Published online Jan 27, 2021. doi: 10.4254/wjh.v13.i1.94
Peer-review started: August 6, 2020
First decision: September 21, 2020
Revised: November 19, 2020
Accepted: December 4, 2020
Article in press: December 4, 2020
Published online: January 27, 2021
Processing time: 172 Days and 21 Hours
The broader use of high-throughput technologies has led to improved molecular characterization of hepatocellular carcinoma (HCC).
To comprehensively analyze and characterize all publicly available genomic, gene expression, methylation, miRNA and proteomic data in HCC, covering 85 studies and 3355 patient sample profiles, to identify the key dysregulated genes and pathways they affect.
We collected and curated all well-annotated and publicly available high-throughput datasets from PubMed and Gene Expression Omnibus derived from human HCC tissue. Comprehensive pathway enrichment analysis was performed using pathDIP for each data type (genomic, gene expression, methylation, miRNA and proteomic), and the overlap of pathways was assessed to elucidate pathway dependencies in HCC.
We identified a total of 8733 abstracts retrieved by the search on PubMed on HCC for the different layers of data on human HCC samples, published until December 2016. The common key dysregulated pathways in HCC tissue across different layers of data included epidermal growth factor (EGFR) and β1-integrin pathways. Genes along these pathways were significantly and consistently dysregulated across the different types of high-throughput data and had prognostic value with respect to overall survival. Using CTD database, estradiol would best modulate and revert these genes appropriately.
By analyzing and integrating all available high-throughput genomic, transcriptomic, miRNA, methylation and proteomic data from human HCC tissue, we identified EGFR, β1-integrin and axon guidance as pathway dependencies in HCC. These are master regulators of key pathways in HCC, such as the mTOR, Ras/Raf/MAPK and p53 pathways. The genes implicated in these pathways had prognostic value in HCC, with Netrin and Slit3 being novel proteins of prognostic importance to HCC. Based on this integrative analysis, EGFR, and β1-integrin are master regulators that could serve as potential therapeutic targets in HCC.
Core Tip: Analyzing all available high-throughput genomic, transcriptomic, miRNA, methylation and proteomic data from human hepatocellular carcinoma tissue, we identified master regulators of key pathways in hepatocellular carcinoma, such as the mTOR, Ras/Raf/MAPK and p53 pathways.
- Citation: Bhat M, Pasini E, Pastrello C, Rahmati S, Angeli M, Kotlyar M, Ghanekar A, Jurisica I. Integrative analysis of layers of data in hepatocellular carcinoma reveals pathway dependencies. World J Hepatol 2021; 13(1): 94-108
- URL: https://www.wjgnet.com/1948-5182/full/v13/i1/94.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i1.94
The molecular basis of hepatocellular carcinoma (HCC) has been elusive, given the significant heterogeneity of this tumor that arises in the context of various chronic liver diseases[1]. HCC remains a high-fatality cancer, despite large-scale efforts to better characterize and therapeutically target this malignancy. Since prevalence of cirrhosis due to hepatitis C and fatty liver disease is increasing in North America, HCC continues to rise[2]. Five-year survival remains poor at 18% due to late diagnosis and inability to tolerate chemotherapy in patients with cirrhosis[2]. Consequently, there is an urgent need to better understand the molecular basis of this highly fatal cancer.
Clinical management of HCC is optimized based on disease stage[3]. Curative treatment with resection, radiofrequency ablation or transplantation is possible in early stage disease[4]. When HCC is diagnosed at a later stage, sorafenib is the first-line chemotherapy, which is directed against the Ras/Raf/MAPK pathway[4]. This is associated with a very modest improvement in overall survival of 3 additional months as compared to placebo (10.7 mo vs 7.9 mo)[5].
The cancer genome atlas (TCGA) is a large-scale project that has enabled improved characterization of cancers with several layers of data. The TCGA multi-platform analysis of 196 HCC tumors described this cancer as highly heterogeneous and difficult to characterize, although certain key pathways did emerge including the Ras/Raf/MAPK, mTOR, Wnt/B-catenin, and Sonic Hedgehog pathways[1,6]. Integration of various types of data has previously been performed to map interaction networks. By integrating genomic, transcriptomic and proteomic data, one can understand potential interactions that contribute to a disease condition or process[7,8]. These interactions may otherwise not be uncovered, on the basis of a single type of data. This systems biology approach has been especially important in cancer, given that alterations in one gene can have a ripple effect on proteins in the rest of a protein-protein interaction network. Therefore, elucidating the layers of data in a disease can provide additional insights into the pathways that drive cancer[9].
In the current study, we aim to characterize the landscape of high-throughput data profiling in HCC and determine the patterns in key dysregulated genes and pathways across these different layers of data. The patterns that emerge could help in better understanding the pathways that drive HCC and could be considered as therapeutic targets.
We downloaded all available high-throughput genomic, transcriptomic, microRNA, methylation, and proteomic datasets related to human HCC samples from published datasets (PubMed, http://www.ncbi.nlm.nih.gov/PubMed and Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo).
Using PubMed, the following search was performed for whole exome sequencing data on HCC: ("carcinoma, hepatocellular" [MeSH Terms] OR ("carcinoma" [All Fields] AND "hepatocellular" [All Fields]) OR "hepatocellular carcinoma" [All Fields] OR ("hepatocellular" [All Fields] AND "carcinoma" [All Fields])) AND (whole [All Fields] AND ("exome" [MeSH Terms] OR "exome" [All Fields]) AND sequencing [All Fields]). The following MeSH terms were used to identify gene expression papers: ("carcinoma, hepatocellular" [MeSH Terms] OR ("carcinoma" [All Fields] AND "hepatocellular" [All Fields]) OR "hepatocellular carcinoma" [All Fields] OR ("hepatocellular" [All Fields] AND "carcinoma" [All Fields])) AND ("gene expression" [MeSH Terms] OR ("gene" [All Fields] AND "expression" [All Fields]) OR "gene expression" [All Fields]) AND ("humans" [MeSH Terms] OR "humans" [All Fields]) AND English [All Fields] NOT ("review" [Publication Type] OR "review literature as topic" [MeSH Terms] OR "reviews" [All Fields]). To identify suitable papers regarding methylation in HCC, we used the following terms: ("methylation" [MeSH Terms] OR "methylation"[All Fields]) AND ("carcinoma, hepatocellular" [MeSH Terms] OR ("carcinoma" [All Fields] AND "hepatocellular" [All Fields]) OR "hepatocellular carcinoma" [All Fields] OR ("hepatocellular" [All Fields] AND "carcinoma" [All Fields]) AND ("humans" [MeSH Terms] AND English [lang]). Proteomics papers were retrieved using the following search: [("proteomics" [MeSH Terms] OR "proteomics" [All Fields]) AND high [All Fields] AND throughput [All Fields]] AND ("carcinoma, hepatocellular" [MeSH Terms]) OR ("carcinoma" [All Fields] AND "hepatocellular" [All Fields]) OR "hepatocellular carcinoma" [All Fields] OR ("hepatocellular"[All Fields] AND "carcinoma"[All Fields]). MicroRNAs reported in HCC were identified using these MeSH terms: ("micrornas" [MeSH Terms] OR "micrornas"[All Fields] OR "mirna" [All Fields]) AND profile [All Fields] AND ("carcinoma, hepatocellular" [MeSH Terms] OR ("carcinoma" [All Fields] AND "hepatocellular" [All Fields]) OR "hepatocellular carcinoma" [All Fields] OR ("hepatocellular" [All Fields] AND "carcinoma" [All Fields]).
