Published online Jun 27, 2024. doi: 10.4254/wjh.v16.i6.932
Revised: April 8, 2024
Accepted: April 28, 2024
Published online: June 27, 2024
Processing time: 148 Days and 17.6 Hours
Hepatocellular carcinoma (HCC) is a primary contributor to cancer-related mortality on a global scale. However, the underlying molecular mechanisms are still poorly understood. Long noncoding RNAs are emerging markers for HCC diagnosis, prognosis, and therapeutic target. No study of LINC01767 in HCC was published.
To conduct a multi-omics analysis to explore the roles of LINC01767 in HCC for the first time.
DESeq2 Package was used to analyze different gene expressions. Receiver operating characteristic curves assessed the diagnostic performance. Kaplan-Meier univariate and Cox multivariate analyses were used to perform survival analysis. The least absolute shrinkage and selection operator (LASSO)-Cox was used to identify the prediction model. Subsequent to the validation of LINC01767 expression in HCC fresh frozen tissues through quantitative real time polymerase chain reaction, next generation sequencing was performed following LINC01767 over expression (GSE243371), and Gene Ontology/Kyoto Encyclopedia of Genes and Genomes/Gene Set Enrichment Analysis/ingenuity pathway analysis was carried out. In vitro experiment in Huh7 cell was carried out.
LINC01767 was down-regulated in HCC with a log fold change = 1.575 and was positively correlated with the cancer stemness. LINC01767 was a good diagnostic marker with area under the curve (AUC) [0.801, 95% confidence interval (CI): 0.751-0.852, P = 0.0106] and an independent predictor for overall survival (OS) with hazard ratio = 1.899 (95%CI: 1.01-3.58, P = 0.048). LINC01767 nomogram model showed a satisfied performance. The top-ranked regulatory network analysis of LINC01767 showed the regulation of genes participating various pathways. LASSO regression identified the 9-genes model showing a more satisfied performance than 5-genes model to predict the OS with AUC > 0.75. LINC01767 was down-expressed obviously in tumor than para-tumor tissues in our cohort as well as in cancer cell line; the over expression of LINC01767 inhibit cell proliferation and clone formation of Huh7 in vitro.
LINC01767 was an important tumor suppressor gene in HCC with good diagnostic and prognostic performance.
Core Tip: LINC01767 was down-regulated based on The Cancer Genome Atlas and GSE dataset. It was positively with the cancer stemness in hepatocellular carcinoma (HCC) based on the single cell sequence data. LINC01767 demonstrate a good diagnostic and diagnostic performance. The 9-gene model demonstrated better performance than the 5-gene model in predicting the overall survival of HCC patients using least absolute shrinkage and selection operator regression, with an area under the curve greater than 0.75. LINC01767 was down regulated obviously in tumor than para-tumor tissues in our cohort. LINC01767 was down regulated in cancer cell line comparing with LO2; the over expression of LINC01767 inhibit the cell proliferation and impede the clone formation of Huh7 in vitro.
- Citation: Zhang L, Cui TX, Li XZ, Liu C, Wang WQ. Diagnostic and prognostic role of LINC01767 in hepatocellular carcinoma. World J Hepatol 2024; 16(6): 932-950
- URL: https://www.wjgnet.com/1948-5182/full/v16/i6/932.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i6.932
Hepatocellular carcinoma (HCC) is the leading liver cancer and ranks fifth among all cancers, resulting in more than 600000 deaths each year globally[1,2]. Especially in Asia, HCC was more prevailing for hepatitis B virus infection. In Western countries, HCC is linked to nonalcoholic fatty liver disease as well[3]. Due to the subtle progression of HCC, the majority of patients are diagnosed at advanced stages[4]. Chemotherapy, radiotherapy, and trans-catheter arterial chemo embolization are the conventional options for treating advanced HCC[5], yet the underlying mechanisms remain largely unknown[6].
Only about 2.3% of the human genome is made up of protein-coding RNAs[7-9]. The majority of primary transcripts consist of noncoding RNAs (ncRNAs), which play a crucial role in the maintenance of normal physiological functions and the development of human cancers[9,10]. Long ncRNAs (lncRNAs) (> 200 nts)[11,12] is a type of regulatory ncRNAs. LncRNAs serve as guides, decoys, scaffolds, and competitive endogenous RNAs, for other molecules, to control the expression of numerous genes and ncRNAs[13].
Numerous lncRNAs have been discovered to have significant functions in the pathology of HCC. Elevated levels of Malat1 have been associated with unfavorable outcomes in different cancer types, such as bladder[14], lung[15], gallbladder[13,16] , and liver cancers[12,17-19]. LncRNAs are utilized in diagnosing HCC and could be targeted for therapy[20,21]. Research indicates that XIST controls the expression of PTEN and enhances the advancement of HCC[22]. Systematic identification of non-coding pharmacogenomic potential in cancer address the importance of lncRNAs[23].
It was part of the diagnostic profile for HCC in a prior investigation[24]. LINC01767, also identified as CRML1 (colorectal metastatic long non-coding RNA, RP4-710M16.2), exhibited increased expression in metastatic colorectal cancer (CRC) and played a crucial part in the spread of CRC to the liver[25]. Nevertheless, there have been no multi-omics assessments or in vitro experiments conducted on it in HCC. Therefore, we conducted a multi-omics analysis to explore the functions of LINC01767 in HCC.
Data from RNA sequencing and patient clinical details were obtained from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/). A total of 422 cases were extracted including 50 normal tissues and 372 HCC patients. Informed certifications were all stated in the TCGA projected. The Gene Expression Omnibus (GEO) data were analyzed via LNCAR website [lnCAR: A comprehensive resource for lncRNAs from cancer arrays (renlab.org)]. Drug sensitivity prediction of regulation related genes was based GSCALite web, and the predicting drug sensitivity details information was listed in previous article.
RNA quantification and qualification for RNA-sequencing: RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, United States). NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, United States) was utilized to create sequencing libraries, adhering to the manufacturer’s guidelines. Index codes were incorporated to assign sequences to individual samples. Index-coded samples were clustered on a cBot Cluster Generation System with TruSeq PE Cluster Kit v3-cBot-HS (Illumia) following the manufacturer’s guidelines. Details can be accessed via genechem company https://www.genechem.com.cn/.Raw DATA is accessible on GSE243371.
Techniques utilized include cell culture, RNA silencing, and quantitative polymerase chain reaction: American Type Culture Collection (Rockville, MD, United States) provided the LO2 normal liver cell line and the HepG2, Hep3B, PLC, SK, 7721, and 293T human liver cancer cell lines. The cell line acquired in August 2022 was tested and authenticated by STR before being used for experiments, with additional testing conducted in March 2023. Subsequently, the cells were cultured in DMEM/DME/F-12/1640 (Invitrogen), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Cell lines were cultured in a 37 °C and 5% CO2 humid environment. The growth media, reagents, and supplements were acquired from the Gibco corporation.
Cells were transfected with 40 nM small interfering RNAs (siRNAs) for RNA interference as directed by the manufacturer. Total RNA was extracted 72 h after treatment with TRIZAL for quantitative real time polymerase chain reaction (RT-qPCR) analysis; cDNAs were generated from 0.5 μg of total RNA using a High-Capacity cDNA Reverse Transcription Kit (Dongyangfang, #FSQ101). The QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) was used to conduct RT-qPCR using Power SYBR Green PCR Master Mix (MonAmp™ ChemoHS qPCR Mix, #MQ00401S). The relative expression of genes was calculated using the ΔΔCt method and normalized to GAPDH.
The siRNAs (sense, antisense) were used as previous described[22]. The primer sequences used for RT-qPCR included LINC01767 forward (GCTAAGGGATTTGCCACTGC) and reverse (GGTTGGAGGATGGACGTTGA), as well as GAPDH forward (GGTGAAGGTCGGAGTCAACG) and reverse (TGGGTGGAATCATATTGGAACA).
