Published online Nov 27, 2024. doi: 10.4254/wjh.v16.i11.1282
Revised: June 23, 2024
Accepted: July 23, 2024
Published online: November 27, 2024
Processing time: 164 Days and 19.8 Hours
Currently, intrahepatic cholangiocarcinoma (ICC) poses a continuing, significant health challenge, but the relationship has yet to be established between ICC and the proteasome 26S subunit non-ATPase 6 (PSMD6).
To investigate the protein expression and clinicopathological significance of PSMD6 in ICC.
The potential impact of the PSMD6 gene on the growth of ICC cell lines was analyzed using clustered regularly interspaced short palindromic repeat knockout screening technology. Forty-two paired specimens of ICC and adjacent non-cancerous tissues were collected. PSMD6 protein expression was determined by immunohistochemistry. Receiver operating characteristic curve analysis was performed to validate PSMD6 expression level, and its association with ICC patients’ various clinicopathological characteristics was investigated.
The PSMD6 gene was found to be essential for the growth of ICC cell lines. PSMD6 protein was significantly overexpressed in ICC tissues (P < 0.001), but showed no significant association with patient age, gender, pathological grade, or tumor-node-metastasis stage (P > 0.05).
PSMD6 can promote the growth of ICC cells, thus playing a pro-oncogenic role.
Core Tip: This study examined the expression and clinicopathological significance of proteasome 26S subunit non-ATPase 6 (PSMD6) in intrahepatic cholangiocarcinoma (ICC). It was discovered that the PSMD6 gene was essential for the prolife
- Citation: Tang ZQ, Tang YL, Qin K, Li Q, Chen G, Huang YB, Li JJ. Overexpression of proteasome 26S subunit non-ATPase 6 protein and its clinicopathological significance in intrahepatic cholangiocarcinoma. World J Hepatol 2024; 16(11): 1282-1289
- URL: https://www.wjgnet.com/1948-5182/full/v16/i11/1282.htm
- DOI: https://dx.doi.org/10.4254/wjh.v16.i11.1282
Intrahepatic cholangiocarcinoma (ICC) is an aggressive tumor and the second most common primary liver cancer. ICC accounts for 10%-20% of primary liver cancer cases[1-4] and its incidence is increasing worldwide[5-8]. Therefore, identifying molecular biomarkers related to ICC is crucial to enhancing the accuracy of early diagnosis, the effectiveness of treatment, and patient prognosis.
The proteasome 26S subunit non-ATPase 6 (PSMD6) gene, located on chromosome 3p14.1, consists of 11 exons and encodes a subunit of the 26S proteasome, which is a member of the ubiquitin-proteasome system. The ubiquitin-protea
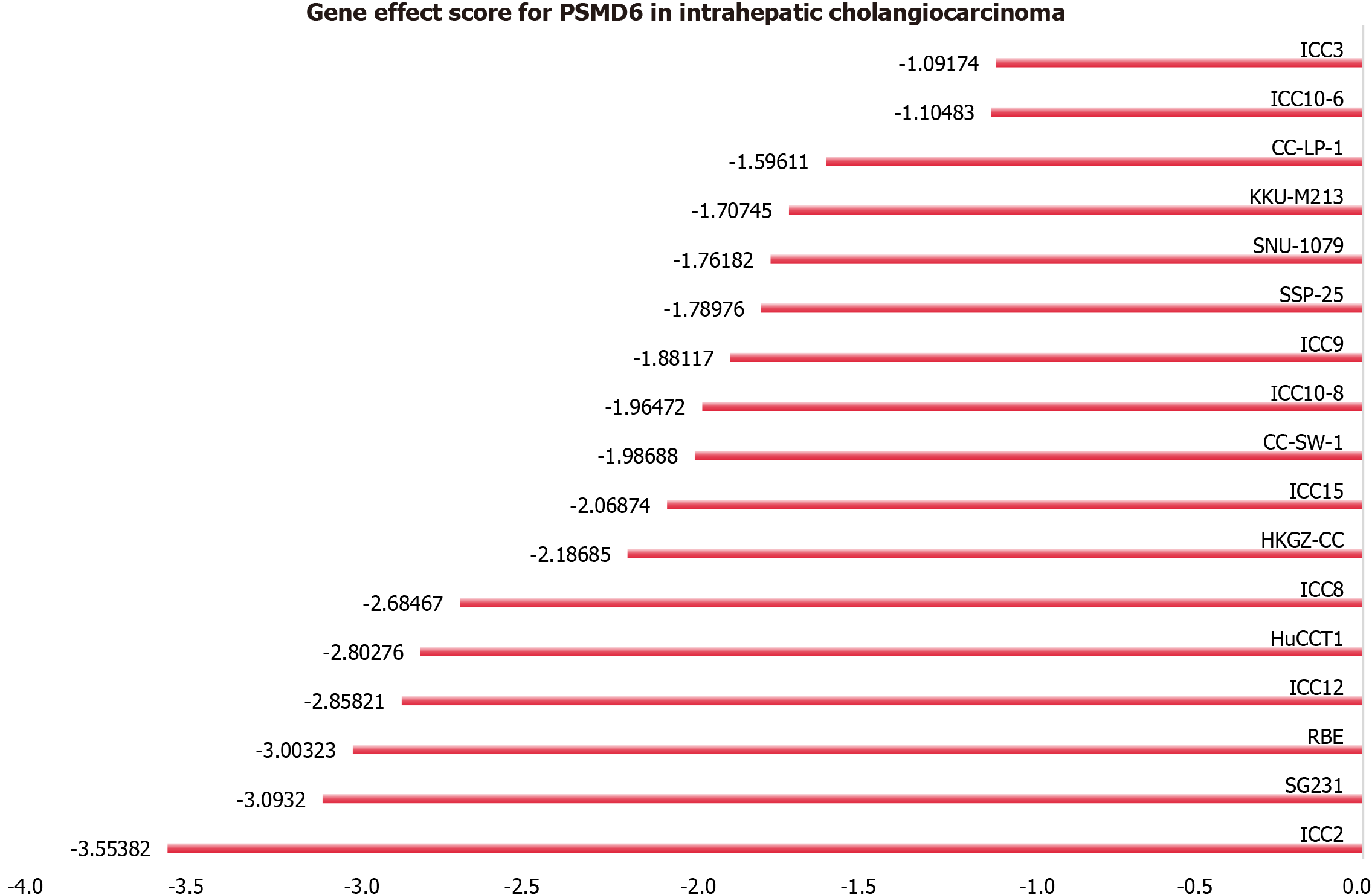
This study employed clustered regularly interspaced short palindromic repeat (CRISPR) knockout screening technology to elucidate the effect of the PSMD6 gene on ICC cell growth. ICC cell lines were subjected to PSMD6 gene knockout, and the CERES algorithm was used to calculate the PSMD6 gene effect scores to evaluate the knockout effects of PSMD6 in ICC cell lines. A negative score indicated inhibited proliferation of ICC cells after PSMD6 gene knockout[12]. In this study, if more than 75% of ICC cell lines had a score less than -1, the gene was deemed as a growth-associated gene for ICC[13].
This study used The Human Protein Atlas (THPA) database (https://www.proteinatlas.org/) and in-house tissue microarrays to explore the PSMD6 protein expression in ICC tissues. We obtained immunohistochemical images of ICC tissue and normal bile duct epithelial tissue from the THPA database[14]. Tissue microarrays (LVC1261) containing 42 pairs of ICC specimens and corresponding adjacent non-cancerous tissues were obtained from Pantomics, Inc. None of the tissue donor patients received any preoperative therapeutic interventions, and cholangiocarcinoma was the pathological type of all specimens. The distance between the ICC tissues and adjacent non-cancerous tissues was at least 3 cm. Relevant clinicopathological characteristics were collected, including age, gender, pathological grade, and tumor-node-metastasis (TNM) stage. The Pantomics, Inc. ethics committee granted ethical approval for this study [Ethics No. Fanpu(2018)23].
Tissue sections of 2 μm thickness were prepared from paraffin-embedded tissue blocks using a microtome. These sections were subsequently incubated at 75 °C for 30 min to remove the wax. After dewaxing, the sections were treated with 3% H2O2 for 15 min, followed by rinsing with distilled water and phosphate-buffered saline (PBS). The sections were then incubated with a polyclonal rabbit anti-PSMD6 antibody (Abcam, catalog No. ab155761) and subsequently washed with PBS. The tissue microarrays were subjected to restaining, dehydration, transparency, and sealing processes. Adjacent tissue sections served as the positive control, whereas PBS alone was used as the negative control.
This study integrated data from the THPA database with the IHC results to summarise and analyse the expression of PSMD6 in ICC tissue and adjacent tissue samples. Two blinded, experienced pathologists independently evaluated the slides. The results were assessed by the proportion of positive cells and staining intensity. The percentage of positive cells was scored as 0, 1, 2, 3, and 4 points, respectively, for no expression, < 10%, 10%-35%, 36%-75%, and > 75%. Staining intensity was scored as 0, 1, 2, and 3 points, respectively, for no or almost no cytoplasmic staining, light yellow, and brown yellow[15-17]. The final score for each specimen was calculated by multiplying the percentage score by the intensity score. If the final scores of two pathologists were inconsistent, the average score was taken.
Data processing was conducted using SPSS 19.0 software. The association between PSMD6 protein expression and patients’ clinicalpathological characteristics was analyzed using the χ2 test and Spearman correlation analysis. The expression differences of the PSMD6 protein in ICC tissues and adjacent non-cancerous tissues were compared using the
This study used the CRISPR knockout screening technology to assess the PSMD6 gene’s impact on the proliferation of ICC cells. The gene effect scores of PSMD6 in 17 ICC cell lines indicate significant growth inhibition in all 17 ICC cell lines after PSMD6 gene knockout (Figure 1). The gene effect scores were all less than -1, suggesting that the PSMD6 gene is a growth-associated gene for ICC cells (17/17, 100%).
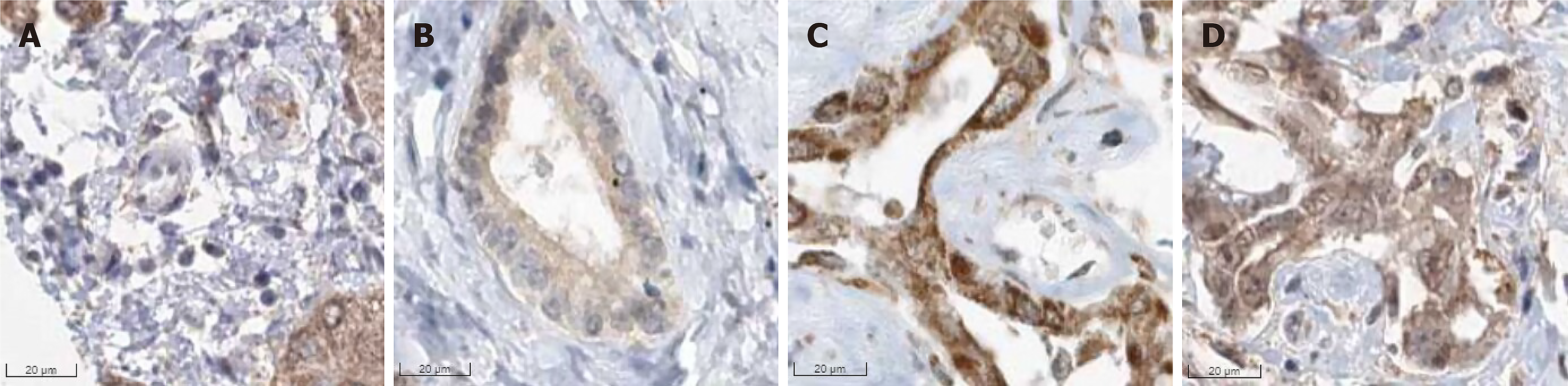
This study preliminarily investigated the expression of PSMD6 protein in ICC tissues using the THPA database. We observed that the protein was distributed in the cell membrane/cytoplasm/nucleus of ICC tissues (antibodies HPA036921 and HPA036922), showing moderately to strongly positive expression in ICC tissues (Figure 2). Due to the limited number of cases, however, statistical analysis could not be performed.
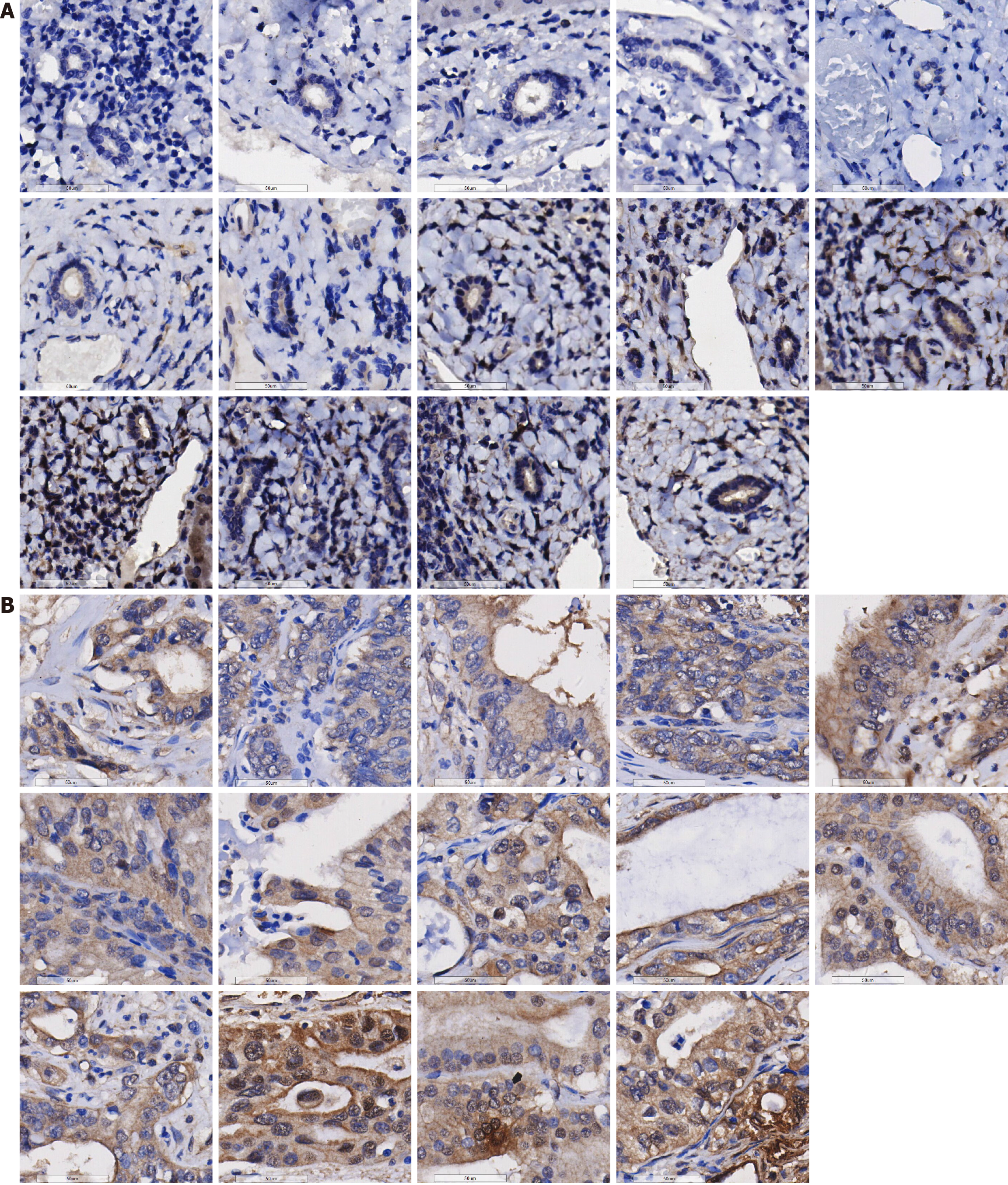
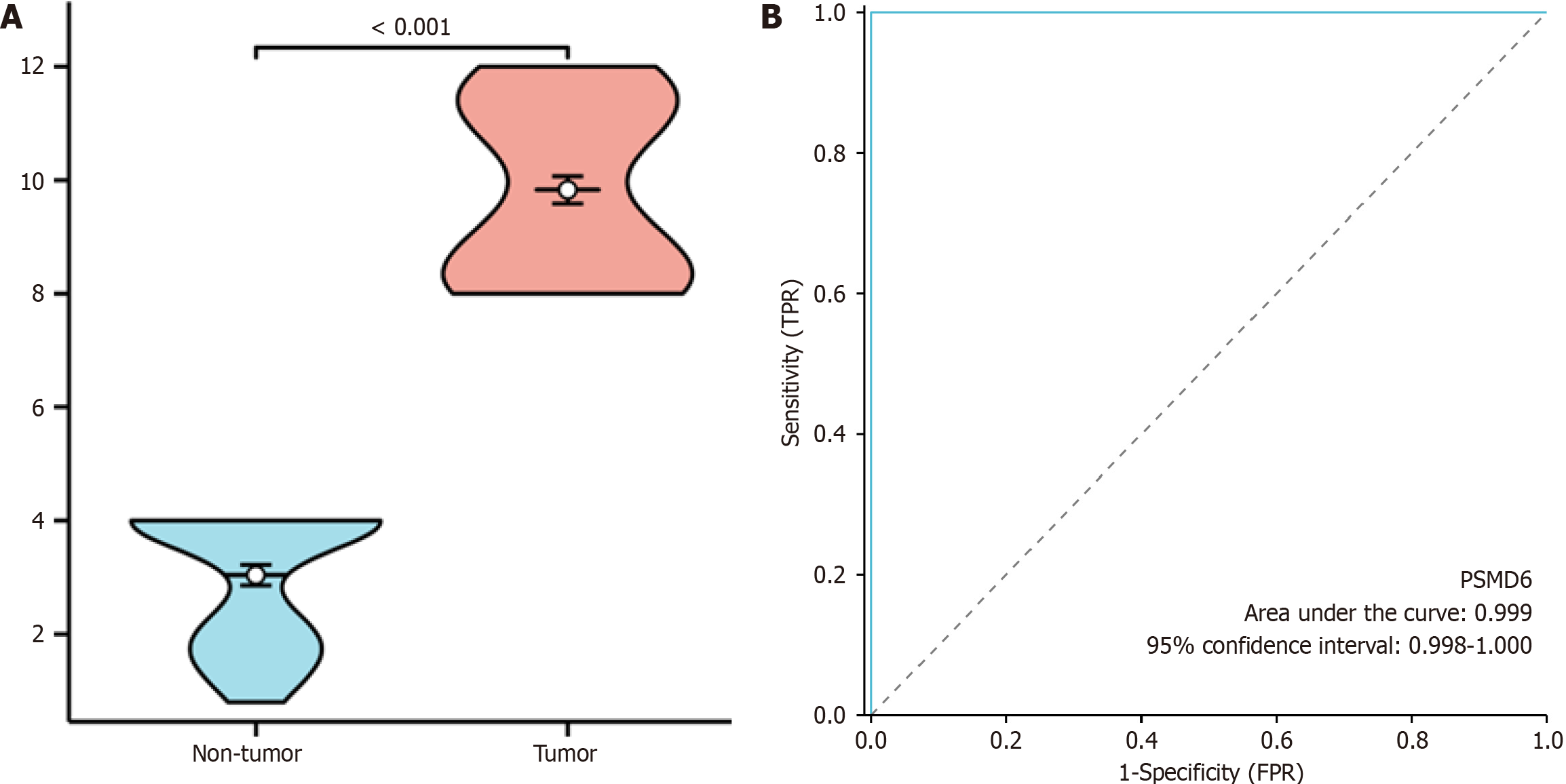
This study validated the expression of the PSMD6 protein in ICC and adjacent non-cancerous tissues. PSMD6 protein staining in adjacent non-cancerous tissues showed negative or weakly positive staining (Figure 3A), whereas it exhibited moderately to strongly positive staining in ICC tissues (Figure 3B). To quantitatively analyse the expression of PSMD6 in ICC tissues, a violin plot and ROC curve were generated. The results indicate significantly higher expression of the PSMD6 protein in ICC tissues than in adjacent non-cancerous tissues (P < 0.001) (Figure 4A). ROC curve analysis confirmed the significant overexpression of PSMD6 in ICC tissues, with an area under the curve of 0.999 (95% confidence interval: 0.998-1.000) (Figure 4B). To elucidate the clinicopathological relevance of PSMD6 protein in ICC patients, this study evaluated the relationship between PSMD6 protein expression and clinicopathological characteristics through univariate analysis. The results showed no significant association between PSMD6 protein expression and patient age, gender, pathological grade, or TNM stage (P > 0.05).
Currently, no studies in the literature report the expression and clinicopathological significance of the PSMD6 protein in ICC. The present research for the first time used the CRISPR knockout screening technology to clarify the impact of the PSMD6 gene on the growth of ICC cells at the cellular level. At the protein level, we also evaluated PSMD6 protein expression in ICC tissues through IHC and explored its association with ICC patients’ clinical pathological characteristics, providing a potential molecular indicator for the diagnosis, treatment, and prognosis assessment of ICC patients.
Previous studies have indicated that PSMD6 is closely associated with the occurrence and progression of several tumours, potentially playing a significant role in these processes. Akbari et al[18] used real-time polymerase chain reaction to assess the gene expression profile in granulosa cells in 33 patients with polycystic ovary syndrome, revealing a significant decrease in PSMD6 expression, which suggests that gene expression changes may have important implications for patient fertility. Zhou et al[19] conducted survival analysis and established a correlation between PSMD6 expression levels and postoperative prognosis in patients with pancreatic ductal adenocarcinoma, using co-expression and interaction network analysis to explore the potential biological role of PSMD6 in postoperative pancreatic ductal adenocarcinoma patients. The findings suggest that PSMD6 may serve as a potential prognostic and diagnostic biomarker for early-stage postoperative pancreatic ductal adenocarcinoma patients and may play a crucial role in disease development through regulating key biological processes, such as tumor protein p53 (TP53), cyclin dependent kinase inhibitor 2A, MYC proto-oncogene, BHLH transcription factor (MYC), DNA repair, KRAS proto-oncogene, GTPase (KRAS), cell cycle checkpoints, nuclear factor kappa B (NF-κB) inducing kinase, the NF-κB signaling pathway, and proteasomes. Zhang et al[20] revealed through gene enrichment analysis that PSMD6 may activate the wingless-related integration site signaling pathway by degrading AXIN protein, providing a new perspective for understanding the molecular mechanisms of PSMD6 in lung adenocarcinoma development.
The CRISPR knockout screening method has become an essential tool in gene function research, combining the
The elevated expression of PSMD6 in ICC suggests its potential role as a key protein promoting the onset and progression of ICC. However, this study was limited by the lack of extensive population data validation, and the association between its identified expression levels and patients’ clinicopathological features requires further investigation. Moreover, the specific biological mechanisms of PSMD6 in ICC have not been thoroughly explored. Therefore, future studies should expand the sample size to delve deeper into the biological functions of PSMD6 in ICC and its potential value as a therapeutic target.
We would like to thank “Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis” for providing technical support.
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 11436] [Article Influence: 3812.0] [Reference Citation Analysis (4)] |
| 2. | Beal EW, Tumin D, Moris D, Zhang XF, Chakedis J, Dilhoff M, Schmidt CM, Pawlik TM. Cohort contributions to trends in the incidence and mortality of intrahepatic cholangiocarcinoma. Hepatobiliary Surg Nutr. 2018;7:270-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 3. | Shichi S, Sugiyama K, Asahi Y, Shirakawa C, Nakamoto H, Kimura S, Wakizaka K, Aiyama T, Nagatsu A, Orimo T, Kakisaka T, Taketomi A. Diacylglycerol kinase alpha is a proliferation marker of intrahepatic cholangiocarcinoma associated with the prognosis. Cancer Med. 2024;13:e7238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 4. | Zhong YJ, Luo XM, Liu F, He ZQ, Yang SQ, Ma WJ, Wang JK, Dai YS, Zou RQ, Hu YF, Lv TR, Li FY, Hu HJ. Integrative analyses of bulk and single-cell transcriptomics reveals the infiltration and crosstalk of cancer-associated fibroblasts as a novel predictor for prognosis and microenvironment remodeling in intrahepatic cholangiocarcinoma. J Transl Med. 2024;22:422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 5. | Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, Calvisi DF, Perugorria MJ, Fabris L, Boulter L, Macias RIR, Gaudio E, Alvaro D, Gradilone SA, Strazzabosco M, Marzioni M, Coulouarn C, Fouassier L, Raggi C, Invernizzi P, Mertens JC, Moncsek A, Ilyas SI, Heimbach J, Koerkamp BG, Bruix J, Forner A, Bridgewater J, Valle JW, Gores GJ. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1555] [Cited by in RCA: 1551] [Article Influence: 310.2] [Reference Citation Analysis (0)] |
| 6. | Massarweh NN, El-Serag HB. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control. 2017;24:1073274817729245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 437] [Article Influence: 54.6] [Reference Citation Analysis (1)] |
| 7. | Saha SK, Zhu AX, Fuchs CS, Brooks GA. Forty-Year Trends in Cholangiocarcinoma Incidence in the U.S.: Intrahepatic Disease on the Rise. Oncologist. 2016;21:594-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 563] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 8. | Zou Y, Xu X, Wu T, Chen Q, Li Z, Yang Z, Wang K, Shen F. Sex disparity in clinical characteristics and long-term prognosis after liver resection for patients with intrahepatic cholangiocarcinoma: A propensity score matching analysis. Heliyon. 2024;10:e29910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 9. | Wong E, Cuervo AM. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb Perspect Biol. 2010;2:a006734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 227] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 10. | Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9 Suppl 1:S7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 143] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 11. | Wing SS. The UPS in diabetes and obesity. BMC Biochem. 2008;9 Suppl 1:S6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 12. | Pacini C, Dempster JM, Boyle I, Gonçalves E, Najgebauer H, Karakoc E, van der Meer D, Barthorpe A, Lightfoot H, Jaaks P, McFarland JM, Garnett MJ, Tsherniak A, Iorio F. Integrated cross-study datasets of genetic dependencies in cancer. Nat Commun. 2021;12:1661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 178] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 13. | Ho KH, Huang TW, Liu AJ, Shih CM, Chen KC. Cancer Essential Genes Stratified Lung Adenocarcinoma Patients with Distinct Survival Outcomes and Identified a Subgroup from the Terminal Respiratory Unit Type with Different Proliferative Signatures in Multiple Cohorts. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7696] [Cited by in RCA: 10544] [Article Influence: 1054.4] [Reference Citation Analysis (0)] |
| 15. | Zhang W, Li GS, Gan XY, Huang ZG, He RQ, Huang H, Li DM, Tang YL, Tang D, Zou W, Liu J, Dang YW, Chen G, Zhou HF, Kong JL, Lu HP. MMP12 serves as an immune cell-related marker of disease status and prognosis in lung squamous cell carcinoma. PeerJ. 2023;11:e15598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 16. | Li SH, Zhai GQ, He RQ, Chen G, Wang SS, Liu JL, Cheng JW, Yan HB, Huang ZG. Down-regulation and clinical significance of Sorbin and SH3 domain-containing protein 1 in bladder cancer tissues. IET Syst Biol. 2023;17:70-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 17. | Liu Z, Gu S, Lu T, Wu K, Li L, Dong C, Zhou Y. IFI6 depletion inhibits esophageal squamous cell carcinoma progression through reactive oxygen species accumulation via mitochondrial dysfunction and endoplasmic reticulum stress. J Exp Clin Cancer Res. 2020;39:144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 18. | Akbari A, Aboutorabi R, Kazemi M, Borzouie Z, Feizi A, Naghshineh E, Mostafavi F. Differential Gene Expressions of CALM1, PSMD6, and AK124742 Long Noncoding RNA in Cumulus Cells from Polycystic Ovary Syndrome Patients versus Normal Control Women. Adv Biomed Res. 2023;12:240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Zhou C, Li H, Han X, Pang H, Wu M, Tang Y, Luo X. Prognostic Value and Molecular Mechanisms of Proteasome 26S Subunit, Non-ATPase Family Genes for Pancreatic Ductal Adenocarcinoma Patients after Pancreaticoduodenectomy. J Invest Surg. 2022;35:330-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 20. | Zhang JY, Shi KZ, Liao XY, Li SJ, Bao D, Qian Y, Li DJ. The Silence of PSMC6 Inhibits Cell Growth and Metastasis in Lung Adenocarcinoma. Biomed Res Int. 2021;2021:9922185. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 21. | Ipsen MB, Sørensen EMG, Thomsen EA, Weiss S, Haldrup J, Dalby A, Palmfeldt J, Bross P, Rasmussen M, Fredsøe J, Klingenberg S, Jochumsen MR, Bouchelouche K, Ulhøi BP, Borre M, Mikkelsen JG, Sørensen KD. A genome-wide CRISPR-Cas9 knockout screen identifies novel PARP inhibitor resistance genes in prostate cancer. Oncogene. 2022;41:4271-4281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 22. | Wenger A, Karlsson I, Kling T, Carén H. CRISPR-Cas9 knockout screen identifies novel treatment targets in childhood high-grade glioma. Clin Epigenetics. 2023;15:80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 23. | Zhang J, Li Y, Liu H, Zhang J, Wang J, Xia J, Zhang Y, Yu X, Ma J, Huang M, Wang J, Wang L, Li Q, Cui R, Yang W, Xu Y, Feng W. Genome-wide CRISPR/Cas9 library screen identifies PCMT1 as a critical driver of ovarian cancer metastasis. J Exp Clin Cancer Res. 2022;41:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 41] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 24. | Cieśla M, Ngoc PCT, Muthukumar S, Todisco G, Madej M, Fritz H, Dimitriou M, Incarnato D, Hellström-Lindberg E, Bellodi C. m(6)A-driven SF3B1 translation control steers splicing to direct genome integrity and leukemogenesis. Mol Cell. 2023;83:1165-1179.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 25. | Xu J, Yang X, Deng Q, Yang C, Wang D, Jiang G, Yao X, He X, Ding J, Qiang J, Tu J, Zhang R, Lei QY, Shao ZM, Bian X, Hu R, Zhang L, Liu S. TEM8 marks neovasculogenic tumor-initiating cells in triple-negative breast cancer. Nat Commun. 2021;12:4413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 26. | Tarantino P, Gandini S, Nicolò E, Trillo P, Giugliano F, Zagami P, Vivanet G, Bellerba F, Trapani D, Marra A, Esposito A, Criscitiello C, Viale G, Curigliano G. Evolution of low HER2 expression between early and advanced-stage breast cancer. Eur J Cancer. 2022;163:35-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 120] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 27. | Qin Y, Liu Y, Xiang X, Long X, Chen Z, Huang X, Yang J, Li W. Cuproptosis correlates with immunosuppressive tumor microenvironment based on pan-cancer multiomics and single-cell sequencing analysis. Mol Cancer. 2023;22:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 82] [Reference Citation Analysis (0)] |