We considered for inclusion all datasets available in PubMed.
The datasets publicly available on the GEO, a public functional genomics data repository of high-throughput array data (https://www.ncbi.nlm.nih.gov/geo) were retrieved and analyzed using GEO2R (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html), a web tool available on the portal, identifying genes differentially expressed between samples of HCC and the non-tumoral liver portion. GEO2R compares original submitter-supplied processed data tables using the GEOquery and limma R packages from the Bioconductor project. Following instructions available online at (https://www.ncbi.nlm.nih.gov/geo/info/geo2r.html), we retrieved all dysregulated genes. Only those with an adjusted P value < 0.05, and expression fold change value below ≤ 0.5 or above ≥ 1.5 were considered for further analysis (Table 1, Supplementary Table 1). The genes included in our list from WES papers were reported as affected by nonsynonymous mutations, and synonymous mutations were not considered. Putative microRNA gene targets were identified using an online database, mirDIP 4.1[10], (http://ophid.utoronto.ca/mirDIP). The most stringent predictive search option (top 1%) was used to obtain the list of putative targets of all differentially expressed miRNAs.
From the selected 11 methylation datasets, raw data from eight studies were available on the GEO website (https://www.ncbi.nlm.nih.gov/geo/). We selected the CpG sites or genes reported to be hyper-or hypo- methylated in these publications. The genomic region was considered differentially methylated between HCC tissue and the adjacent non-tumoral sample, if the FDR corrected P value < 0.01. Furthermore, we filtered out everything that did not satisfy the criteria: ∆β ≥ 0.20 or ∆β ≤ -0.20, where ∆β = βHCC - βadjacent was the difference in methylation between above specified groups. When the CpG sites were considered, the Illumina HumanMethylation450K and 27K platforms were used for mapping to the genes. When multiple sites or genes were found to have the same sense of differential methylation, the mean value of ∆β was calculated. Only the CpGs in the 5’UTR, 1st Exon, TSS200, TSS1500 or in CpG islands were considered in our analysis. Proteomic results were retrieved and included only if protein abundance was reported as different in HCC liver samples compared to control samples.
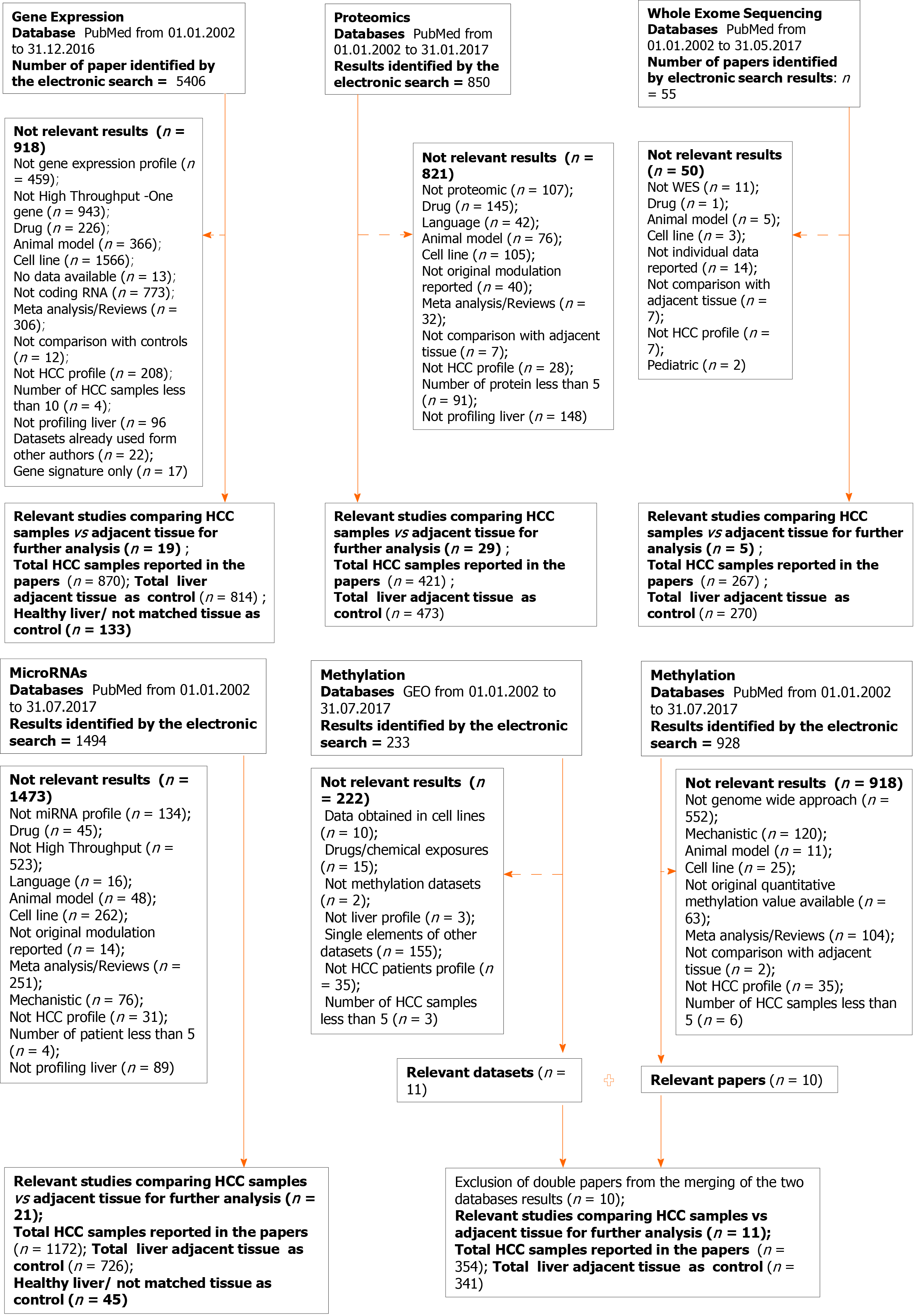
Figure 1 outlines our study workflow. Papers were excluded from each specific search for the following reasons: Data from cell lines, or animal models, studying efficacy or drugs, or the presence of long non-coding RNA, mechanistic studies not performing high-throughput or evaluating the role of one molecule, papers focused on liver diseases but not HCC or liver tissue, not original data such as review articles, or those studies using already selected datasets, not reporting the modulation of the molecules, and papers without data available.