Cell proliferation assay: The Thiazolyl Blue (MTT) experimental assay was utilized to generate cell growth curves. 1000 cells were plated in 96-well dishes and incubated for six days to measure cell survival. The experiments were conducted three times separately.
Transwell migration and invasion assays: Huh7 cells were placed in the top compartments of a 24-well transwell plate from Corning in Beijing, China. Matrigel was applied to the membranes during the invasion assay conducted by BD Biosciences. Cells were placed in the upper chamber with serum-free media. The bottom compartments were filled with the culture medium containing 10% FBS. After being treated with 4% paraformaldehyde for 15 min, the cells were then exposed to crystal violet following a 48-h incubation period. Five randomly chosen microscopic fields per filter were used to evaluate the cell counts, which were then observed using microscopy.
Wound-healing assay: Huh7 cells were cultured for 48 h until they reached 80% confluence, at which point a linear artificial wound was created using a 200 μL pipette tip. The cell migration ability was measured by photographing the distance at 0 h and 24 h.
Clone formation assay: After transfection, the 400-1000 cells/well (according to the cell generation) were seeded in each experimental group in 6-well plate culture plates. The seeded cells are cultured in the incubator for 14 d or the number of cells is greater than 50 cells in most individual clones change the fluid every 3 d and observe the cell state. Before the experiment is terminated, photograph the cell clone under fluorescence microscopy and wash the cells once with phosphate buffered saline (PBS). Add 1 mL of 4% paraformaldehyde per well, fix the cells for 30-60 min, and wash the cells once with PBS. Apply 1000 μL of pure crystal violet staining solution for 10-20 min. ddH2O was used to wash the cells, then microscopy was used to take pictures, and count the clones.
Cell cycle and apoptosis assay: PI-FACS cell cycle assay experimental was carried out as the procedures from the (PI Sigma P4170, Triton X-100 Sigma SLBT4524, RNase A Thermo Fisher Scientific EN0531). For adherent cells, once the 6 cm dish cells in each experimental group reach a coverage rate of approximately 80% (before entering the growth plateau), the cells will be digested and resuspended into a cell suspension, ensuring that the number of cells on the machine is adequate (≥ 106/group). Spin at 1300 r/min for 5 min, remove the liquid above the sediment, and rinse the cells with PBS (pH = 7.2-7.4) chilled at 4 degrees Celsius. Repeat the precipitation process once. Centrifuge at 1300 r/min for 5 min. Prepare the cell staining solution. Stain the cells by adding an appropriate volume of the staining solution (0.6-1 mL) based on the cell quantity and resuspending. The cell throughput rate is 300-800 cells/s. Apoptosis assay test using Annexin V-APC mono staining method was carried out as instruction of eBioscience kit (88-8007).
Development of a predictive signature associated with LINC01767 using the least absolute shrinkage and selection operator Cox regression model: In our study, the LINC01767 solely and its regulated pathway genes were used as candidate biomarkers. Using the ‘glmnet’ package in R, we applied least absolute shrinkage and selection operator (LASSO) Cox regression analysis to develop a precise prognostic signature for LIHC samples with the selected biomarkers. The risk score for each sample was determined by calculating the relative expression of each prognostic factor in the LINC01767 gene signature and its corresponding coefficient. The signature’s risk score was determined by summing the products of each gene’s relative expression and its corresponding LASSO coefficient.
Patients from the TCGA datasets were evenly split into two groups, low risk and the high risk, determined by the risk score of the gene signature associated with LINC01767. Next, we utilized time-sensitive receiver operating characteristic (ROC) curve assessment to determine the accuracy of our model’s prediction by calculating the area under the curve (AUC) for overall survival (OS) at 1 year, 3 years, and 5 years. This analysis was conducted using the ’survivalROC’ tool in R.
The RNA-sequence data from TCGA were normalized using TPM (Transcripts Per Million). Then different genes expression was analyzed by DESeq2 Package (Limma.R). To compare the expression levels of LINC01767 between the tumor and para-tumor groups using the two tailed unpaired t test, and between the different characteristic subgroup using ANOVA. Adjusted P values were calculated using Benjamini and Hochberg’s method to control the false rate. DESeq2 identified genes as differentially expressed if they had an adjusted P value < 0.05 and a log fold change (FC) > 1.5. Perform enrichment analysis based on differential expressed genes (DEGs) using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The Cluster Profiler R package was used to conduct GO enrichment analysis on DEGs. An analysis using Gene Set Enrichment Analysis (GSEA) was performed to evaluate the pathways that were enriched in the LINC01767-low and LINC01767-high groups.
ROC curves were employed to assess the diagnostic performance for HCC and the predictive ability of the nomogram and LASSO model. When AUC is greater than 0.5, the closer AUC is to 1, the more effective this variable is in predicting outcomes. AUC exhibits decreased accuracy between 0.5 and 0.7, demonstrates a satisfactory level of accuracy between 0.7 and 0.9, and achieves exceptional accuracy when exceeding 0.9. When AUC is equal to 0.5, the variable has no diagnostic value.
Kaplan-Meier analysis and Cox proportional hazards models were utilized to identify significant prognostic factors. Survival curves were generated with the Kaplan-Meier technique, and curves were compared using the log-rank test. A multivariate Cox regression analysis was conducted on the variables that achieved a significance level of P < 0.05 in the initial univariate analysis. When a notable impact was seen in the Cox model, independent predictors of prognosis were identified (P < 0.05). The final model’s variables were chosen through a stepwise process of backward regression based on the Akaike information criterion. The nomogram model was built using the final Cox proportional hazard regression model and the rms package in R version 3.8.1 (http//www.r-project.org/). The nomogram underwent internal verifications, with an evaluation of the model’s discrimination and calibration. Discrimination assessment in this article relied on the concordance index (C-index). A C-index value of 0.5 signified that the model did not have any predictive impact. A C-index of 1 showed that the model’s predicted outcomes matched perfectly with the observed outcomes. A higher C-index value indicates more accurate predictions from the model. ROC curve was plotted to indicate the precision of prediction of 1-year, 3-year, 5-year OS rate. AUC greater than 0.7 stand a promising prediction performance. All data were processed with R 3.8.1 and MedCalc software 19.4.2.0. A P value less than 0.05 indicates statistical significance.
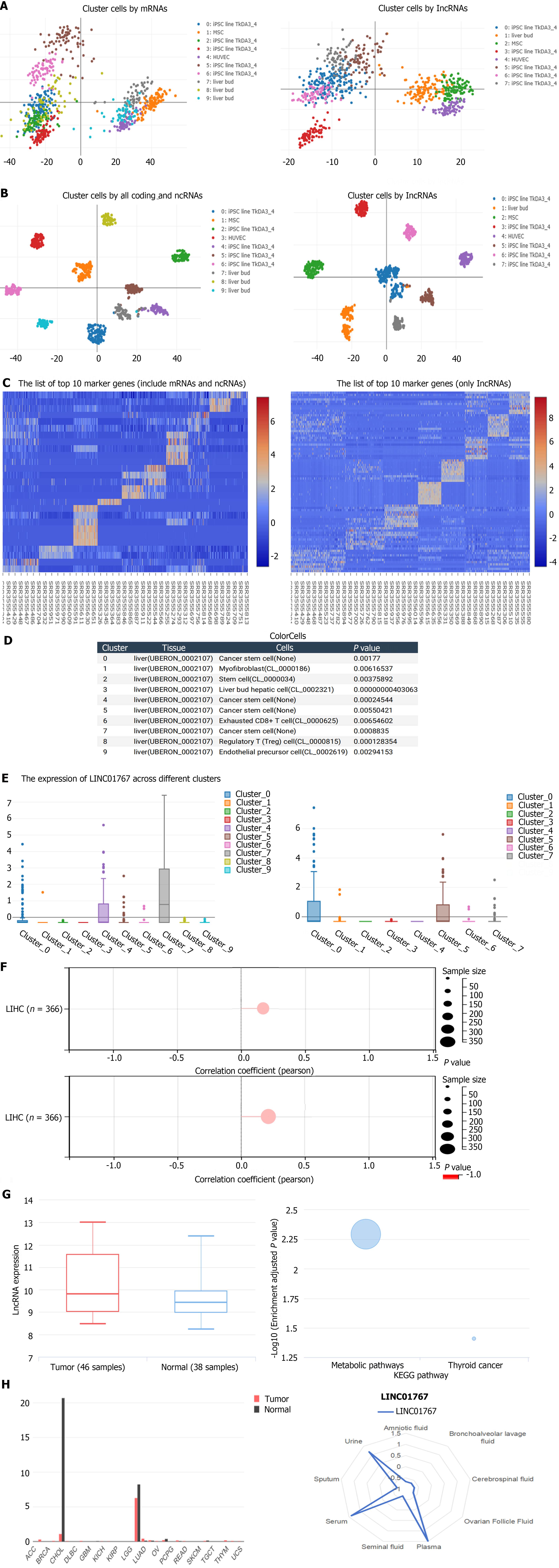
We analyze the expression of LINC01767 in single-cell sequencing data GSE81252 from web (https://rna.sysu.edu.cn/colorcells/index.php). Principal components analysis results showed that the 9-cluster cell clustered by all coding and ncRNAs and 7-cluster cell by ncRNAs solely were identified Figure 1A. The t-SNE Nonlinear dimensionality reduction showed the corresponding dimensionality reduction Figure 1B. The differential genes among different cell-cluster were shown in Figure 1C. The 9-clusters cell subgroups were classified as below: (1) Cluser 0/4/5/7 - cancer stem cell; (2) Cluser 1 - myofibroblast; cluster 2 - stem cell; (3) Cluser 3 - liver bud hepatic called cancer stem cell; (4) Cluster 6 - exhausted CD8+ T cell; (5) Cluster; (6) 8-regulatory T cell; and (7) Cluster 9 - endothelial precursor cell (Figure 1D). The expression of LINC01767 in different cluster was shown in Figure 1F, it was up regulated in cancer stem cell (cluster 7). And the differential gene enrichment of cluster 7 was shown in Figure 1E. In our analysis of liver carcinoma, we utilized the Stemness group’s DNA methylation-based Stemness Scores, specifically the Stem cell signature probes (219 probes), to assess the correlation between LINC01767 and the stem score using Pearson correlation. We found a notable correlation in LIHC (n = 366, r = 0.213, P = 0.000039). Additionally, the RNAss, which represents the RNA-based Stemness Scores from the Stemness group, confirmed the link between LINC01767 and cancer stem cells. there was a significant positive association in LIHC (n = 366, lR = 0.221, P = 0.0000206) (Figure 1F). All the above results showed that the LINC07167 was positively correlated with the cancer stem cell in liver cancer.
GSE78160 (serum lncRNAs) data showed that the serum LINC01767 were up regulated in HCC patients with a logFC 0.699, P = 0.026524 (Figure 1G). Besides, the GO/KEGG analysis from lnCAR website showed that the LINC01767 is associated with the metabolic Figure 1G. The LINC01767 was expressed in liver and bile duct with a significantly higher level than other tissues Figure 1H. LINC01767 demonstrate a high expression level of serum, urine, and plasma LINC01767 (Figure 1H). Therefore, investigating the significance of LINC01767 in liver cancer was crucial.
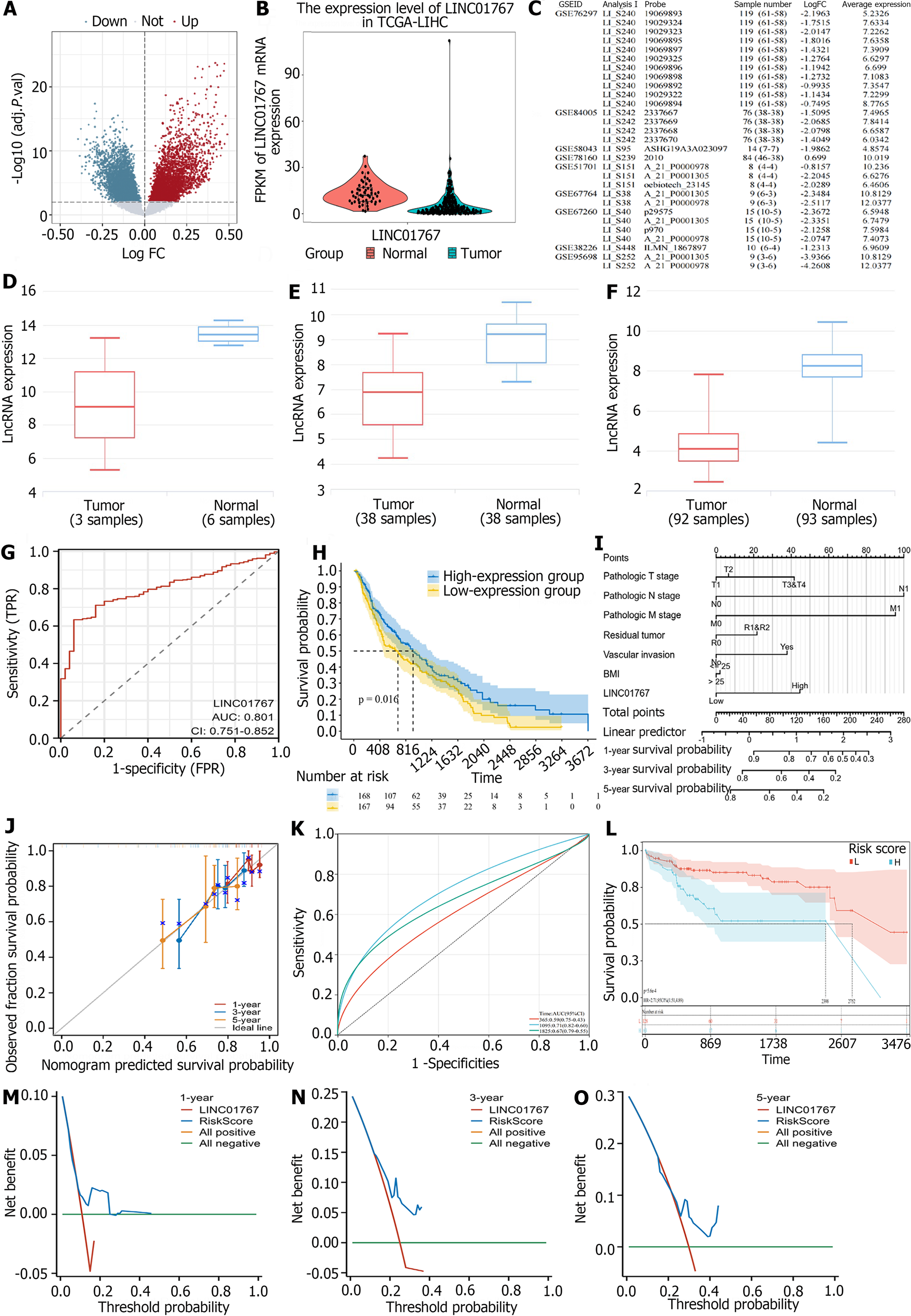
A total of 3529 up regulated and 250 down regulated lncRNAs was identified in TCGA liver cancer. Among them, the previous identified lncRNAs (Figure 2A), such as HULC (upregulation), FENDRR (downregulation) were included as well. Additionally, we were intrigued by the LINC01767 due to its logFC of -1.575 and P < 0.0001 in Figure 2B. Furthermore, we confirmed the expression of LINC01767 in the GEO data set, revealing down regulation in GSE76297, GSE84005, GSE58043, and GSE51701 with fold changes between -0.815 and -2.196 (Figure 2C-F).
ROC curves were used to evaluate the diagnostic accuracy of LINC01767 in our study. The findings indicated that LINC01767 exhibited excellent diagnostic accuracy, with an AUC of 0.801 [95% confidence interval (CI): 0.751-0.852, P = 0.0106], specificity of 94.00% (95%CI: 83.5-98.7), and sensitivity of 61.19% (95%CI: 56.0-66.2), at the optimal threshold of TPM 2.6729 (see Figure 2G). Higher LINC01767 was correlated with higher alpha-fetoprotein (AFP) (P = 0.038), but it had no significant correlation with other characteristics (Supplementary Table 1). LINC01767 could be a potentially sensitive diagnostic marker for HCC better than AFP.
After omitting the dead cases within 30 d a total of 342 cases from TCGA were included. The study cohort had OS rates of 85.09% at 1 year, 71.64% at 3 years, and 66.67% at 5 years, along with a median OS of 20.745 months. The cohort had disease-free survival (DFS) rates of 65.44%, 47.65%, and 44.30% at 1, 3, and 5 years, with a median DFS of 14.45 months. The clinical and pathological viable influencing OS and DFS are listed in Table 1. In univariate viables survival analysis using Kaplan-Meier plot, significant predictors of OS were body mass index (BMI) (P = 0.008), risk factors (P = 0.007), the surgery method (P < 0.001), American Joint Committee on Cancer (AJCC) 7th tumor-node-metastasis (TNM) classification (PT < 0.0001, PN = 0.0325, Pstage < 0.0001), margin status (P = 0.0002), vascular invasion (P = 0.0326). While the LINC01767 expression level stands as a predictor for OS (P = 0.016) (Figure 2H). Significant predictors of DFS were BMI (P = 0.0295), risk factors (P = 0.007), surgery method (P = 0.001), AJCC 7th TNM classification (PT < 0.0001, Pstage < 0.0001), vascular invasion (P = 0.0004). And LINC01767 expression level did not showed as the predictor for DFS (P = 0.7114), and the margin status did not either (with borderline significance, P = 0.0806).
| Variable | Number | Event/total | MST months | P value | Number | Event/total | MST months | P value |
| DFS | OS | |||||||
| Gender | 0.429 | 0.227 | ||||||
| Male | 208 | 116/208 | 21.55 (17.64-33.05) | 233 | 74 | 81.67 (53.29-NA) | ||
| Female | 90 | 57/90 | 18.59 (14.85-35.84) | 109 | 49 | 48.95 (37.29-80.68) | ||
| Age | 0.644 | 0.488 | ||||||
| ≤ 40 | 25 | 17/25 | 12.91 (8.61-NA) | 29 | 9 | NA (20.11-NA) | ||
| 40-60 | 113 | 61/113 | 25.3 (14.85-67.58) | 128 | 44 | 69.51 (40.37-NA) | ||
| > 60 | 100 | 55/100 | 24.77 (18.59-47.04) | 1113 | 35 | 80.68 (55.35-NA) | ||
| > 70 | 52 | 35/52 | 21.16 (14.59-33.05) | 64 | 34 | 45.53 (33.02-70.01) | ||
| Race | 0.145 | 0.392 | ||||||
| White | 146 | 96 | 19.19 (14.956-23.424) | 168 | 71 | 5.117 (35.861-55.919) | ||
| Asian | 133 | 69 | 27.2 (4.977-49.423) | 148 | 42 | NA | ||
| Black | 11 | 4 | 67.58 (NA) | 15 | 5 | 16.286 (5.829-69.671) | ||
| BMI | 0.029a | 0.008a | ||||||
| < 18.5 | 16 | 9 | 13.07 (9.13-NA) | 19 | 6 | 107.03 (20.8-NA) | ||
| 18.2-23 | 77 | 50 | 12.84 (9.49-29.66) | 89 | 40 | 35.74 (22.77-NA) | ||
| 23-25 | 49 | 22 | 50.03 (21.16-NA) | 55 | 12 | 81.67 (81.67-NA) | ||
| 25-30 | 78 | 40 | 21.62 (16.13-NA) | 88 | 28 | 70.01 (37.68-NA) | ||
| > 30 | 57 | 36 | 25.3 (15.7-NA) | 64 | 20 | 69.51 (45.89-NA) | ||
| Gene expression | 0.711 | 0.016a | ||||||
| High | 133 | 72 | 28.88 (19.45-47.73) | 168 | 103 | 53.29 (39.75-107.03) | ||
| Low | 132 | 67 | 16.49 (13.07-23.62) | 167 | 122 | 69.51 (51.25-NA) | ||
| Family history of cancer | 0.515 | 0.439 | ||||||
| Yes | 91 | 53 | 18.33 (13.14-37.12) | 104 | 39 | 69.51 (43.47-95.55) | ||
| No | 168 | 99 | 23.95 (18.59-35.58) | 194 | 71 | 55.65 (33.384-77.916) | ||
| Risks of HCC | 0.724 | 0.007a | ||||||
| No | 60 | 35 | 18.59 (12.514-24.66) | 83 | 44 | 33.02 (23.447 42.593) | ||
| Yes | 219 | 125 | 23.030 (16.229-29.831) | 241 | 73 | 80.68 (50.109-111.251) | ||
| AFP | 0.736 | 0.698 | ||||||
| < 400 ng/mL | 183 | 99 | 27.2 (20.99-36.7) | 199 | 56 | 80.68 (55.35-NA) | ||
| ≥ 400 ng/mL | 51 | 29 | 15.7 (8.64-NA) | 61 | 21 | 81.67 (33.02-NA) | ||
| CHIl-phu grade | 0.658 | 0.220 | ||||||
| A | 187 | 101 | 23.03 (18.59-42.04) | 204 | 52 | 102.66 (70.01-NA) | ||
| B | 17 | 10 | 23.95 (8.64-NA) | 20 | 8 | 33.02 (19.74-NA) | ||
| C | 1 | 1 | 35.58 (NA-NA) | 1 | 1 | 53.35 (NA-NA) | ||
| Surgery | 0.0001a | < 0.0001a | ||||||
| 1 | 121 | 79 | 13.14 (9.72-19.25) | 129 | 48 | 39.75 (30.58-NA) | ||
| 2 | 18 | 12 | 15.41 (6.24-NA) | 25 | 14 | 37.29 (17.58) | ||
| 3 | 75 | 34 | 47.04 (28.8-71.06) | 81 | 13 | 81.67 (69.51) | ||
| 4 | 68 | 39 | 29.96 (18.33-71.91) | 79 | 32 | 70.01 (46.75) | ||
| 5 | 1 | 0 | NA (NA-NA) | 1 | 0 | NA (NA-NA) | ||
| 6 | 15 | 9 | 19.65 (8.28-NA) | 25 | 14 | 19.58 (4.24-NA) | ||
| Tumor histological grade | 0.656 | 0.693 | ||||||
| G1 | 47 | 25 | 29.66 (19.45-73.62) | 53 | 17 | 69.51 (46.75-NA) | ||
| G2 | 138 | 80 | 20.99 (16.36-28.88) | 161 | 58 | 55.65 (41.75-NA) | ||
| G3 | 99 | 59 | 15.74 (12.61-47.04) | 111 | 39 | 53.29 (45.07-NA) | ||
| G4 | 10 | 5 | 7.36 (4.17-NA) | 12 | 5 | NA (13.47-NA) | ||
| T-stage | < 0.0001a | < 0.0001a | ||||||
| T1 | 151 | 65 | 42.02 (33.06-71.06) | 168 | 41 | 80.68 (69.51-NA) | ||
| T2 | 73 | 49 | 15.7 (11.17-23.62) | 84 | 28 | 60.84 (37.75-NA) | ||
| T3 | 62 | 49 | 9.94 (8.54-14.85) | 74 | 43 | 25.3 (18.27-58.84) | ||
| T4 | 9 | 8 | 7.85 (5.91-NA) | 13 | 10 | 18.33 (8.94-NA) | ||
| Lymph node status | 0.357 | 0.033a | ||||||
| Negative | 211 | 118 | 23.03 (16.49-40.37) | 239 | 2 | 83.51 (60.84-NA) | ||
| Positive | 3 | 2 | 8.64 (6.6-NA) | 3 | 77 | 33.02 (9.86-NA) | ||
| X | 84 | 53 | 14.13-28.88 | 99 | 43 | 37.68 (27.5-55.65) | ||
| Margin | 0.0806 | 302 | 104 | 69.51 (53.29-102.66) | 0.0002a | |||
| R0 | 270 | 154 | 21.62 | 15 | 8 | 37.29 (27.5-NA) | ||
| R1 | 13 | 11 | 15.6 | 1 | 1 | 7.33 (NA-NA) | ||
| Vascular invasion | 0.0004a | 0.0326a | ||||||
| Negative | 164 | 77 | 35.84 (28.88-67.58) | 187 | 54 | 80.68 (55.65-NA) | ||
| Positive | 73 | 47 | 13.63 (10.81-24.77) | 27 | 27 | 81.67 (45.89-NA) | ||
| NA | 13 | 9 | 8.74 (4.14-NA) | 8 | 8 | 48.95 (13.96-NA) | ||
| Adjacent hepatic tissue inflammation | 0.3787 | 0.772 | ||||||
| No | 100 | 56 | 23.95 (18.59-40.37) | 112 | 35 | 80.68 (55.65-NA) | ||
| Mild | 86 | 48 | 21.55 (18.17-53.55) | 93 | 25 | NA (45.53-NA) | ||
| Severe | 15 | 10 | 18.33 (9.72-NA) | 17 | 5 | NA (35.74-NA) | ||
| Stage | < 0.0001a | < 0.0001a | ||||||
| 1 | 145 | 62 | 40.37 (29.96-71.91) | 161 | 37 | 83.18 (70.01-NA) | ||
| 2 | 66 | 43 | 18.17 (9.72-29.3) | 77 | 24 | 60.84 (45.53-NA) | ||
| 3 | 67 | 52 | 9.76 (8.34-14.22) | 80 | 45 | 25.3 (20.11-58.84) | ||
| 4 | 2 | 2 | 7.49 (5.49-NA) | 3 | 3 | 18.33 (7.33-NA) | ||
Further, we conduct the multiple factor analysis using Cox regression. There were 373 samples in the original data, 182 samples with missing variable information, and the final number of samples was 191. LINC01767 was an independent predictor for OS in HCC when included the above factors in Cox regression with hazard ratio (HR) = 1.899 (95%CI: 1.01-3.58, P = 0.048) (Table 2).
| Characteristics | Beta | HR | 95%CI | P value |
| Pathologic T stage | ||||
| T1 | Reference | |||
| T2 | 0.085 | 1.089 | 0.455-2.603 | 0.848 |
| T3 | 0.608 | 1.838 | 0.857-3.943 | 0.118 |
| T4 | 0.364 | 1.440 | 0.1684-12.317 | 0.739 |
| Pathologic N stage | ||||
| N0 | Reference | |||
| N1 | 1.413 | 4.108 | 0.528-31.958 | 0.177 |
| Pathologic M stage | ||||
| M0 | Reference | |||
| M1 | 1.580 | 4.855 | 0.378-62.357 | 0.225 |
| BMI | ||||
| ≤ 25 | Reference | |||
| > 25 | -0.013 | 0.987 | 0.532-1.830 | 0.966 |
| Residual tumor | ||||
| R0 | Reference | |||
| R1 | 0.289 | 1.334 | 0.172-10.360 | 0.782 |
| R2 | 0.0602 | 1.062 | 0.0340-33.231 | 0.972 |
| Vascular invasion | ||||
| No | Reference | |||
| Yes | 0.556 | 1.743 | 0.829-3.664 | 0.143 |
| LINC01767 | ||||
| Low | Reference | |||
| High | 0.642 | 1.89944998718678 | 1.006-3.585 | 0.048a |
A predictive nomogram was created using important factors identified in the Cox analysis to predict the outcome of HCC, such as T/N/Residual tumor, vascular invasion, BMI, and LINC01767 shown in Figure 2I. In this case, a patient who had a BMI greater than 5 (which earned 2 points), underwent extensive surgery (earning 0 points), and the postoperative pathology revealed no lymph node metastasis (also earning 0 points), Elevated levels of LINC01767 expression (44 points) was calculated. The overall score is 46 points, with a 1-year survival rate of approximately 95%, a 3-year survival rate of around 85%, and a 5-year survival rate of roughly 75%. Internal validation demonstrated the nomogram’s ability to effectively predict OS, with a C-index of 0.620 (0.573-0.667), indicating strong agreement. The calibration graph indicated a strong agreement between the projected and actual outcomes for 1-year and 3-year OS rates (Figure 2J). ROC was used to assess prediction performance, yielding an AUC of 0.59 (95%CI: 0.75-0.43) for 1-year accuracy, 0.71 (95%CI: 0.82-0.60) for 3-year accuracy, and 0.67 (95%CI: 0.79-0.55) for 5-year accuracy. These results indicate good predictive performance, as shown in Figure 2K. In Figure 2L, the TCGA high-risk group had a poorer prognosis with a median survival time of 2398 d compared to 2752 d in the low-risk group, with a HR of 2.71 (95%CI: 1.51-4.89, P = 5.6e-4). The nomogram model demonstrated superior DCA performance compared to LINC01767 alone, with a value above the line 0 in Figure 2M-O. Internal validation confirmed the nomogram’s ability to accurately predict the C-index of OS, which was 0.620 (95%CI: 0.573-0.667) with strong concordance. ROC was used to assess the prediction performance, yielding an AUC of 0.59 (95%CI: 0.75-0.43) for 1-year accuracy, 0.71 (95%CI: 0.82-0.60) for 3-year accuracy, and 0.67 (95%CI: 0.79-0.55) for 5-year accuracy, indicating strong predictive capabilities. Cox regression is applied on the premise that the independent variables are required to meet the proportional risk hypothesis (P > 0.05), and if the GLOBAL satisfies P > 0.05, it can be considered that the multi-factor model satisfies the proportional risk hypothesis in Supplementary Table 2. The VIF can be utilized to examine variables within a model for problems related to multicollinearity. It is generally believed that when 0 < VIF < 10, there is no multicollinearity. The findings indicated that the LINC01767 nomogram’s Cox model met the PH hypothesis (P > 0.05) and the VIF confirmed the independence of these factors. These results suggest that the LASSO model of LINC01767 demonstrated a strong prognostic ability (Supplementary Table 3).
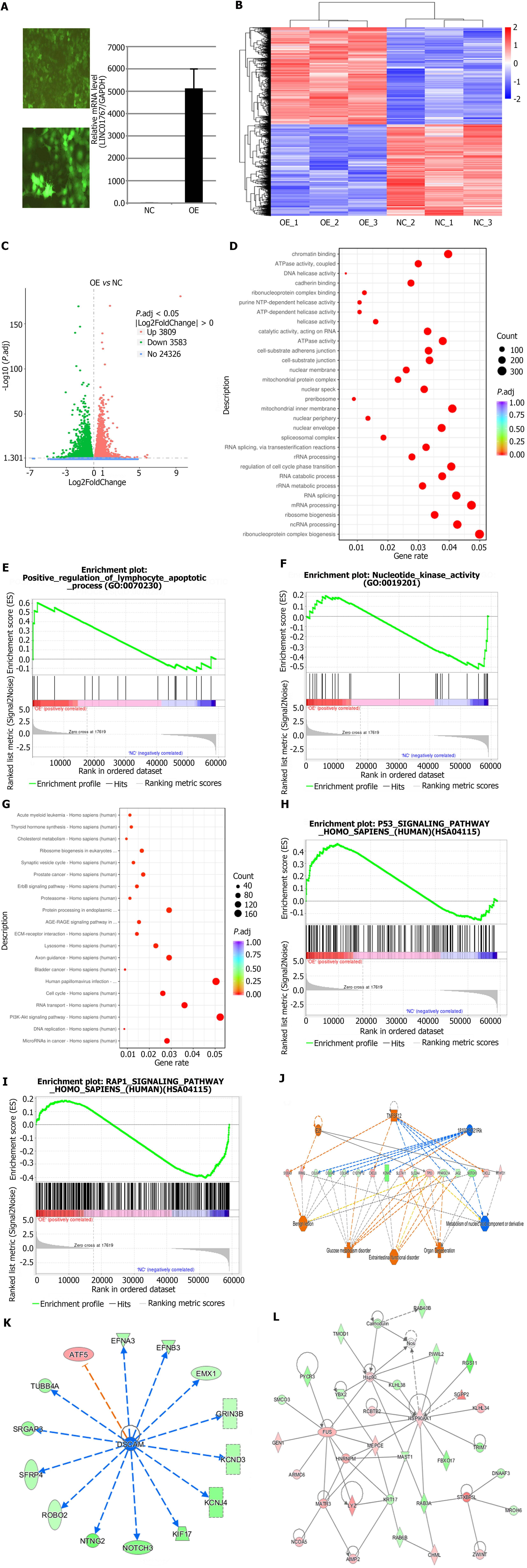
After transfecting of Huh7 cell lines with lentivirus - LINC017670-OE plasmid, RNA transcriptome sequencing were performed for preliminary DEGs analysis (Figure 3A), the sequencing data quality was valid, the differential expression volcano plot is presented using the logFC > 1.5, and P < 0.05 (Figure 3B and C). The colors green and red indicate genes that are down regulated and up regulated, respectively.
GO and KEGG pathway analysis for deferentially expressed genes was performed. The top 10 most significantly enriched GO (Figure 3D) and KEGG terms (Figure 3G) were presented, respectively. The GO analysis identified 498 GO categories that were significantly enriched, including ‘carbohydrate biosynthetic process’, ‘gluconeogenesis’, and ‘cellular carbohydrate metabolic process’ shown in Figure 3D. 21 KEGG pathways were identified with P and Q-values less than 0.05. GSEA analysis indicated that the go term of LINC01767 is associated with positive regulation of lymphocyte apoptosis, nucleotide kinase activity, and more other pathways (Figure 3E and F). Additionally, KEGG pathway analysis revealed that LINC01767 is linked to P53 signaling, RAP1 signaling, and other significant pathways (Figure 3H and I).
The P < 0.05 and the differential gene of |FC| > 2 was adopted for ingenuity pathway analysis (IPA) analysis. Classical path analysis in pathogenesis of LINC01767 based on IPA shows the significant enrichment of differential genes in disease and function. Figure 3J shows the interaction between genes, regulators and functions in the dataset. The top-ranked regulatory network shows that the regulation net involved in regulators 1810019D21Rik, E2f, TNF superfamily member 12 (TNFSF12) through C1q and TNF related 12 (C1QTNF12), collagen type VI alpha 1 (COL6A1), COL6A2, COL9A3, CXC motif chemokine ligand 2 (CXCL2), CXCL8, Jagged2 (JAG2), KCNH2, methylenetetrahydrofolate dehydrogenase-cyclohydrolase 1 (MTHFD1), NOTCH3, peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PPARGC1A), ribonucleotide reductase regulatory subunit M2 (RRM2), S100A9, solute carrier 2A4 (SLC2A4), SLC7A11, TP53, and other genes can activate Benign lesion, extraintestinal functional disorder, glucose metabolism disorder, organ degeneration, and inhibit metabolism of nucleic acid component or derivative. Figure 3K and L showed the protein-protein interaction of the LINC01767 related genes.
LASSO Cox regression analysis was performed with LINC01767 related genes. The model identified 9 genes: S100A9, RRM2, COL6A1, CXCL8, KCNH2, SLC7A11, TP53, PPARGC1A, and CXCL2. The ROC curve demonstrated that the 9-gene model had better performance in predicting OS of liver cancer patients compared to the 5-gene model. The best cut-off value for LASSO risk score was calculated to be 0.68567, dividing patients into high- and low- risk groups. Prognostic differences between the groups were analyzed using the log-rank test, showing a significant difference (P = 3.4e-14). Additionally, using a 5 genes signature, the optimal cutoff was 0.875627, with a significant prognostic difference between groups (P = 7.9e-14) (Supplementary Figure 1).
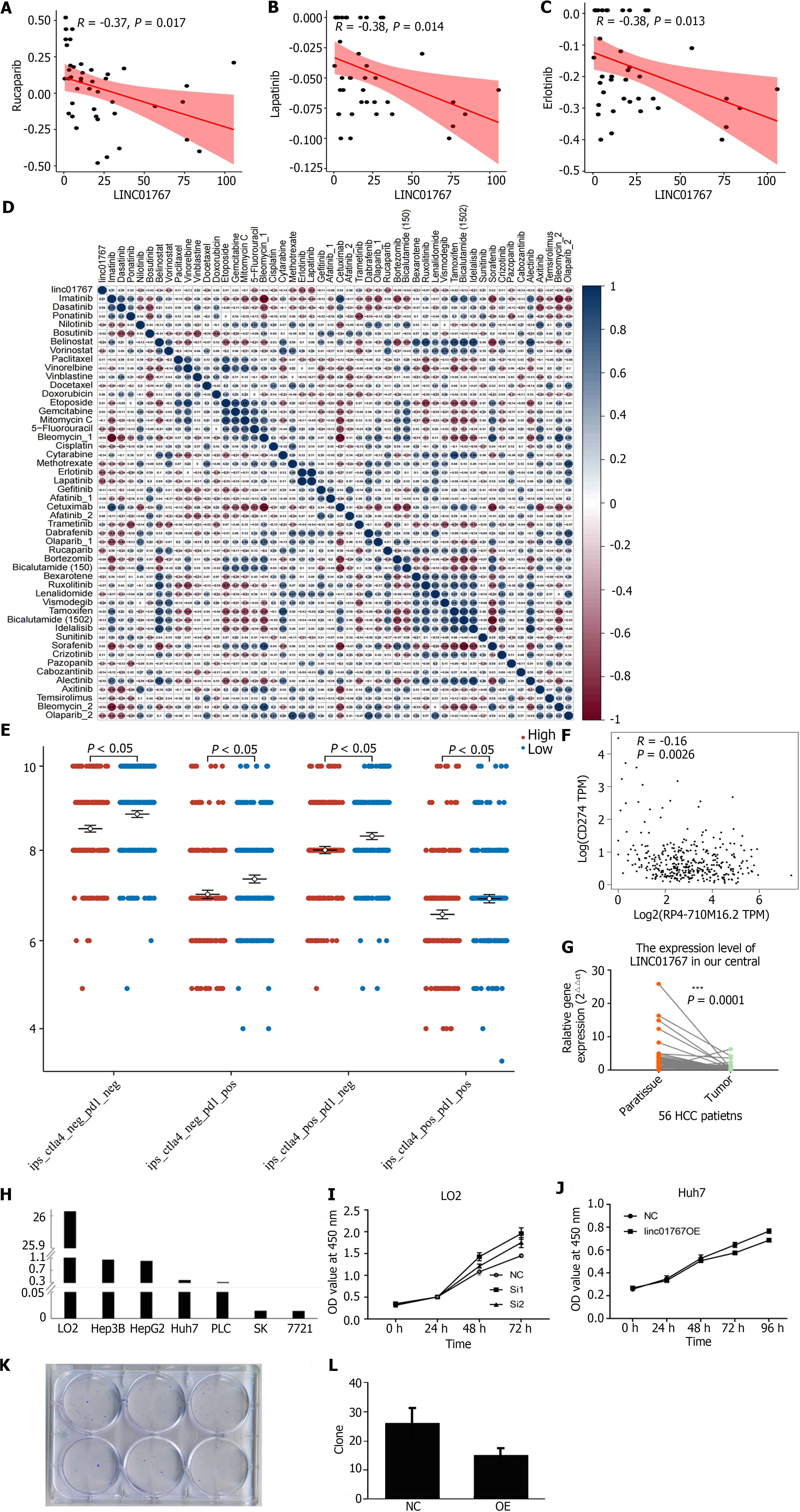
A previous study developed a lncRNA profile to forecast drug responsiveness in TCGA HCC patients[24]. Upon re-evaluation, it was found that LINC01767 exhibited a negative correlation with the sensitivity to Rucapanib (R = -0.37, P = 0.017) as shown in Figure 4A, lapatinib (R = -0.38, P = 0.014) as shown in Figure 4B, and erlotinib (R = -0.38, P = 0.013) as shown in Figure 4C. Figure 4D displayed the correlation analysis between the mRNA expression of LINC01767 and the IC50 values of the candidate drugs.
Recent research has indicated that the immunophenoscore (IPS), which is based on immunogenicity, can be used to predict how patients will respond to immunotherapy. The correlation between IPS and LINC01767 expression was examined. We used IPS, IPS-CTLA4-programmed cell death ligand 1 (PDL1)+, IPS-CTLA4+-PDL1-, IPS-CTLA4+-PDL1+ to evaluate the potential role of LINC01767 prediction of immune checkpoint inhibitor. The levels of IPS, IPS-PD1/PD-L1/PD-L2, and IPS-CTLA4 were notably elevated in the high LINC01767 group compared to the low LINC01767 groups (all P < 0.05) as shown in Figure 4E. The spearman correlation analysis between LINC01767 mRNA and PDL1 mRNA from TCGA was conducted, the results showed LINC01767 was negatively correlated with PDL1, P = 0.0026 (Figure 4F).
Additionally, we examine the possible role of LINC01767 in liver cancer immune cells using ImmReg (hrbmu.edu.cn), with a correlation coefficient of over 0.5 and a P < 0.05. The findings indicate a positive association between LINC01767 and Macrophage, and a negative relationship with B cell, CD4+ cell, neutrophil, myeloid dendritic cell, and macrophage M2 cell. What’s more, it was negatively correlated with PDL1 rather than CTLA4, P = 0.0026, R = -0.16 (Supplementary Figure 2).
The RT-qPCR of tumor tissue and para-tumor tissue from HCC patients in our cohort showed that the LINC01767 was down regulated obviously in tumor cases (Figure 4G). The LINC01767 was down regulated in cancer cell line comparing with LO2 (Figure 4H). The down-regulation of LINC01767 in LO2 cell showed the growth of the LO2 was impeded (Figure 4I); Huh7 was transfected with LINC01767 over expression lentivirus, and then MTT method and formation of clones was conducted, the over expression of LINC01767 inhibit cell proliferation (Figure 4J) and impede the clone formation of Huh7 (Figure 4K and L). The results of in vitro experiment showed the LINC01767 has no significant influence on the cell migration/invasion/cell cycle or apoptosis in Huh7 cell (Supplementary Figure 3).
We firstly identified the down regulated non-coding RNA LINC01767 in HCC based on TCGA data , GEO data and our patient cohort. And we firstly analyze the diagnostic and prognostic performance of LINC01767 in HCC via bio-informatics analysis and its correlation with clinical characteristics. The underlying mechanism of LINC01767 in HCC was explored and displayed via RNA sequencing and bioinformatics analysis. LINC01767 was correlated with the cancer stem cell in HCC as well as in pan-cancer. The gene regulation net of LINC01767 was firstly identified, as following, E2f, TNFSF12, C1QTNF12, COL6A1, COL6A2, COL9A3, CXCL2, CXCL8, JAG2, KCNH2, MTHFD1, NOTCH3, PPARGC1A, RRM2, S100A9, SLC2A4, SLC7A11, and TP53. Additionally, a nomogram was developed using LINC01767 and a LASSO model with genes related to LINC01767 to accurately predict OS, demonstrating strong predictive capabilities. Previous studies were used to predict the drug sensitivity of HCC, revealing a negative association between LINC01767 and the sensitivity of rucapanib, lapatinib, and erlotinib, as well as a negative correlation with the immune score IPS and PDL1. The in vitro experiment showed the LINC01767 negatively regulated the cell growth ability and clone formation ability. All the above evidence draws out that LINC01767 was an important and pivotal tumor suppressor gene in progression of HCC.
A prior study demonstrated that LINC01767 (ENSG00000223956, RP4-710M16.2), referred to as CRLM1, was found to be increased in metastatic CRC, leading to the inhibition of apoptosis in CRC cells and the promotion of liver metastasis in BALB/C nude mice. The CRLM1 facilitates liver metastasis by physically interacting with the hnRNPK protein and promoting its nuclear localization. CRLM1 significantly increases binding to the hnRNPK promoter and controls the activation of a group of genes related to metastasis[25]. We here add another potential mechanism from the single cell sequencing analysis showing the LINC01767 participates in the cancer stemness which may participate in metastasis.
AFP serves as the diagnostic and screening indicator for HCC. However, approximately 30%-40% of all patients with HCC exhibit AFP levels within the normal range, which is less than 20 ng/mL. AFP has a sensitivity of 41%-65% and specificity of 80%-90%, while DCP has a sensitivity of 56%-77% and specificity of 82%-87%. AFP-L3 has a sensitivity of 28%-56% and specificity of 92%-97% for diagnosing HCC[26]. Other tumor markers like GP73, GPC-3, PIVKAII, and certain microRNAs have been suggested, but none have demonstrated adequate sensitivity and/or specificity for early HCC diagnosis. We here demonstrated higher LINC01767 was correlated with higher AFP, LINC01767 could be a potentially diagnostic marker for HCC with sensitivity[27,28] better than AFP. GSE78160 showed that the serum LINC01767 was up regulated in HCC patients. However, why the level of LINC01767 down regulated in the tumor tissue but up regulated in serum was unclear, which was very interesting subject calling for further and deep exploration. This may be explained by the tumor heterogeneity or tumor cell subgroup heterogeneity, such as cluster7 which stands for the tumor stem cell was with a higher level of LINC01767 and it may secret LINC01767 into serum via exosome then demonstrating a higher level of serum LINC01767. It can be detected in urine as well, which gives the possible diagnosis HCC via the urine test as a non-invasive testing.
LINC01767 was a good predictor for OS rather than DFS. And the Cox regression verified the good performance of it in OS of HCC. A predictive nomogram was created using key factors identified in the Cox analysis to forecast the outcome of HCC, such as T/N/Residual tumor, vascular invasion, BMI, and LINC01767. After identified the LINC01767 related genes via next generation sequencing and bioinformatics analysis, we further construct a prognostic model via LASSO Cox regression analysis. The LASSO model’s performance was evaluated using the ROC curve, indicating that the 9-genes model successfully predicted the OS of liver cancer patients, achieving an AUC greater than 0.75. This verified the prognostic performance of LINC01767, and also provide the potential mechanism of it in HCC initiation and progression.
The underlying mechanism how lncRNAs participate in the initiation and progression in HCC remains unexplored, we here provide an RNA-sequencing after over expression of LINC01767 and bioinformatics analysis hoping to intriguer the interest in this specific lncRNA[29,30]. We conducted potential co-related coding genes of LINC01767 via RNA-sequencing and IPA analysis, the results showed the related genes E2f, TNFSF12 through C1QTNF12, COL6A1, COL6A2, COL9A3, CXCL2, CXCL8, JAG2, KCNH2, MTHFD1, NOTCH3, PPARGC1A, RRM2, S100A9, SLC2A4, SLC7A11, TP53 participating in multiple process in cancers. The regulatory genes showed that the FMR1 was predicted to be strongly activated with 10 uniformly activated genes of LINC01767 (not shown). FMR1, also known as FMRP, has the ability to attach to RNA and is linked to polysome, showing high levels of expression in HCC tissues. Knocking down FMRP effectively inhibits HCC metastasis both in vitro and in vivo. The encoded protein plays a role in transporting mRNA from the nucleus to the cytoplasm. Fragile X syndrome is caused by an expansion of 55-230 repeats of a trinucleotide sequence (CGG) in the 5’ untranslated region (UTR), which is typically found at 6-53 copies[31]. FMRP aids in the localization of STAT3 mRNA to protrusions by interacting with its 3’ UTR. FMRP could promote the interleukin-6-mediated translation of STAT3 via phosphorylation at serine 114 of FMRP. Although some potential target genes or regulatory genes did not show aberrant expression in HCC, such as KCNH2 which was involved in lipid metabolism[32]. Our research adds a fundamental evidence for the potential regulatory genes involved in the metabolism indicating a role of LINC01767 in HCC[33].
The findings from the in vitro study indicated that LINC01767 does not have a notable impact on cell migration, invasion, cell cycle, or apoptosis in Huh7 cells. Besides, the difference on the growth of cell after knockdown or over expression of LINC01767 was not so obvious. This may result from that only the over expression of LINC01767 in specific cell subgroup such as tumor stem cell can influence the function of tumor cell significantly, another potential hypothesis was that the LINC01767 exosome derived from tumor cell will function as the signal to influence other cells, such as immune cell. Thus, the role of LINC01767 in immune microenvironment was analyzed. The results showed the LINC01767 was positively correlated with the macrophage, and negatively with the B cell, CD4+ cell, neutrophil, myeloid dendritic cell, and macrophage M2 cell, what’s more, it was negatively correlated with PDL1 rather than CTLA4; and the differential IPS in high-and low-LINC01767 group verified the correlation of LINC01767 with immunity in HCC. Besides, the sole dysregulation of LINC01767 may be rescued too fast in cell to show an obvious influence thus a gene set influenced by LINC01767 warrants further exploration in the future.
Our research has constraints. Our study is lacking biological research on the underlying mechanisms for specific conditions. Additionally, the initial examination of clinical features was conducted without additional analysis in a separate cohort due to the absence of public data sets and the lack of prognosis information in our cohort. Additionally, the mechanism or pathways was not investigated. Further investigation and testing are needed to confirm the effectiveness of diagnostic tests and treatments based on LINC01767. Furthermore, the impact of deregulation of LINC01767 on cancer diversity and evaluation of intratumoral clonality is still unclear. The therapeutic development the verification of in vivo bioactivity and cancer-cell-specific delivery of oligonucleotides.
LINC01767 is an important suppressor gene in HCC with good diagnostic and prognostic performance.
We thanks for the colleagues from Shandong University.
| 1. | Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 9901] [Article Influence: 4950.5] [Reference Citation Analysis (2)] |
| 2. | Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345-1362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1284] [Cited by in RCA: 1214] [Article Influence: 404.7] [Reference Citation Analysis (41)] |
| 3. | Younossi ZM, Otgonsuren M, Henry L, Venkatesan C, Mishra A, Erario M, Hunt S. Association of nonalcoholic fatty liver disease (NAFLD) with hepatocellular carcinoma (HCC) in the United States from 2004 to 2009. Hepatology. 2015;62:1723-1730. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 495] [Cited by in RCA: 617] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 4. | Shariff MI, Cox IJ, Gomaa AI, Khan SA, Gedroyc W, Taylor-Robinson SD. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev Gastroenterol Hepatol. 2009;3:353-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 226] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 6. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1102] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 7. | CRICK FH. On protein synthesis. Symp Soc Exp Biol. 1958;12:138-163. [PubMed] |
| 8. | International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3224] [Cited by in RCA: 2882] [Article Influence: 137.2] [Reference Citation Analysis (0)] |
| 9. | Fang XY, Pan HF, Leng RX, Ye DQ. Long noncoding RNAs: novel insights into gastric cancer. Cancer Lett. 2015;356:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 166] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 10. | Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 278] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 11. | Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, Diekhans M, Fujita PA, Goldman M, Gravell RC, Harte RA, Hinrichs AS, Kirkup VM, Kuhn RM, Learned K, Maddren M, Meyer LR, Pohl A, Rhead B, Wong MC, Zweig AS, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40:D912-D917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 12. | Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775-1789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3516] [Cited by in RCA: 3957] [Article Influence: 329.8] [Reference Citation Analysis (0)] |
| 13. | Cai Q, Wang S, Jin L, Weng M, Zhou D, Wang J, Tang Z, Quan Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol Cancer. 2019;18:82. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 149] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 14. | Ren D, Li H, Li R, Sun J, Guo P, Han H, Yang Y, Li J. Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol Lett. 2016;11:1621-1630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 15. | Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zörnig M, MacLeod AR, Spector DL, Diederichs S. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180-1189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1087] [Cited by in RCA: 1298] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 16. | Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng MZ, Zhou D, Wang JD, Quan ZW. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7:37857-37867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 17. | Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, Wu LM, Chen LM, Zheng SS. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 477] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 18. | Chen L, Yao H, Wang K, Liu X. Long Non-Coding RNA MALAT1 Regulates ZEB1 Expression by Sponging miR-143-3p and Promotes Hepatocellular Carcinoma Progression. J Cell Biochem. 2017;118:4836-4843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 19. | Yuan P, Cao W, Zang Q, Li G, Guo X, Fan J. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem Biophys Res Commun. 2016;478:1067-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 140] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 20. | Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577-4587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 717] [Cited by in RCA: 843] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 21. | Zhang P, Cao P, Zhu X, Pan M, Zhong K, He R, Li Y, Jiao X, Gao Y. Upregulation of long non-coding RNA HOXA-AS2 promotes proliferation and induces epithelial-mesenchymal transition in gallbladder carcinoma. Oncotarget. 2017;8:33137-33143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 111] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 23. | Wang Y, Wang Z, Xu J, Li J, Li S, Zhang M, Yang D. Systematic identification of non-coding pharmacogenomic landscape in cancer. Nat Commun. 2018;9:3192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 69] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 24. | Xia Q, Shu Z, Ye T, Zhang M. Corrigendum: Identification and Analysis of the Blood lncRNA Signature for Liver Cirrhosis and Hepatocellular Carcinoma. Front Genet. 2021;12:692400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 25. | Wang Z, Chen J, Sun F, Zhao X, Dong Y, Yu S, Li J, Liang H. LncRNA CRLM1 inhibits apoptosis and promotes metastasis through transcriptional regulation cooperated with hnRNPK in colorectal cancer. Cell Biosci. 2022;12:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 26. | Yıldırım HÇ, Kavgaci G, Chalabiyev E, Dizdar O. Advances in the Early Detection of Hepatobiliary Cancers. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 27. | Du Z, Liu X, Wei X, Luo H, Li P, Shi M, Guo B, Cui Y, Su Z, Zeng J, Si A, Cao P, Zhou G. Quantitative proteomics identifies a plasma multi-protein model for detection of hepatocellular carcinoma. Sci Rep. 2020;10:15552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 28. | Wang K, Guo WX, Li N, Gao CF, Shi J, Tang YF, Shen F, Wu MC, Liu SR, Cheng SQ. Serum LncRNAs Profiles Serve as Novel Potential Biomarkers for the Diagnosis of HBV-Positive Hepatocellular Carcinoma. PLoS One. 2015;10:e0144934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B, Zhang T, Zhou Y, Chen Q, Wei C, Wang W, Wang Z. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. 2017;8:e3092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 30. | Zhao W, Ma X, Liu L, Chen Q, Liu Z, Zhang Z, Ma S, Wang Z, Li H, Wang Z, Wu J. SNHG20: A vital lncRNA in multiple human cancers. J Cell Physiol. 2019;234:14519-14525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 31. | Shen Z, Liu B, Wu B, Zhou H, Wang X, Cao J, Jiang M, Zhou Y, Guo F, Xue C, Wu ZS. FMRP regulates STAT3 mRNA localization to cellular protrusions and local translation to promote hepatocellular carcinoma metastasis. Commun Biol. 2021;4:540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 32. | Feng NN, Du XY, Zhang YS, Jiao ZK, Wu XH, Yang BM. Overweight/obesity-related transcriptomic signature as a correlate of clinical outcome, immune microenvironment, and treatment response in hepatocellular carcinoma. Front Endocrinol (Lausanne). 2022;13:1061091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 33. | Zhu P, Li FF, Zeng J, Tang DG, Chen WB, Guo CC. Integrative analysis of the characteristics of lipid metabolism-related genes as prognostic prediction markers for hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2021;25:116-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |