ± 20), Bacteroidetes (24 ± 26), Proteobacteria (7 ± 15); (2) Family: Bacteroidetes-Bacteroidaceae (18.2 ± 25.2), Bacteroidetes_Porphyromonadaceae (3.9 ± 7.7), Bacteroidetes-Prevotellaceae (2.9 ± 8), Bacteroidetes-Rikenelleaceae (0 ± 2.4), Firmicutes_Lactobacillaceae (4 ± 9.5), Firmicutes-Enterococcaceae (3.6 ± 10.9), Firmicutes-Clostridiales XIV (1.4 ± 3.5), Firmicutes-Lachnospiraceae (6.3 ± 11), Firmicutes-Ruminococcaceae (3.0 ± 5.4), Firmicutes-Veillonellaceae (2. 0 ± 4.8), and Proteobacteria_Enterobacteriaceae (5.5 ± 14.8); and (3) N/A | |||||
| 56.2 ± 13.5 | 16/69 | 15.5 ± 5.2 | N/A | HCV (27)/alcohol (16)/HCV + alcohol (20)/NASH (12)/others (10) | N/A | ||
| Cirrhosis without prior HE (n = 62): (1) Phylum: Firmicutes (28 ± 19), Bacteroidetes (33 ± 30), and Proteobacteria (3 ± 6); (2) Family: Bacteroidetes-Bacteroidaceae (22.2 ± 24), Bacteroidetes-Porphyromonadaceae (2.1 ± 5.2), Bacteroidetes-Prevotellaceae (4.5 ± 13.5), Bacteroidetes-Rikenelleaceae (2.0 ± 5.5), Firmicutes-Lactobacillaceae (2.0 ± 8.5), Firmicutes-Enterococcaceae (1.1 ± 6.7), Firmicutes-Clostridiales XIV (3.2 ± 4.7), Firmicutes-Lachnospiraceae (10.5 ± 8.4), Firmicutes-Ruminococcaceae (5.5 ± 6.2), Firmicutes-Veillonellaceae (1.7 ± 2.9), and Proteobacteria-Enterobacteriaceae (2.4 ± 5.9); and (3) N/A | |||||||
| 54.2 ± 11.6 | 46/16 | 11.0 ± 4.2 | N/A | HCV (21)/alcohol (15)/HCV + alcohol (3)/NASH (15)/others (8) | N/A | ||
| Zhang et al[17], 2013, China | N/A | Cirrhosis with MHE (n = 26): Higher of S. salivarius; Chao 1 (276 ± 104) | |||||
| 53 ± 12 | 13/13 | N/A | Child A: 3; Child B: 16; Child C: 7 | AIH (1)/HBV (13)/PBC (7)/PSC (0)/alcohol (5) | N/A | ||
| Cirrhosis without MHE (n = 25): Lower of S. salivarius; Chao 1 (250 ± 139) | |||||||
| 59 ± 11 | 16/9 | N/A | Child A: 12; Child B: 11; Child C: 2 | AIH (2)/HBV (12)/PBC (3)/PSC (1)/alcohol (7) | N/A | ||
| Bajaj et al[61], 2012, United State | N/A | Cirrhosis with HE (n = 19): Incertae Sedis XIV_Blautia (1.6%), Incertae Sedis XIV_other (1.4%), and Fusobacteriaceae_other (1.1%); N/A | |||||
| N/A | N/A | N/A | N/A | N/A | N/A | ||
| Cirrhosis without HE (n = 17): Leuconostocaceae_Leuconostoc (1.0%), Bacteroidales incertae sedis other (1.0%), and Alcaligenaceae other (0.8%); N/A | |||||||
| N/A | N/A | N/A | N/A | N/A | N/A | ||
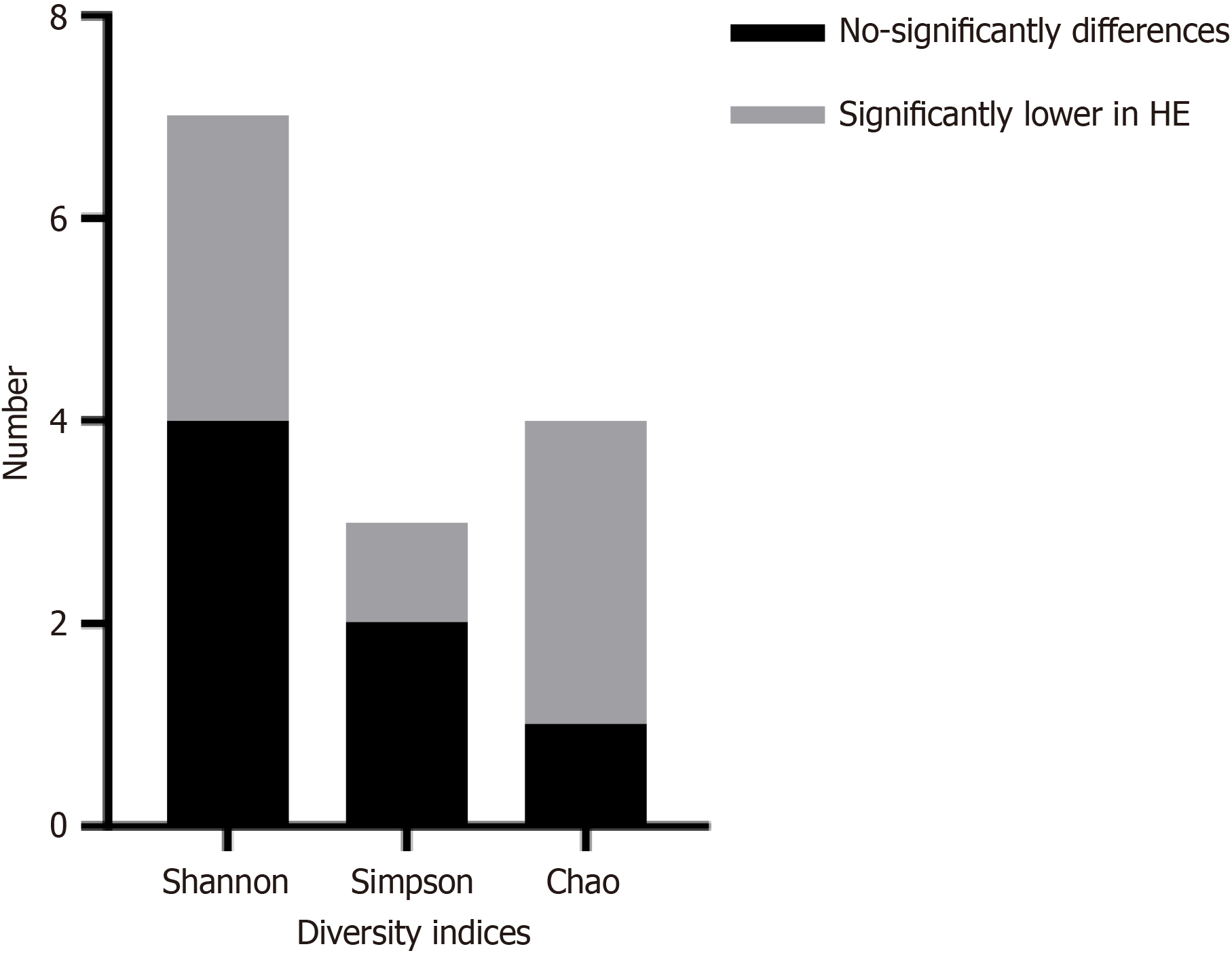
The alpha diversity of the gut microbiota can be represented by the Shannon, Simpson, and Chao 1 indices. A total of 7 studies compared the Shannon index between HE and non-HE in cirrhosis (4 studies reported no significant difference between the two groups, and 3 studies reported lower values in cirrhosis with HE), 3 studies reported the Simpson index between HE and non-HE in cirrhosis (1 study reported no significant difference between the two groups, and 2 studies reported lower values in cirrhosis with HE), and 5 studies reported the Chao 1 index between cirrhosis with HE and without HE (1 study reported no significant difference between the two groups, 3 studies reported lower values in cirrhosis with HE, and 1 study reported higher values in cirrhosis with HE), as shown in Figure 2.
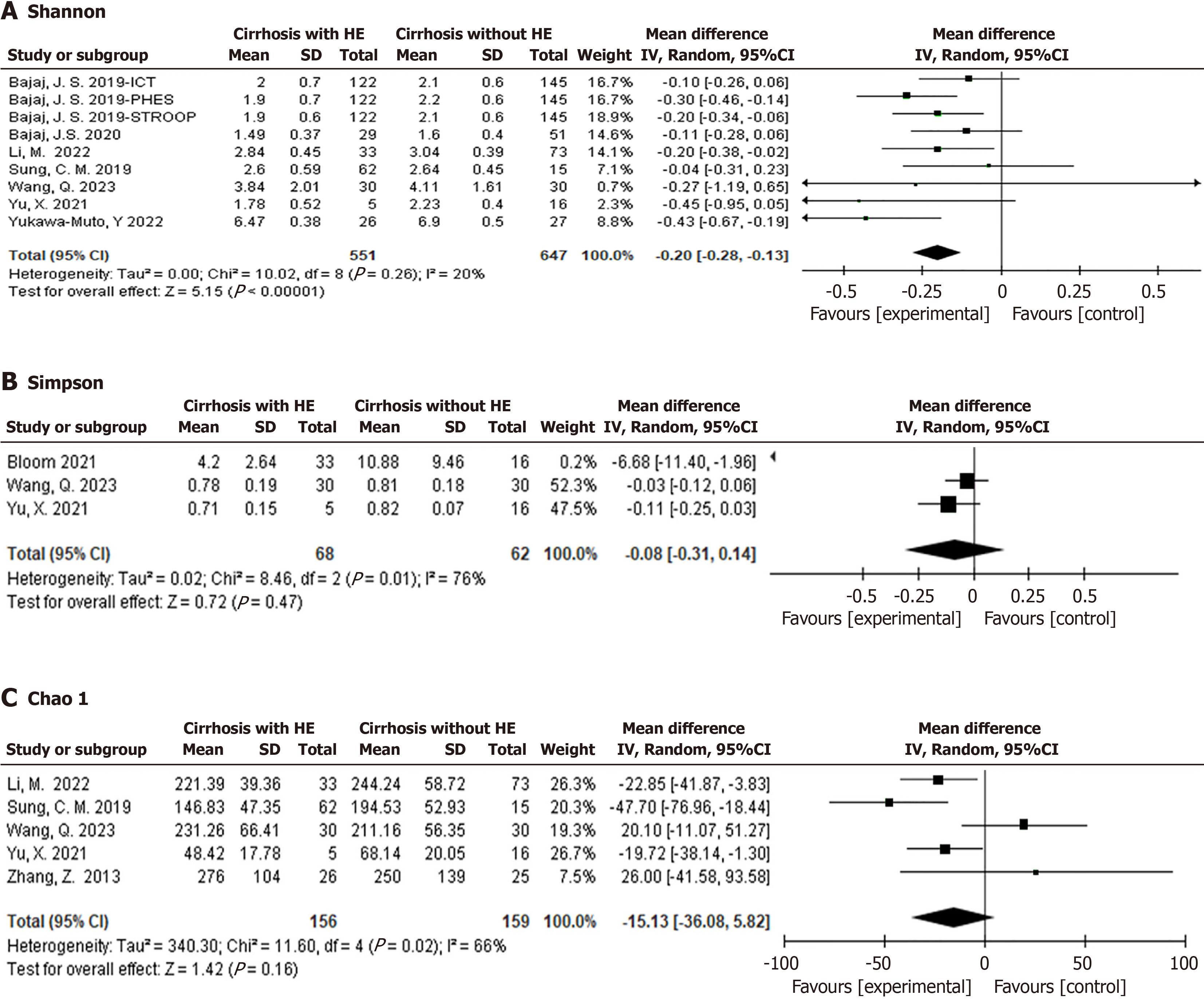
Seven studies extracted Shannon data, in one study, three different results were obtained based on different diagnostic criteria for minimal HE (MHE). The final analysis revealed that the Shannon index in cirrhosis patients with HE was significantly lower than that in patients without HE (P < 0.00001), and there was good consistency between studies (I2 = 20%), as shown in Figure 3A. Three studies extracted Simpson data, and the analysis results revealed no significant difference between patients with liver cirrhosis with HE and those without HE (P = 0.47), but there was significant heterogeneity among the studies (I2 = 76%) as shown in Figure 3B. Given the significant heterogeneity, we conducted a sensitivity analysis by omitting one study at a time. These analyses indicate that the heterogeneity contribution of the Bloom et al[8] to the Simpson meta-analysis was the greatest. When we excluded this study from the analysis, there was still no significant difference in Simpson between the two groups (P = 0.16, I2 = 0%, n = 2 studies). As shown in Supplementary Figure 1. Five studies extracted the Chao 1 index, and the results revealed no significant difference in the Chao 1 index between patients with liver cirrhosis with HE and those without HE (P = 0.16), but there was a certain degree of difference among the studies (I2 = 66%), as shown in Figure 3C. Given the significant heterogeneity, using the same method as before, Wang et al[9] contributed the most to heterogeneity, and after deletion, heterogeneity significantly decreased (I2 = 38%). However, when we excluded this study from the analysis, the Chao 1 index of cirrhosis patients with HE was significantly lower than that of cirrhosis patients without HE (P = 0.004, n = 4 studies), as shown in Supplementary Figure 2.
Based on the linear discriminant analysis effect size or relative abundance data of cirrhosis patients with or without HE mentioned in the literature, the same bacteria are summarized in Table 3. Ruminococcaceae, Lachnospiraceae, Prevotellaceae, and Bacteroidetes tended to decrease in patients with liver cirrhosis with HE. The abundances of Enterococcus, Proteobac
| Bacterium | Ref.1 | Ref.2 |
| Ruminococcaceae | / | [8,12,13,16,56,60] |
| Lachnospiraceae | [57,58] | [8,12-14,16,60] |
| Clostridiales XIV | / | [13,16] |
| Prevotellaceae | / | [13,16,60] |
| Bacteroidaceae | [13,16,60] | [8,14,56] |
| Eubacteriaceae | [14] | [14,18] |
| Bacteroidetes | [11] | [9,14-16] |
| Lactobacillus | [15,59] | / |
| Enterococcus | [9,15,59] | / |
| Veillonella | [15,59] | [58] |
| Lactobacillaceae | [16,60] | / |
| Veillonella | [15,59] | [58] |
| Ruminococcus | [57] | [9] |
| Firmicutes | [11,15] | [9,16] |
| Proteobacteria | [9,11,15,16] | / |
| Coprococcus | [57,58] | / |
| Actinobacteria | [9,15] | / |
| Bacteroides | [15] | [9] |
| Prevotella | [15] | [9] |
| Escherichia-shigella | [9,59] | / |
| Erysipelotrichales | [55] | [8] |
| Enterococcaceae | [8,13,16] | / |
| Enterobacteriaceae | [8,14,16,60] | [13] |
| Streptococcaceae | [14,60] | / |
| Fusobacteriaceae | [13] | [12] |
| Pasteurellaceae | [58] | [14] |
| Pasteurellales | [58] | [14] |
| Veillonellaceae | [16] | [8] |
The subjects in the 7 studies had previously used proton pump inhibitors (PPIs), ranging from 7% to 94%, but only one study reported the effect of PPIs on the gut microbiota of patients with cirrhosis[10]. Cirrhosis patients without HE who used PPIs experienced an increase in the abundance of Streptococcus salivarius (S. salivarius) compared with that in liver cirrhosis patients with HE who used PPIs. Three studies reported the effects of rifaximin or lactulose on the gut mi
Yu et al[11] analyzed the differential metabolic pathways of the gut microbiota in cirrhosis patients with and without HE that were involved mainly in the synthesis of phosphopeptidic acid, peptidoglycans, amino acids, and fatty acids; the degradation of alcohols, rockulose and rhamnose; and the synthesis and degradation of coenzymes. Wang et al[9] also analyzed metabolites in feces and reported no difference in the concentration of short-chain fatty acids between the HE and non-HE groups. However, the average concentrations of acetic acid, propionic acid, butyric acid, and nonanoic acid in the feces of cirrhosis patients with HE is lower than those in normal patients. Moreover, the concentration of try
Wang et al[9] reported that metabolites such as acetic acid, propionic acid, and butyric acid can be used to distinguish between the HE and healthy groups, with area under the curve values of 0.923, 0.917, and 0.865, respectively. It can also be used to distinguish between the cirrhosis and the healthy groups, with area under the curve values of 0.947, 0.89, and 0.927, respectively. Bajaj et al[14] reported via logistic regression that the Lachnospiraceae genus (Ruminococcus and Clostridium XIVb) in feces was associated with good cognition but not with clinical variables. Sung et al[15] reported that Bacteroides, Clostridium XI, and others are associated with the recurrence and overall survival of HE patients within 1 year in patients with liver cirrhosis. Another study by Bloom et al[8] revealed that eight bacterial species, including Clostridium species and Ruminococcus species, were associated with a history of HE. Yu et al[11] reported that Prevotellaceae, Ruminococcaceae, and Lachnospiraceae can reduce hospitalization risk regardless of the presence of model for end-stage liver disease (MELD) and ascites. Prevotellaceae can also reduce the risk of MHE. Ahluwalia et al[16] reported that Lachnospiraceae, Ruminococcaeae, and Incertae Sedis XIV were negatively correlated with MELD. Enterococcus was positively co
HE is still a common complication and cause of death in patients with end-stage liver disease, affecting the quality of life of patients and their families. HE can be divided into covert HE and overt HE (OHE), with covert HE including MHE[2]. Compared with patients without MHE, cirrhosis patients with MHE have more frequent episodes of OHE[18], and the mortality rate of OHE patients is also greater. Without liver transplantation, the 1-year survival rate after the first episode is 42%, and the 3-year survival rate is 23%[19]. The gut-liver axis has become a focus of chronic liver disease, prompting more research on the role of the gut microbiota in cirrhosis[20]. In patients with cirrhosis, changes in the structure and function of the gut microbiota are closely related to clinical prognosis[5,21,22] and are also closely associated with the occurrence of HE[6,23-25]. Changes in the gut-liver-brain axis are considered core pathogenic factors in the cognitive impairment associated with liver cirrhosis. Most treatments related to HE focus on the gut[14,26,27].
Therefore, this study searched for studies on the relationship between the gut microbiota and HE. During the screening process, two studies were excluded because one focused on patients with alcoholic hepatitis and the other focused on patients with chronic acute liver failure. Eventually, 17 studies were included. First, the data on the alpha diversity of the gut microbiota were analyzed, and 7 studies reported the Shannon index. A meta-analysis revealed that it was significantly reduced in patients with liver cirrhosis with HE, and the heterogeneity was small. After the Chao 1 and Simpson indices were compared, there was no significant difference between the two groups, and there was a significant heterogeneity, which may be attributed to the limited research reporting on these two indices.
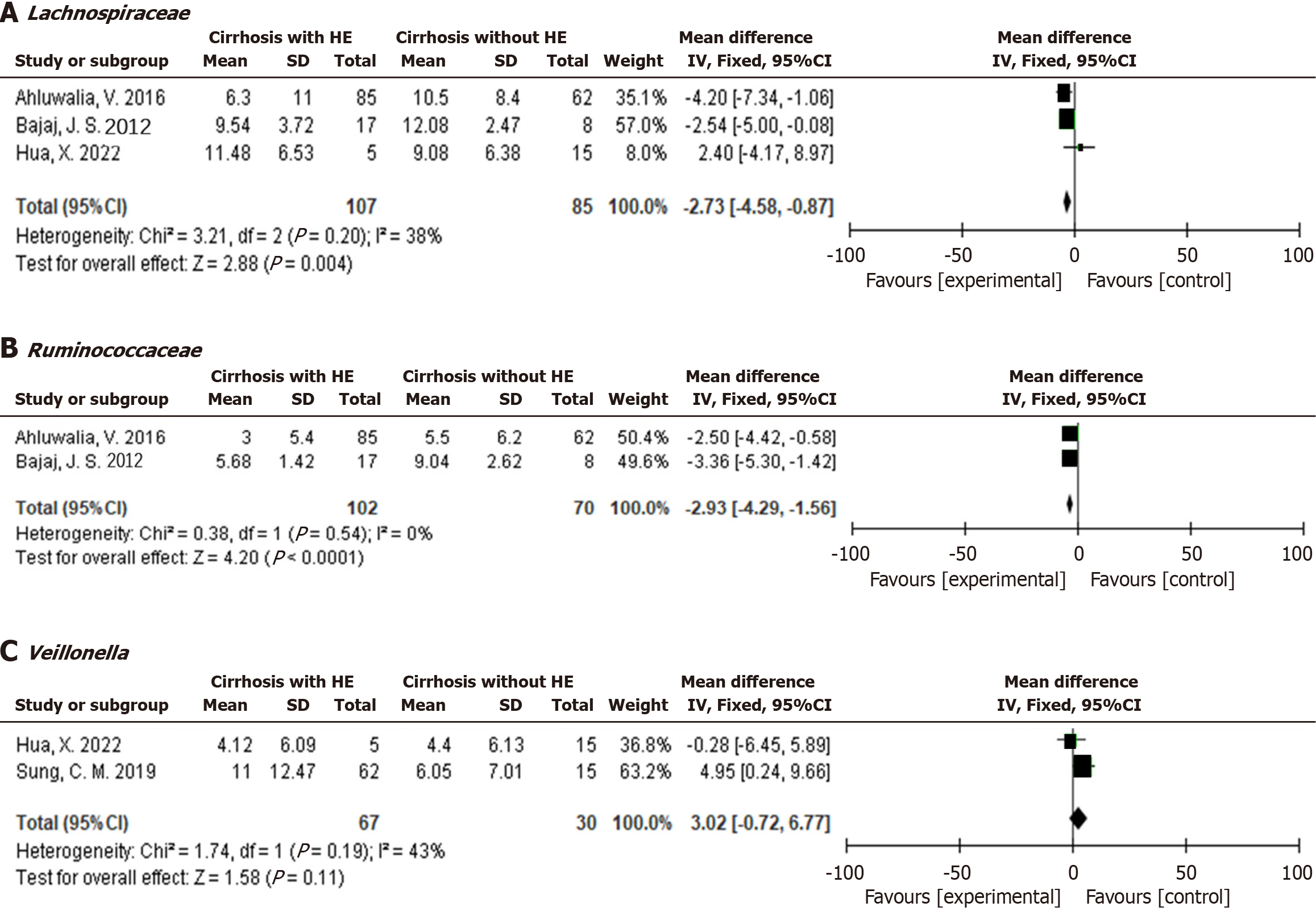
Owing to the lack of detailed information on the quantitative relative abundance of various gut bacteria between patients with liver cirrhosis with HE and those without HE in some studies, data on linear discriminant analysis effect size were available. Therefore, we summarized the gut microbiota mentioned in the included literature. Several intestinal bacteria, including Ruminococcaceae, Lachnospiraceae, Prevotellaceae, and Bacteroidetes, which showed a decreasing trend in patients with liver cirrhosis with HE, were consistent across multiple studies. The abundances of Enterococcus, Proteobacteria, Enterococcaceae, and Enterobacteriaceae increased in HE patients with liver cirrhosis. Afterwards, we analyzed the intestinal bacteria that could extract accurate data and found that Lachnospiraceae (-2.72, 95%CI: -4.58 to -0.87, I2 = 38%) and Ruminococcaceae (-2.93, 95%CI: -4.29 to -1.56, I2 = 0%) showed lower levels in liver cirrhosis patients with HE at the family level, but Veillonella at the genus level was not significantly different between the two groups (3.02, 95%CI: -0.72 to 6.77, I2 = 43%).
Lachnospiraceae, Ruminococcaceae, and Clostridiales XIV are beneficial local taxa that can produce short-chain fatty acids and increase the content of Bifidobacterium and Lactobacillus bacteria, maintaining the integrity of the intestinal barrier. In previous studies, these bacteria were generally found to be relatively abundant in patients with good cognition[28]. Other studies have also shown that the use of materials from healthy donors rich in Lachnospiraceae and Ruminococcaceae for fecal transplantation can effectively reduce the recurrence of HE and the occurrence of serious adverse events[29], which are negatively correlated with the Child-Pugh score[30] and increase the level of short-chain fatty acids in plasma[31]. Therefore, increasing these beneficial bacteria can contribute to the overall improvement of cognitive function and may prevent hospitalization related to the liver. However, these bacteria were reduced in patients with liver cirrhosis with HE, and their reduction also led to relative overgrowth of potential pathogenic taxa. Enterobacteriaceae and Enterococcus are increased in patients with liver cirrhosis with HE, and they increase intestinal permeability, which is related to disease progression and endotoxemia[5,30,32]. Moreover, Fusobacteriaceae, Veillonellaceae, and Enterobacteriaceae are positively correlated with inflammation[33].
With respect to the metabolic differences between HE patients and non-HE patients with in cirrhosis, all three articles mentioned short-chain fatty acids, but there were some differences in the research results. Previous studies have shown that cirrhosis patients have a decreased ability to produce butyric acid bacteria[34]. Based on the previous analysis of the differences in the gut microbiota between cirrhosis patients with HE and without HE, bacteria that produce short-chain fatty acids are reduced in liver cirrhosis patients with HE, and supplementation with bacteria that produce short-chain fatty acids can help improve cognitive function. Therefore, it was speculated that the level of short-chain fatty acids was lower in patients with liver cirrhosis with HE. The addition of short-chain fatty acids to strengthen the intestinal barrier and reduce the relative abundance of potential pathogenic taxa may be used to treat HE. Some studies have also shown that the gut microbiota is influenced by bile acid metabolism[35].
Owing to the inclusion of very few studies on the impact of PPIs on the gut microbiota, no meta-analysis has been conducted. The literature mentioned that after applying PPIs, the abundance of S. salivarius increased, and this bacterium contained urease operons encoding ammonia-producing enzymes. This may be due to reduced gastric acid secretion in liver cirrhosis, making it easier for S. salivarius to transfer from the mouth to the intestine through the weakened gastric acid barrier[36-38]. Previous studies revealed that the use of PPIs increased the abundance of Streptococcaceae in both cirrhotic and noncirrhotic patients[4,39,40], and that certain Streptococcal species can cause spontaneous bacterial pe
Previous studies have shown that lactulose and rifaximin have almost no effect on the gut microbiota[45-47]. In mouse studies, rifaximin did not affect the composition of the gut microbiota but rather beneficially altered the production of intestinal ammonia by regulating the expression of intestinal glutamine enzymes[48]. However, the two articles included in this study revealed that rifaximin can affect the abundance of certain intestinal bacteria to some extent, such as re
Although some studies have suggested that the gut microbiota is relatively stable within 6 months[5], the influence of the gut microbiota is significant, and there are significant differences in reported microbiota in the literature, with some inconsistencies in research results. However, several bacteria still presented consistent trends, indicating that the gut microbiota may become a biomarker for predicting the occurrence and prognosis of liver cirrhosis with HE[51-54]. Some studies have also suggested that these microbial changes are not related to the diagnostic criteria for HE; however, the existence of specific bacterial taxa is related to the understanding of this population[14]. This study has limitations in terms of heterogeneity: The number of included studies was limited, and the data that can be extracted for quantitative analysis were also limited. Therefore, subgroup analysis was not conducted on the basis of demographic characteristics such as race, age, or degree of HE.
The diversity of the gut microbiota in patients with liver cirrhosis with HE was reduced. There are characteristic changes in the gut microbiota in patients with liver cirrhosis with HE, and these changes in the gut microbiota composition and function can help predict the occurrence of HE and predict the prognosis of patients to some extent. However, there were significant differences between studies, and more large-scale studies are needed.
The authors would like to thank all the individuals who participated in this study.
| 1. | Bajaj JS, O'Leary JG, Tandon P, Wong F, Garcia-Tsao G, Kamath PS, Maliakkal B, Biggins SW, Thuluvath PJ, Fallon MB, Subramanian RM, Vargas HE, Lai J, Thacker LR, Reddy KR. Hepatic Encephalopathy Is Associated With Mortality in Patients With Cirrhosis Independent of Other Extrahepatic Organ Failures. Clin Gastroenterol Hepatol. 2017;15:565-574.e4. [PubMed] [DOI] [Full Text] |
| 2. | Elsaid MI, Rustgi VK. Epidemiology of Hepatic Encephalopathy. Clin Liver Dis. 2020;24:157-174. [PubMed] [DOI] [Full Text] |
| 3. | Schmidt TSB, Raes J, Bork P. The Human Gut Microbiome: From Association to Modulation. Cell. 2018;172:1198-1215. [PubMed] [DOI] [Full Text] |
| 4. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [PubMed] [DOI] [Full Text] |
| 5. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [PubMed] [DOI] [Full Text] |
| 6. | Chen Z, Ruan J, Li D, Wang M, Han Z, Qiu W, Wu G. The Role of Intestinal Bacteria and Gut-Brain Axis in Hepatic Encephalopathy. Front Cell Infect Microbiol. 2020;10:595759. [PubMed] [DOI] [Full Text] |
| 7. | Hassouneh R, Bajaj JS. Gut Microbiota Modulation and Fecal Transplantation: An Overview on Innovative Strategies for Hepatic Encephalopathy Treatment. J Clin Med. 2021;10. [PubMed] [DOI] [Full Text] |
| 8. | Bloom PP, Luévano JM Jr, Miller KJ, Chung RT. Deep stool microbiome analysis in cirrhosis reveals an association between short-chain fatty acids and hepatic encephalopathy. Ann Hepatol. 2021;25:100333. [PubMed] [DOI] [Full Text] |
| 9. | Wang Q, Chen C, Zuo S, Cao K, Li H. Integrative analysis of the gut microbiota and faecal and serum short-chain fatty acids and tryptophan metabolites in patients with cirrhosis and hepatic encephalopathy. J Transl Med. 2023;21:395. [PubMed] [DOI] [Full Text] |
| 10. | Yukawa-Muto Y, Kamiya T, Fujii H, Mori H, Toyoda A, Sato I, Konishi Y, Hirayama A, Hara E, Fukuda S, Kawada N, Ohtani N. Distinct responsiveness to rifaximin in patients with hepatic encephalopathy depends on functional gut microbial species. Hepatol Commun. 2022;6:2090-2104. [PubMed] [DOI] [Full Text] |
| 11. | Yu X, Jin Y, Zhou W, Xiao T, Wu Z, Su J, Gao H, Shen P, Zheng B, Luo Q, Li L, Xiao Y. Rifaximin Modulates the Gut Microbiota to Prevent Hepatic Encephalopathy in Liver Cirrhosis Without Impacting the Resistome. Front Cell Infect Microbiol. 2021;11:761192. [PubMed] [DOI] [Full Text] |
| 12. | Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168-G175. [PubMed] [DOI] [Full Text] |
| 13. | Bajaj JS, Betrapally NS, Hylemon PB, Heuman DM, Daita K, White MB, Unser A, Thacker LR, Sanyal AJ, Kang DJ, Sikaroodi M, Gillevet PM. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260-1271. [PubMed] [DOI] [Full Text] |
| 14. | Bajaj JS, Fagan A, White MB, Wade JB, Hylemon PB, Heuman DM, Fuchs M, John BV, Acharya C, Sikaroodi M, Gillevet PM. Specific Gut and Salivary Microbiota Patterns Are Linked With Different Cognitive Testing Strategies in Minimal Hepatic Encephalopathy. Am J Gastroenterol. 2019;114:1080-1090. [PubMed] [DOI] [Full Text] |
| 15. | Sung CM, Lin YF, Chen KF, Ke HM, Huang HY, Gong YN, Tsai WS, You JF, Lu MJ, Cheng HT, Lin CY, Kuo CJ, Tsai IJ, Hsieh SY. Predicting Clinical Outcomes of Cirrhosis Patients With Hepatic Encephalopathy From the Fecal Microbiome. Cell Mol Gastroenterol Hepatol. 2019;8:301-318.e2. [PubMed] [DOI] [Full Text] |
| 16. | Ahluwalia V, Betrapally NS, Hylemon PB, White MB, Gillevet PM, Unser AB, Fagan A, Daita K, Heuman DM, Zhou H, Sikaroodi M, Bajaj JS. Impaired Gut-Liver-Brain Axis in Patients with Cirrhosis. Sci Rep. 2016;6:26800. [PubMed] [DOI] [Full Text] |
| 17. | Zhang Z, Zhai H, Geng J, Yu R, Ren H, Fan H, Shi P. Large-scale survey of gut microbiota associated with MHE Via 16S rRNA-based pyrosequencing. Am J Gastroenterol. 2013;108:1601-1611. [PubMed] [DOI] [Full Text] |
| 18. | Montgomery JY, Bajaj JS. Advances in the evaluation and management of minimal hepatic encephalopathy. Curr Gastroenterol Rep. 2011;13:26-33. [PubMed] [DOI] [Full Text] |
| 19. | Prakash R, Mullen KD. Mechanisms, diagnosis and management of hepatic encephalopathy. Nat Rev Gastroenterol Hepatol. 2010;7:515-525. [PubMed] [DOI] [Full Text] |
| 20. | Zhu X, Zhou Z, Pan X. Research reviews and prospects of gut microbiota in liver cirrhosis: a bibliometric analysis (2001-2023). Front Microbiol. 2024;15:1342356. [PubMed] [DOI] [Full Text] |
| 21. | Yamamoto K, Honda T, Inukai Y, Yokoyama S, Ito T, Imai N, Ishizu Y, Nakamura M, Kawashima H. Identification of the Microbiome Associated with Prognosis in Patients with Chronic Liver Disease. Microorganisms. 2024;12. [PubMed] [DOI] [Full Text] |
| 22. | Wu Z, Zhou H, Liu D, Deng F. Alterations in the gut microbiota and the efficacy of adjuvant probiotic therapy in liver cirrhosis. Front Cell Infect Microbiol. 2023;13:1218552. [PubMed] [DOI] [Full Text] |
| 23. | Bajaj JS. The role of microbiota in hepatic encephalopathy. Gut Microbes. 2014;5:397-403. [PubMed] [DOI] [Full Text] |
| 24. | Won SM, Oh KK, Gupta H, Ganesan R, Sharma SP, Jeong JJ, Yoon SJ, Jeong MK, Min BH, Hyun JY, Park HJ, Eom JA, Lee SB, Cha MG, Kwon GH, Choi MR, Kim DJ, Suk KT. The Link between Gut Microbiota and Hepatic Encephalopathy. Int J Mol Sci. 2022;23. [PubMed] [DOI] [Full Text] |
| 25. | Zhu R, Liu L, Zhang G, Dong J, Ren Z, Li Z. The pathogenesis of gut microbiota in hepatic encephalopathy by the gut-liver-brain axis. Biosci Rep. 2023;43. [PubMed] [DOI] [Full Text] |
| 26. | Vilstrup H, Amodio P, Bajaj J, Cordoba J, Ferenci P, Mullen KD, Weissenborn K, Wong P. Hepatic encephalopathy in chronic liver disease: 2014 Practice Guideline by the American Association for the Study of Liver Diseases and the European Association for the Study of the Liver. Hepatology. 2014;60:715-735. [PubMed] [DOI] [Full Text] |
| 27. | Kang DJ, Betrapally NS, Ghosh SA, Sartor RB, Hylemon PB, Gillevet PM, Sanyal AJ, Heuman DM, Carl D, Zhou H, Liu R, Wang X, Yang J, Jiao C, Herzog J, Lippman HR, Sikaroodi M, Brown RR, Bajaj JS. Gut microbiota drive the development of neuroinflammatory response in cirrhosis in mice. Hepatology. 2016;64:1232-1248. [PubMed] [DOI] [Full Text] |
| 28. | Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189-200. [PubMed] [DOI] [Full Text] |
| 29. | Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, Williams R, Sikaroodi M, Fuchs M, Alm E, John B, Thacker LR, Riva A, Smith M, Taylor-Robinson SD, Gillevet PM. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology. 2017;66:1727-1738. [PubMed] [DOI] [Full Text] |
| 30. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [PubMed] [DOI] [Full Text] |
| 31. | Bajaj JS, Gavis EA, Fagan A, Wade JB, Thacker LR, Fuchs M, Patel S, Davis B, Meador J, Puri P, Sikaroodi M, Gillevet PM. A Randomized Clinical Trial of Fecal Microbiota Transplant for Alcohol Use Disorder. Hepatology. 2021;73:1688-1700. [PubMed] [DOI] [Full Text] |
| 32. | Quigley EM, Stanton C, Murphy EF. The gut microbiota and the liver. Pathophysiological and clinical implications. J Hepatol. 2013;58:1020-1027. [PubMed] [DOI] [Full Text] |
| 33. | Dhiman RK. Gut microbiota and hepatic encephalopathy. Metab Brain Dis. 2013;28:321-326. [PubMed] [DOI] [Full Text] |
| 34. | Jin M, Kalainy S, Baskota N, Chiang D, Deehan EC, McDougall C, Tandon P, Martínez I, Cervera C, Walter J, Abraldes JG. Faecal microbiota from patients with cirrhosis has a low capacity to ferment non-digestible carbohydrates into short-chain fatty acids. Liver Int. 2019;39:1437-1447. [PubMed] [DOI] [Full Text] |
| 35. | Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949-955. [PubMed] [DOI] [Full Text] |
| 36. | Tsai CF, Chen MH, Wang YP, Chu CJ, Huang YH, Lin HC, Hou MC, Lee FY, Su TP, Lu CL. Proton Pump Inhibitors Increase Risk for Hepatic Encephalopathy in Patients With Cirrhosis in A Population Study. Gastroenterology. 2017;152:134-141. [PubMed] [DOI] [Full Text] |
| 37. | Scobie BA, Summerskill WH. Reduced gastric acid output in cirrhosis: Quantitation and relationships. Gut. 1964;5:422-428. [PubMed] [DOI] [Full Text] |
| 38. | Lodato F, Azzaroli F, Di Girolamo M, Feletti V, Cecinato P, Lisotti A, Festi D, Roda E, Mazzella G. Proton pump inhibitors in cirrhosis: tradition or evidence based practice? World J Gastroenterol. 2008;14:2980-2985. [PubMed] [DOI] [Full Text] |
| 39. | Bajaj JS, Acharya C, Fagan A, White MB, Gavis E, Heuman DM, Hylemon PB, Fuchs M, Puri P, Schubert ML, Sanyal AJ, Sterling RK, Stravitz TR, Siddiqui MS, Luketic V, Lee H, Sikaroodi M, Gillevet PM. Proton Pump Inhibitor Initiation and Withdrawal affects Gut Microbiota and Readmission Risk in Cirrhosis. Am J Gastroenterol. 2018;113:1177-1186. [PubMed] [DOI] [Full Text] |
| 40. | Minalyan A, Gabrielyan L, Scott D, Jacobs J, Pisegna JR. The Gastric and Intestinal Microbiome: Role of Proton Pump Inhibitors. Curr Gastroenterol Rep. 2017;19:42. [PubMed] [DOI] [Full Text] |
| 41. | Bert F, Valla D, Moreau R, Nicolas-Chanoine MH. Viridans group streptococci causing spontaneous bacterial peritonitis and bacteremia in patients with end-stage liver disease. Liver Transpl. 2008;14:710-1; author reply 712. [PubMed] [DOI] [Full Text] |
| 42. | Zhang J, Zhang C, Zhang Q, Yu L, Chen W, Xue Y, Zhai Q. Meta-analysis of the effects of proton pump inhibitors on the human gut microbiota. BMC Microbiol. 2023;23:171. [PubMed] [DOI] [Full Text] |
| 43. | Yamamoto K, Ishigami M, Honda T, Takeyama T, Ito T, Ishizu Y, Kuzuya T, Hayashi K, Goto H, Hirooka Y. Influence of proton pump inhibitors on microbiota in chronic liver disease patients. Hepatol Int. 2019;13:234-244. [PubMed] [DOI] [Full Text] |
| 44. | Dam G, Vilstrup H, Watson H, Jepsen P. Proton pump inhibitors as a risk factor for hepatic encephalopathy and spontaneous bacterial peritonitis in patients with cirrhosis with ascites. Hepatology. 2016;64:1265-1272. [PubMed] [DOI] [Full Text] |
| 45. | Vanhoutte T, De Preter V, De Brandt E, Verbeke K, Swings J, Huys G. Molecular monitoring of the fecal microbiota of healthy human subjects during administration of lactulose and Saccharomyces boulardii. Appl Environ Microbiol. 2006;72:5990-5997. [PubMed] [DOI] [Full Text] |
| 46. | Bajaj JS, Gillevet PM, Patel NR, Ahluwalia V, Ridlon JM, Kettenmann B, Schubert CM, Sikaroodi M, Heuman DM, Crossey MM, Bell DE, Hylemon PB, Fatouros PP, Taylor-Robinson SD. A longitudinal systems biology analysis of lactulose withdrawal in hepatic encephalopathy. Metab Brain Dis. 2012;27:205-215. [PubMed] [DOI] [Full Text] |
| 47. | Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, Fuchs M, Ridlon JM, Daita K, Monteith P, Noble NA, White MB, Fisher A, Sikaroodi M, Rangwala H, Gillevet PM. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8:e60042. [PubMed] [DOI] [Full Text] |
| 48. | Kang DJ, Kakiyama G, Betrapally NS, Herzog J, Nittono H, Hylemon PB, Zhou H, Carroll I, Yang J, Gillevet PM, Jiao C, Takei H, Pandak WM, Iida T, Heuman DM, Fan S, Fiehn O, Kurosawa T, Sikaroodi M, Sartor RB, Bajaj JS. Rifaximin Exerts Beneficial Effects Independent of its Ability to Alter Microbiota Composition. Clin Transl Gastroenterol. 2016;7:e187. [PubMed] [DOI] [Full Text] |
| 49. | Ponziani FR, Gerardi V, Pecere S, D'Aversa F, Lopetuso L, Zocco MA, Pompili M, Gasbarrini A. Effect of rifaximin on gut microbiota composition in advanced liver disease and its complications. World J Gastroenterol. 2015;21:12322-12333. [PubMed] [DOI] [Full Text] |
| 50. | Ruszkowski J, Witkowski JM. Lactulose: Patient- and dose-dependent prebiotic properties in humans. Anaerobe. 2019;59:100-106. [PubMed] [DOI] [Full Text] |
| 51. | Bajaj JS, Betrapally NS, Hylemon PB, Thacker LR, Daita K, Kang DJ, White MB, Unser AB, Fagan A, Gavis EA, Sikaroodi M, Dalmet S, Heuman DM, Gillevet PM. Gut Microbiota Alterations can predict Hospitalizations in Cirrhosis Independent of Diabetes Mellitus. Sci Rep. 2015;5:18559. [PubMed] [DOI] [Full Text] |
| 52. | Bajaj JS, Fagan A, Sikaroodi M, White MB, Sterling RK, Gilles H, Heuman D, Stravitz RT, Matherly SC, Siddiqui MS, Puri P, Sanyal AJ, Luketic V, John B, Fuchs M, Ahluwalia V, Gillevet PM. Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transpl. 2017;23:907-914. [PubMed] [DOI] [Full Text] |
| 53. | Chen Y, Guo J, Qian G, Fang D, Shi D, Guo L, Li L. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol. 2015;30:1429-1437. [PubMed] [DOI] [Full Text] |
| 54. | Bajaj JS, Vargas HE, Reddy KR, Lai JC, O'Leary JG, Tandon P, Wong F, Mitrani R, White MB, Kelly M, Fagan A, Patil R, Sait S, Sikaroodi M, Thacker LR, Gillevet PM. Association Between Intestinal Microbiota Collected at Hospital Admission and Outcomes of Patients With Cirrhosis. Clin Gastroenterol Hepatol. 2019;17:756-765.e3. [PubMed] [DOI] [Full Text] |
| 55. | Bajaj JS, Matin P, White MB, Fagan A, Deeb JG, Acharya C, Dalmet SS, Sikaroodi M, Gillevet PM, Sahingur SE. Periodontal therapy favorably modulates the oral-gut-hepatic axis in cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2018;315:G824-G837. [PubMed] [DOI] [Full Text] |
| 56. | Saboo K, Petrakov NV, Shamsaddini A, Fagan A, Gavis EA, Sikaroodi M, McGeorge S, Gillevet PM, Iyer RK, Bajaj JS. Stool microbiota are superior to saliva in distinguishing cirrhosis and hepatic encephalopathy using machine learning. J Hepatol. 2022;76:600-607. [PubMed] [DOI] [Full Text] |
| 57. | Li M, Li K, Tang S, Lv Y, Wang Q, Wang Z, Luo B, Niu J, Zhu Y, Guo W, Bai W, Wang E, Xia D, Wang Z, Li X, Yuan J, Yin Z, Trebicka J, Han G. Restoration of the gut microbiota is associated with a decreased risk of hepatic encephalopathy after TIPS. JHEP Rep. 2022;4:100448. [PubMed] [DOI] [Full Text] |
| 58. | Hua X, Feng H. Changes in intestinal microbiota of HBV-associated liver cirrhosis with/without hepatic encephalopathy. Medicine (Baltimore). 2022;101:e29935. [PubMed] [DOI] [Full Text] |
| 59. | Bajaj JS, Fagan A, McGeorge S, Sterling RK, Rogal S, Sikaroodi M, Gillevet PM. Area Deprivation Index and Gut-Brain Axis in Cirrhosis. Clin Transl Gastroenterol. 2022;13:e00495. [PubMed] [DOI] [Full Text] |
| 60. | Bajaj JS, Torre A, Rojas ML, Fagan A, Nandez IE, Gavis EA, De Leon Osorio O, White MB, Fuchs M, Sikaroodi M, Gillevet PM. Cognition and hospitalizations are linked with salivary and faecal microbiota in cirrhosis cohorts from the USA and Mexico. Liver Int. 2020;40:1395-1407. [PubMed] [DOI] [Full Text] |
| 61. | Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675-G685. [PubMed] [DOI] [Full Text] |