| Gene expression | |||||
| No. | year | PMID | HCC (n) | Controls (n) | GEO dataset |
| 1 | 2004 | 17393520 | 35 | 13 | GSE6764 |
| 2 | 2008 | 18504433 | 11 | 2 | GSE6222 |
| 3 | 2008 | 18923165 | 80 | 82 | GSE10143 |
| 4 | 2009 | 19098997 | 47 | 58 | GSE14323 |
| 5 | 2009 | 19861515 | 16 | 47 | GSE17967 |
| 6 | 2011 | 21320499 | 34 | 34 | GSE20140 (GSE10141, GSE10140) |
| 7 | 2011 | 21712445 | 40 | 40 | GSE28248 |
| 8 | 2013 | 23691139 | 15 | 15 | GSE17548 |
| 9 | 2013 | 23800896 | GSE36376_276; GSE25097_211 | GSE36376_247; GSE25097_283 | GSE36376, GSE25097 |
| 10 | 2014 | 24498002 | 46 | 46 | GSE47595 |
| 11 | 2014 | 24564407 | 45 | 45 | GSE45114 |
| 12 | 2014 | 25093504 | 39 | 40 | GSE57958 |
| 13 | 2014 | 25141867 | 11 | 11 | GSE55092 |
| 14 | 2014 | 25376302 | 18 | 18 | GSE60502 |
| 15 | 2014 | 25536056 | 72 | 72 | GSE39791 |
| 16 | 2015 | 25666192 | 132 | 132 | GSE54236 |
| 17 | 2015 | 25645722 | 228 | 168 | GSE63898 |
| 18 | 2016 | 27499918 | 60 | 60 | GSE64041 |
| 19 | 2016 | 25964079 | 26 | 20 | GSE54238 |
| Proteomics | |||||
| No. | year | PMID | HCC (n) | Controls (n) | |
| 1 | 2004 | 14726492 | 8 | 8 | |
| 2 | 2008 | 19003864 | 12 | 12 | |
| 3 | 2005 | 15759316 | 10 | 10 | |
| 4 | 2005 | 16097030 | 14 | 14 | |
| 5 | 2007 | 17627933 | 12 | 12 | |
| 6 | 2014 | 23621634 | 3 | 3 | |
| 7 | 2009 | 19562805 | 3 | 3 | |
| 8 | 2016 | 26709725 | 24 | 12 | |
| 9 | 2013 | 23589362 | 20 | 20 | |
| 10 | 2012 | 22813877 | 10 | 10 | |
| 11 | 2012 | 22082227 | 11 | 11 | |
| 12 | 2011 | 21631109 | 69 | 123 | |
| 13 | 2010 | 20230046 | 5 | 5 | |
| 14 | 2010 | 19956837 | 20 | 20 | |
| 15 | 2009 | 19715608 | 18 | 18 | |
| 16 | 2009 | 19535095 | 3 | 3 | |
| 17 | 2009 | 19161326 | 80 | 80 | |
| 18 | 2004 | 15221772 | 20 | 20 | |
| 19 | 2003 | 14673798 | 21 | 21 | |
| 20 | 2003 | 14654528 | 21 | 21 | |
| 21 | 2002 | 12481271 | 11 | 11 | |
| 22 | 2013 | 23462207 | 7 | 7 | |
| 23 | 2005 | 16335951 | 8 | 8 | |
| 24 | 2006 | 16342242 | 10 | 10 | |
| 25 | 2011 | 22034872 | 3 | 3 | |
| 26 | 2005 | 15852300 | 7 | 7 | |
| 27 | 2011 | 21913717 | 3 | 3 | |
| 28 | 2007 | 17203974 | 25 | 28 | |
| 29 | 2007 | 17586277 | 10 | 10 | |
| Whole exome sequencing | |||||
| No. | year | PMID | HCC (n) | Controls (n) | GEO dataset |
| 1 | 2013 | 23912677 | 3 | 3 | N/A |
| 2 | 2014 | 24055508 | 4 | 7 | N/A |
| 3 | 2017 | 28323123 | 5 | 5 | N/A |
| 4 | 2014 | 24798001 | 231 | 231 | GSE54504 |
| 5 | 2012 | 22561517 | 24 | 24 | N/A |
| Epigenetic_miRNAs | |||||
| No. | year | PMID | HCC (n) | Controls (n) | GEO dataset |
| 1 | 2015 | 26190160 | 9 | 7 | N/A |
| 2 | 2014 | 24789420 | 10 | 9 | GSE31383 |
| 3 | 2014 | 24564407 | 45 | 45 | GSE10694 |
| 4 | 2011 | 21298008 | 73 | 73 | GSE21362 |
| 5 | 2008 | 18649363 | 78 | 10 | N/A |
| 6 | 2012 | 22135159 | 20 | 20 | N/A |
| 7 | 2011 | 21319996 | 94 | 94 | N/A |
| 8 | 2009 | 19473441 | 20 | 20 | N/A |
| 9 | 2009 | 19173277 | 35 | N/A | |
| 10 | 2007 | 18171346 | 10 | 10 | N/A |
| 11 | 2006 | 16331254 | 25 | 25 | N/A |
| 12 | 2015 | 26062888 | 30 | 30 | N/A |
| 13 | 2015 | 26046780 | 327 | 43 | N/A |
| 14 | 2015 | 25861255 | 66 | 66 | GSE54751 |
| 15 | 2015 | 25500075 | 6 | 6 | GSE54537 |
| 16 | 2014 | 24875649 | 24 | 24 | |
| 17 | 2013 | 23812667 | 166 | 166 | GSE31384 |
| 18 | 2013 | 23390000 | 9 | 17 | GSE40744 |
| 19 | 2012 | 23082062 | 18 | 18 | N/A |
| 20 | 2014 | 24586785 | 29 | 29 | N/A |
| 21 | 2013 | 24417970 | 78 | 78 | N/A |
| Epigenetic methylation | |||||
| No. | year | PMID | HCC (n) | Controls (n) | GEO dataset |
| 1 | 2011 | 21500188 | 13 | 12 | N/A |
| 2 | 2014 | 24306662 | 45 | 45 | N/A |
| 3 | 2014 | 25376292 | 22 | 22 | N/A |
| 4 | 2015 | 25945129 | 8 | 8 | GSE59260 |
| 5 | 2011 | 21747116 | 12 | 12 | GSE29720 |
| 6 | 2010 | 20165882 | 20 | 20 | GSE18081 |
| 7 | 2012 | 22234943 | 62 | 62 | GSE37988 |
| 8 | 2013 | 24012984 | 20 | 8 | GSE44970 |
| 9 | 2013 | 23208076 | 66 | 66 | GSE54503 |
| 10 | 2014 | 25093504 | 59 | 59 | GSE57956 |
| 11 | 2014 | 25294808 | 27 | 27 | GSE60753 |
Available patient data, including etiology of liver disease (hepatitis C, hepatitis B, alcohol, fatty liver disease) on the basis of which the HCC tumors developed, presence of cirrhosis, the Model for End-stage Liver Disease score (MELD score, an assessment of the severity of liver dysfunction), tumor histology, stage of cancer, alpha-fetoprotein level, overall and recurrence-free survival following treatment were also documented (Supplementary Table 2).
The key dysregulated genes from each type of data (genomic, miRNA, methylation, transcriptomic, and proteomic) were fed into the Integrated Interactions Database[11] (IID, http://ophid.utoronto.ca/iid), to obtain a list of the protein-protein interactions. For the miRNA dataset, we determined the target genes of the differentially expressed miRNAs in tumors using the miRNA Data Integration Portal mirDIP v4.1[10]. The individual lists derived from each type of data were then fed into the pathway Data Integration Portal, pathDIP v3.0 (http://ophid.utoronto.ca/pathDIP)[12], in order to determine the significantly dysregulated pathways in HCC. pathDIP integrates data from 20 major pathway databases, and computationally predicts gene association to curated pathways using protein-protein interactions from IID significance of their connectivity[12]. We used this comprehensive pathway enrichment analysis portal to obtain a list of significantly enriched pathways using literature curated (core) pathway memberships P value (FDR: BH-method) less than 0.05.
The lists of pathways from each type of data were then assessed for overlap using Venny 2.1, an online tool for Venn diagram design (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
In order to determine whether key differentially expressed genes along the overlapping pathways had prognostic value, we used KMplotter, a web-based tool that enables survival analysis across multiple cancers and datasets[13]. Patient samples were split into two groups per autoselection of the best cutoff for each gene, in order to assess its prognostic value. We ran multivariate overall survival analysis based on the high vs low expression of each gene in HCC tumors. The two groups were compared by a Kaplan-Meier survival plot, and the hazard ratio with 95% confidence intervals and log-rank P value were calculated.
The identification of putative therapeutic agents able to revert the modulation of genes of interest based on their modulation associated with a worse prognosis was obtained using the online Comparative Toxicogenomics Database http://ctdbase.org[14]. This database provides manually curated information about chemical–gene/protein interactions, chemical–disease and gene–disease relationships.
We identified a total of 8733 abstracts retrieved by the search on PubMed on HCC for the different layers of data on human HCC samples, published until December 2016. The flow chart outlining the selection process is detailed in Figure 1.
The number of samples included in our analysis are as follows: (1) Whole exome sequencing: 267 HCC and 270 control samples; (2) Gene expression: 870 HCC and 814 control samples; (3) miRNA: 1172 HCC and 771 control samples; (4) Methylation: 354 HCC and 341 control samples; and (5) Proteomics: 421 HCC and 473 control samples. The methodologies and platforms used to obtain these high-throughput data are reported by type of data (genomic, transcriptomic, miRNA, methylation and proteomic) in Table 1. Clinical data, regarding etiology of liver disease (hepatitis C, hepatitis B, alcohol, fatty liver disease) were frequently reported, on the other side serum levels of liver enzymes, AST and ALT, frequently used to assess liver functions were not available. Pathological details relative to differentiation or stage were frequently absent as well as other crucial variables in the clinic setting, such as Child Pugh/MELD score (Supplementary Table 2).
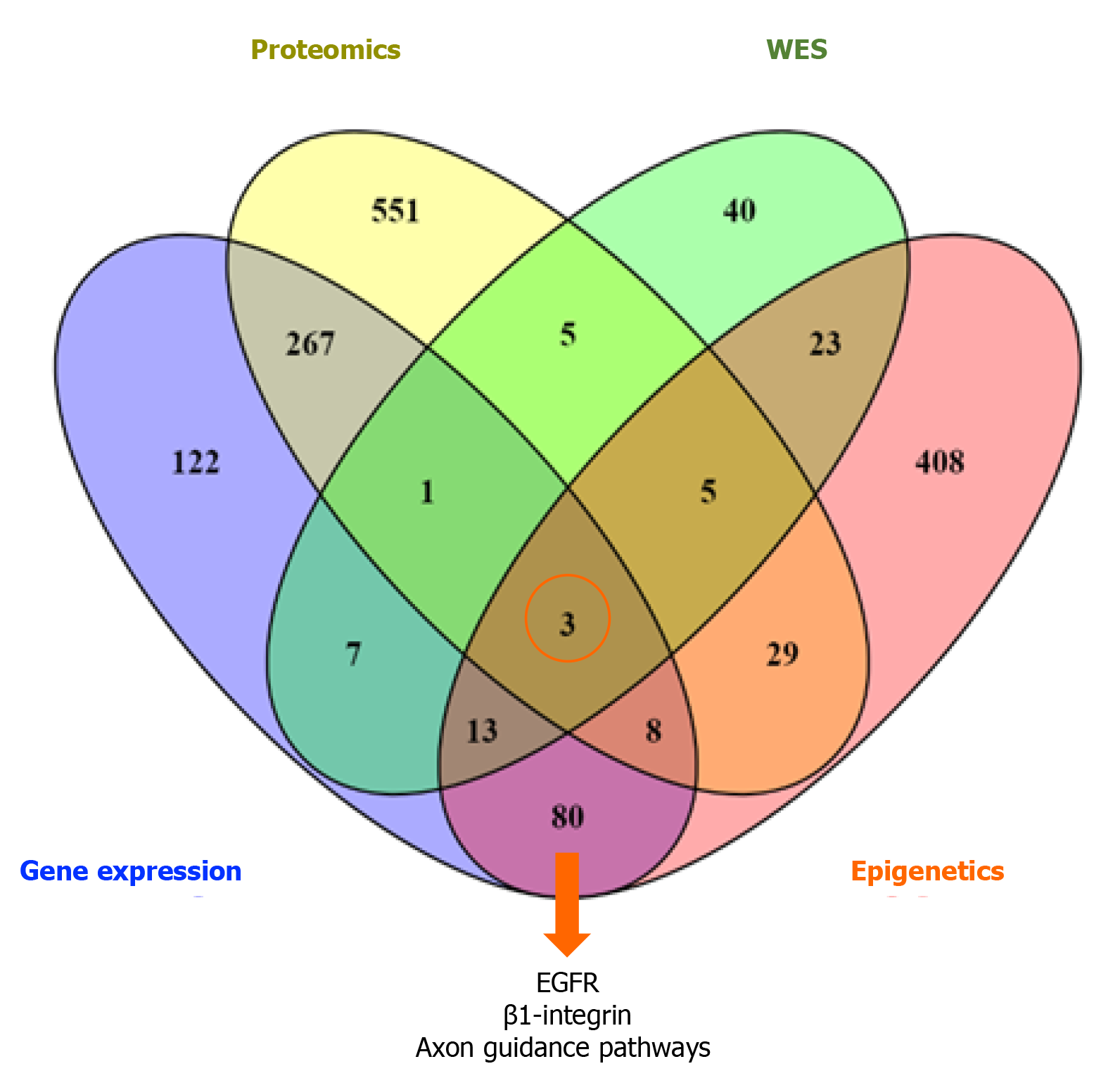
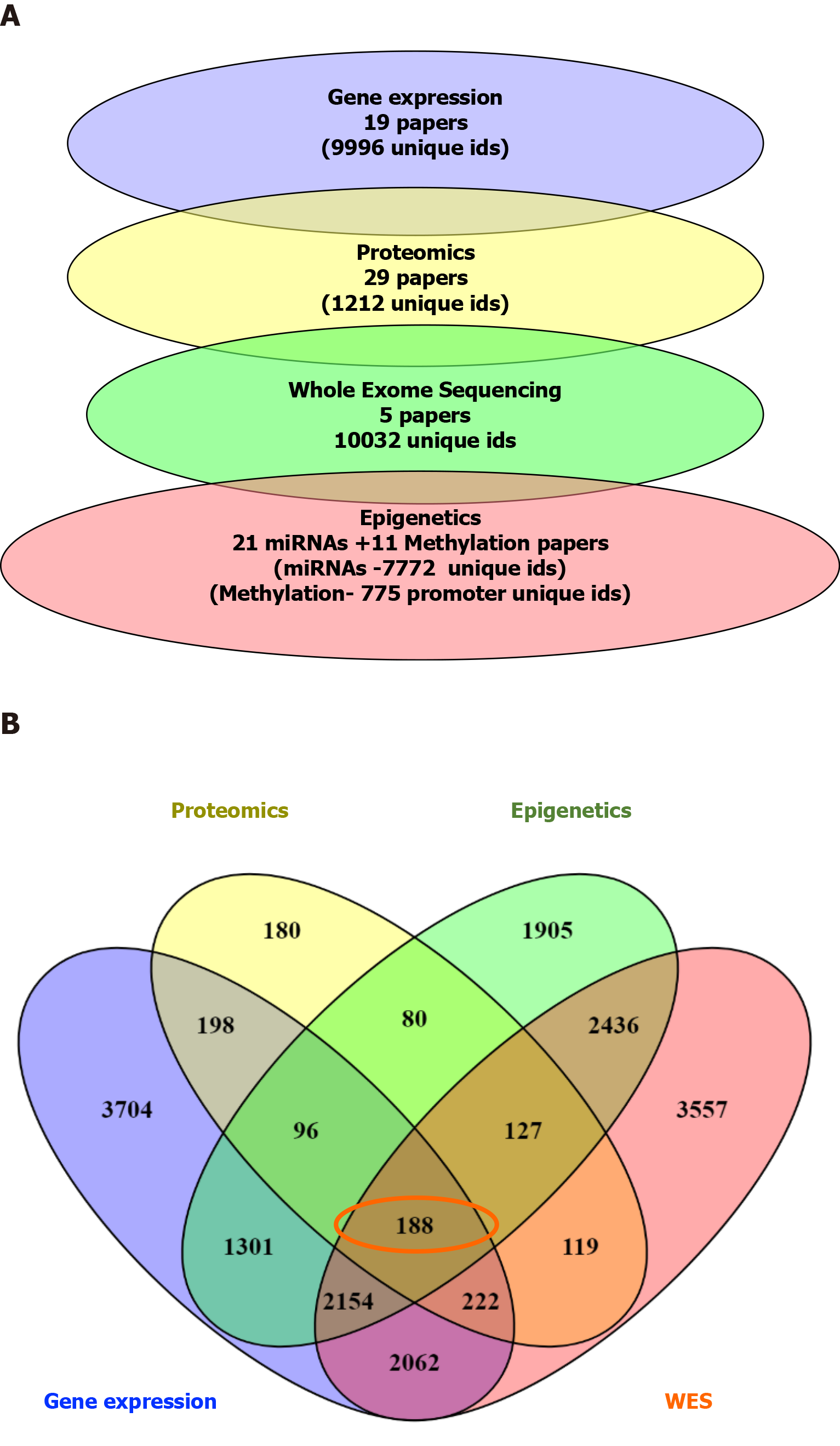
There were 188 overlapping dysregulated genes/proteins across the different types of data. Independently for each type of data, we obtained a list of pathways using pathDIP. We merged the list of dysregulated pathways in miRNA and methylation, given that these epigenetically regulate gene expression, in order to assess for overlapping pathways across the datasets.
This resulted in a list of 3 common, overlapping pathways among the different types of data: EGFR, β1-integrin, and axon guidance pathways, as depicted in Figure 2. From the previous list of 188 common dysregulated elements in all different layers of data (Figure 3), we were able to identify 35/188 genes that were involved in these 3 shared pathways across the layers of data (Supplementary Table 1).
We then examined the prognostic value of the deregulated genes associated to pathways of interest in HCC using TCGA RNA seq dataset, as listed in Table 2. Median survival of 364 patients in the TCGA, which was used for validation purposes regarding the prognostic value is reported. KMplotter HR results from TCGA RNA seq data reflected the altered modulation identified for these 9 genes in the 19 HCC papers relative to the gene expression data (Table 2). Among the five upregulated genes associated with positive HR values, CDK5, was reported with the highest HR value (1.85, P = 0.0035) and involved in cell cycle (Table 3). The other 4/9 genes reported as upregulated, COL2A1, LAMC1, RPS6KA3 and ITGB1 were identified with positive HR value by KM plotter analysis and involved in cellular migration (Table 2 and Table 3).
| Gene | Modulation in the 19 HCC papers | Probe-ID | HR | CI | Log-Rank P value | Median survival low (mo) | Median survival high (mo) | Estradiol gene modulation predicted by CTD |
| COL2A1 | Up | 1280 | 1.49 | 1.05-2.11 | 0.0229 | 61.7 | 54.1 | N/A |
| FGA | Down | 2243 | 0.52 | 0.35-0.77 | 0.0009 | 49.7 | 70.5 | + |
| FGG | Down | 2266 | 0.56 | 0.39-0.79 | 0.0009 | 38.3 | 70.5 | + |
| LAMC1 | Up | 3915 | 1.43 | 0.98-2.09 | 0.06 | 56.5 | 38.3 | N/A |
| CDK5 | Up | 1020 | 1.85 | 1.22-2.81 | 0.0035 | 81.9 | 6.2 | N/A |
| EPHB1 | Down | 2047 | 0.72 | 0.048-1.08 | 0.1135 | 54.1 | 70.5 | N/A |
| RPS6KA3 | Up | 6197 | 1.2 | 0.8-1.78 | 0.3743 | 54.1 | 56.5 | - |
| EGFR | Down | 1956 | 0.61 | 0.43-0.89 | 0.0085 | 31 | 70.5 | + |
| ITGB1 | Up | 3688 | 1.37 | 0.95-1.97 | 0.0924 | 82.9 | 49.7 | N/A |
| Gene | Modulation in the 19 HCC papers | PMID | Mutation in HCC (PMID) | Role in cancer (PMID) |
| COL2A1 | Up (2/19) | 23800896/25666192 | (rs3917) polymorphism is associated with higher risk of HCC (21665180) | COL2A1 promotes migration in HCC (29858962) |
| FGA | Down (9/19) | 21320499/23800896/25093504/25536056/25141867/ | Deleted in HCC patients (27511114) | FGA is a positive predictor of survival in gastric cancer patients (15756001) |
| FGG | Down 8/19 | 21320499/23800896/25093504/25536056/25141867/ | Allelic loss (16980951) | FGG is involved in amino acid and redox metabolism pathway in HCC (28089356) |
| LAMC1 | Up (4/19) | 23800896/25536056/25141867/25645722 | Not identified | LAMC1 promotes tumor cell invasion and migration in HCC (28928891) |
| CDK5 | Up (2/19) | 25141867/25376302 | Not identified | CDK5 promotes proliferation in HCC (29312535) |
| EPHB1 | Down (2/19) | 23800896/25141867 | Missense mutation (19469653) | EPHB1 inhibits cell migration(22242939) |
| RPS6KA3 | Up 1/19 | 25141867 | Somatic mutation and copy number variations (22561517) | RPS6KA3 increases cell proliferation (15833840) |
| EGFR | Down (2/19) | 19098997/25141867 | Missense mutation (26436086) | EGFR promotes cell adhesion (31465839) |
| ITGB1 | Up (1/19) | 25141867 | Somatic number variations (24512821) | ITGB1 promotes migration (30664185) |
Four out of 9 genes were reported as downmodulated in the 19 HCC gene expression papers. Among these four, two genes, FGA and FGG, were identified as the top statistically significantly (P = 0.0009) associated with a protective role in HCC (HR values 0.52 and 0.59, respectively). FGA and FGG were consistently reported as downmodulated in about 45% of our 19 selected gene expression papers (Table 3). The other two downmodulated genes, EPHB1 and EFGR with negative HR values (Table 2) are reported to be affected by missense mutation leading to a loss of their protective role against cell migration.
Using CTD, we found that estradiol was able to appropriately down- or upmodulate 4 out of 9 cancer-related genes (Table 2). Particularly, CTD reported estradiol capabilities to upregulated FGA, FGG and EGFR reported downmodulated in HCC (Table 2) and counteracting the upregulation of RPS6KA3 in HCC, suggesting a possible role for this hormone in HCC treatment.
In this study, we evaluate the molecular pathogenesis of HCC using a unique approach, that of combining all publicly available high-throughput data from patient HCC tumors. This encompasses all miRNA, methylation, genomic, transcriptomic and proteomic profiling data present in the literature, and represents the first effort to derive a consensus molecular model of HCC through analysis of these different types of data. Although these datasets originated from different patient cohorts, presented integrative analysis offers the opportunity to explore common key pathway dependencies of HCC. Starting with the initial generation of genomics and whole exome sequencing data, previous high-throughput studies have brought forth different lists of dysregulated genes, depending on the type of data evaluated. Dysregulated genes may affect different parts of a pathway. Therefore, a pathway-based approach when evaluating different types of high-throughput data offers the ability to assess the pathways most commonly affected in a given cancer. Additionally, the integrative analysis in our study encompasses a large number of patient samples.
Using this integrative approach, we confirm the importance of EGFR, β1-integrin and axon guidance as pathways critical in hepatocarcinogenesis. EGFR activates the signaling cascades of the Ras/Raf/MAPK and mTOR pathways, two pathways that were identified as key to HCC pathogenesis in the TCGA study[6]. The identification of β1-integrin as being commonly dysregulated in HCC is novel, and its significance is confirmed through its consistent dysregulation across types of data. β1-integrin is a cell surface receptor that senses the extracellular matrix, thereby modulating the hallmarks of cancer such as proliferative signaling with continuous activated cell replication, evasion of growth suppressors, resistance to angiogenesis as well as cancer cell invasion and metastasis[14]. Ras/Raf/MAPK and mTOR are established pathways in hepatocarcinogenesis, and are integrin-dependent signaling pathways[15]. Additionally, β1-integrin is known to crosstalk with EGFR. In fact, the downregulation of β1-integrin was found to decrease phosphorylation of EGFR and c-Met in hepatocytes during liver regeneration[16]. A synergistic relationship between integrins and EGFR has also been demonstrated in tumor progression[17]. The finding of axon guidance pathway-related proteins as being dysregulated across types of data, thereby establishing consistent dysregulation of this pathway in HCC, is also novel. Netrin-1 is the best studied protein in the axon guidance pathway, and is known to be overexpressed in various cancers[13]. It is responsible for regulation of apoptosis, with increased presence of netrin-1 leading to inhibition of apoptosis. The tumor suppressor p53, frequently mutated in the TCGA HCC study, regulates the cell cycle through netrin-1. The axon guidance pathway has previously been identified as a pathway that is significantly mutated in HCC based on integration of all genomic data in HCC[18]. This analysis revealed mutations along the axon guidance pathway as being prognostic of a higher rate of HCC metastasis. We were able to additionally validate the prognostic importance of dysregulated proteins in these pathways proteins using TCGA data.
HCC is a cancer that develops in the context of various chronic liver diseases, which may influence the molecular characteristics of HCC. Additionally, the underlying cirrhosis and liver dysfunction that are often concurrent may influence HCC development and behavior[2]. Patients are often diagnosed at an advanced stage of disease, when it is too late for curative treatment. A unique consideration in HCC is the inability to tolerate hepatotoxic chemotherapy in patients with liver dysfunction, as it is often patients with cirrhosis who develop HCC[19,20]. Therefore, liver function must be considered prior to, during, and after any form of treatment for HCC.
Thus, especially for HCC, it has been suggested that a multi-pronged approach to HCC therapy jointly targeting different pathways be adopted.
Omics technologies are essential in the progress towards elucidating the molecular basis of HCC. The current study represents the largest integration of all publicly available genomic, gene expression, methylation, miRNA and proteomic data in HCC, covering 85 studies and 3355 patient sample profiles. We identified consistently deregulated pathways associated with hepatocarcinogenesis across different types of data using integrative analysis tools, thereby confirming the importance of these genes in HCC pathogenesis. EGFR (activator of Ras/Raf/MAPK and mTOR) and β1-integrin (also modulator of the aforementioned pathways) were clearly identified as pivotal to HCC[5,21-23]. This is in keeping with the efficacy of the Ras/Raf/MAPK inhibitors sorafenib and regorafenib in HCC[24].
Even beyond this, we found these consistently deregulated genes across pathways to be appropriately modulated by estradiol. HCC is less common in women, and there have been clinical studies demonstrating that hormone therapy and female sex are protective against HCC as described earlier in this thesis.
Other integrative multi-omics studies have been recently performed for other tumors with high mortality such as breast and ovarian cancer[6,25]. Several breast cancer studies emphasizing how data integration of genomic/transcriptomic and proteomic has improved the molecular characterization of subtypes of breast cancer and elucidate its heterogeneity and its interaction with the microenvironment and aggressiveness[26,27]. A single source of data was used in the ovarian cancer multi-omics mathematical integration performed by Bhardwaj et al[25]. Copy number variation gene expression and methylation data from TCGA data portal were integrated using mathematical algorithm and identified 32 co-expressed genes and 6 pathways associated with survival.
The main limitation of our study is the different patient samples represented by the various types of data. Nonetheless, there is a large amount of high-throughput data, which allowed us to detect pathway dependency patterns that are compatible with the current HCC literature. Additionally, HCC tumors arise in the setting of various chronic liver diseases. We could not assess for etiology-specific genes and pathways in this study, given that the clinical and genetic data to evaluate these differences were not fully available for all the studies. Therefore, we could only evaluate gene differences over whole datasets, rather than individual patients, due not complete individual annotation of the samples available on GEO for each specific dataset. The HCC samples in this integrative analysis all came from patients who had undergone hepatectomy. There were no specimens from patients who were candidates for ablation therapy (early stage), those who were undergoing liver transplantation, or those with advanced HCC. One might anticipate that the molecular features of such tumors differ, given the different stages of HCC captured, but there is unfortunately scarcity of data in this regard.
In conclusion, our study represents the largest integrative analysis of all publicly available data in HCC, spanning different types of high-throughput data. Pathway enrichment analysis elucidated EGFR, β1-integrin and axon guidance as pathway dependencies in HCC. These are proteins known to serve as master regulators of key pathways in HCC such as Ras/Raf/MAPK, Wnt/β-catenin and mTOR[28], and may serve as potential overarching therapeutic targets in HCC. The axon guidance pathway was identified as being of potential importance to HCC for the first time, with prognostic value suggested in patient sample validation with TCGA. Estradiol affects a large number of deregulated genes across data with appropriate modulation and may be a therapeutic agent that helps in HCC. A combined therapeutic approach conjointly targeting different pathways may be more optimal in the treatment of HCC, especially when underlying hepatic dysfunction compromises the ability to tolerate optimal chemotherapeutic doses.
Hepatocellular carcinoma (HCC) is highly heterogeneous, difficult to characterize and the molecular basis of HCC has been elusive.
The Cancer Genome Atlas is a large-scale project that has enabled improved characterization of cancers with several layers of data. Elucidating the layers of data in a disease can provide additional insights into the pathways that drive cancer.
A novel integrative approach of all publicly available high-throughput data from patient HCC tumors was used to delineate critical pathway dependencies in HCC.
A comprehensive analysis and characterization of all publicly available genomic, gene expression, methylation, miRNA and proteomic data in HCC covered 85 studies and 3355 patient sample profiles and identified the key overlapping dysregulated genes and pathways affected.
We identified the prognostic value of these genes in HCC genes, specifically with Netrin and Slit3 being novel proteins of prognostic importance to HCC.
Our large integrative analysis of all publicly available data in HCC and our pathway enrichment analysis has elucidated epidermal growth factor, β1-integrin, and axon guidance as pathway dependencies in HCC.
Based on our integrative analysis, epidermal growth factor, and β1-integrin are master regulators that could be considered as potential therapeutic targets in HCC.
The authors thank undergraduate students Sujitha Srinathan, Emily Chen, Bishoy Lawendy, Nangi Suo and Amira Abdallah for their help in data curation.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Canada
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Troncoso MF S-Editor: Zhang L L-Editor: A P-Editor: Wang LL
| 1. | Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989-5005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 680] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4264] [Article Influence: 236.9] [Reference Citation Analysis (2)] |
| 3. | Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, Zhu AX, Murad MH, Marrero JA. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2107] [Cited by in RCA: 3023] [Article Influence: 431.9] [Reference Citation Analysis (3)] |
| 4. | Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5972] [Cited by in RCA: 6572] [Article Influence: 469.4] [Reference Citation Analysis (1)] |
| 5. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Häussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J; SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9016] [Cited by in RCA: 10261] [Article Influence: 603.6] [Reference Citation Analysis (2)] |
| 6. | Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017; 169: 1327-1341. e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1578] [Cited by in RCA: 1729] [Article Influence: 216.1] [Reference Citation Analysis (1)] |
| 7. | Wilk G, Braun R. Integrative analysis reveals disrupted pathways regulated by microRNAs in cancer. Nucleic Acids Res. 2018;46:1089-1101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 8. | Srivastava A, Kumar S, Ramaswamy R. Two-layer modular analysis of gene and protein networks in breast cancer. BMC Syst Biol. 2014;8:81. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, Zhou JY, Petyuk VA, Chen L, Ray D, Sun S, Yang F, Chen L, Wang J, Shah P, Cha SW, Aiyetan P, Woo S, Tian Y, Gritsenko MA, Clauss TR, Choi C, Monroe ME, Thomas S, Nie S, Wu C, Moore RJ, Yu KH, Tabb DL, Fenyö D, Bafna V, Wang Y, Rodriguez H, Boja ES, Hiltke T, Rivers RC, Sokoll L, Zhu H, Shih IM, Cope L, Pandey A, Zhang B, Snyder MP, Levine DA, Smith RD, Chan DW, Rodland KD; CPTAC Investigators. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell. 2016;166:755-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 752] [Article Influence: 83.6] [Reference Citation Analysis (0)] |
| 10. | Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360-D370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 408] [Article Influence: 68.0] [Reference Citation Analysis (0)] |
| 11. | Kotlyar M, Pastrello C, Sheahan N, Jurisica I. Integrated interactions database: tissue-specific view of the human and model organism interactomes. Nucleic Acids Res. 2016;44:D536-D541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 192] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 12. | Rahmati S, Abovsky M, Pastrello C, Jurisica I. pathDIP: an annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucleic Acids Res. 2017;45:D419-D426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 13. | Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004;4:978-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 203] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 14. | Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res. 2017;45:D972-D978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 401] [Cited by in RCA: 398] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 15. | Griffiths GS, Grundl M, Leychenko A, Reiter S, Young-Robbins SS, Sulzmaier FJ, Caliva MJ, Ramos JW, Matter ML. Bit-1 mediates integrin-dependent cell survival through activation of the NFkappaB pathway. J Biol Chem. 2011;286:14713-14723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | Speicher T, Siegenthaler B, Bogorad RL, Ruppert R, Petzold T, Padrissa-Altes S, Bachofner M, Anderson DG, Koteliansky V, Fässler R, Werner S. Knockdown and knockout of β1-integrin in hepatocytes impairs liver regeneration through inhibition of growth factor signalling. Nat Commun. 2014;5:3862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 17. | Ivaska J, Heino J. Cooperation between integrins and growth factor receptors in signaling and endocytosis. Annu Rev Cell Dev Biol. 2011;27:291-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 195] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 18. | Zhang Y, Qiu Z, Wei L, Tang R, Lian B, Zhao Y, He X, Xie L. Integrated analysis of mutation data from various sources identifies key genes and signaling pathways in hepatocellular carcinoma. PLoS One. 2014;9:e100854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 19. | Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2-S6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 879] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 20. | Fitzmorris P, Shoreibah M, Anand BS, Singal AK. Management of hepatocellular carcinoma. J Cancer Res Clin Oncol. 2015;141:861-876. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 21. | Zhu AX, Abrams TA, Miksad R, Blaszkowsky LS, Meyerhardt JA, Zheng H, Muzikansky A, Clark JW, Kwak EL, Schrag D, Jors KR, Fuchs CS, Iafrate AJ, Borger DR, Ryan DP. Phase 1/2 study of everolimus in advanced hepatocellular carcinoma. Cancer. 2011;117:5094-5102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 162] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 22. | Zhou Q, Lui VW, Yeo W. Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 2011;7:1149-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 186] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 23. | Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12:436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 133] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 24. | Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, Gerolami R, Masi G, Ross PJ, Song T, Bronowicki JP, Ollivier-Hourmand I, Kudo M, Cheng AL, Llovet JM, Finn RS, LeBerre MA, Baumhauer A, Meinhardt G, Han G; RESORCE Investigators. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2160] [Cited by in RCA: 2709] [Article Influence: 338.6] [Reference Citation Analysis (0)] |
| 25. | Bhardwaj A, Van Steen K. Multi-omics Data and Analytics Integration in Ovarian Cancer. In: Maglogiannis I, Iliadis L, Pimenidis E, editors. Artificial Intelligence Applications and Innovations. 2020;347-57. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 26. | Wagner J, Rapsomaniki MA, Chevrier S, Anzeneder T, Langwieder C, Dykgers A, Rees M, Ramaswamy A, Muenst S, Soysal SD, Jacobs A, Windhager J, Silina K, van den Broek M, Dedes KJ, Rodríguez Martínez M, Weber WP, Bodenmiller B. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 2019; 177: 1330-1345. e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 551] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 27. | Bhatia S, Monkman J, Blick T, Duijf PH, Nagaraj SH, Thompson EW. Multi-Omics Characterization of the Spontaneous Mesenchymal-Epithelial Transition in the PMC42 Breast Cancer Cell Lines. J Clin Med. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Bhat M, Sonenberg N, Gores GJ. The mTOR pathway in hepatic malignancies. Hepatology. 2013;58:810-818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |