Published online Apr 26, 2016. doi: 10.4252/wjsc.v8.i4.118
Peer-review started: September 10, 2015
First decision: October 8, 2015
Revised: January 21, 2016
Accepted: February 14, 2016
Article in press: February 16, 2016
Published online: April 26, 2016
Processing time: 216 Days and 19.5 Hours
Recent and advanced protocols are now available to derive human induced pluripotent stem cells (hiPSCs) from patients affected by genetic diseases. No curative treatments are available for many of these diseases; thus, hiPSCs represent a major impact on patient’ health. hiPSCs represent a valid model for the in vitro study of monogenic diseases, together with a better comprehension of the pathogenic mechanisms of the pathology, for both cell and gene therapy protocol applications. Moreover, these pluripotent cells represent a good opportunity to test innovative pharmacological treatments focused on evaluating the efficacy and toxicity of novel drugs. Today, innovative gene therapy protocols, especially gene editing-based, are being developed, allowing the use of these cells not only as in vitro disease models but also as an unlimited source of cells useful for tissue regeneration and regenerative medicine, eluding ethical and immune rejection problems. In this review, we will provide an up-to-date of modelling monogenic disease by using hiPSCs and the ultimate applications of these in vitro models for cell therapy. We consider and summarize some peculiar aspects such as the type of parental cells used for reprogramming, the methods currently used to induce the transcription of the reprogramming factors, and the type of iPSC-derived differentiated cells, relating them to the genetic basis of diseases and to their inheritance model.
Core tip: With the development of human induced pluripotent stem cells (hiPSCs) deriving from patients, we can begin to understand the molecular mechanisms underlying monogenic diseases and consequently identify new drugs for their treatment. hiPSCs can differentiate into many disease-relevant cell types, providing in this way to innovative applications in the field of cell replacement therapy, disease modelling, drug testing and drug discovery.
- Citation: Spitalieri P, Talarico VR, Murdocca M, Novelli G, Sangiuolo F. Human induced pluripotent stem cells for monogenic disease modelling and therapy. World J Stem Cells 2016; 8(4): 118-135
- URL: https://www.wjgnet.com/1948-0210/full/v8/i4/118.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v8.i4.118
For many years the research and experimentation on stem cells have been made, taking advantage of their extraordinary ability to divide and self-renew into undifferentiated cells. Pluripotent stem cells are able to differentiate into all cell types. These characteristics offer the possibility of different applications from the use of stem cells for model disease, for cell therapy and tissue regeneration and pharmacological and toxicological tests.
Researchers have always worked with both embryonic and adult stem cells for the study of disease and for gene therapy. Despite the unique characteristic of embryonic stem cells (ESCs), there are controversies moral, ethical and legal regarding their use.
Adult stem cells have limited differentiation potentiality, so this aspect reduces the options for their use.
Thus, induced pluripotent stem cells (iPSCs), derived from somatic cells, have equal characteristics of ESCs. It is possible reprogramming cells from patients with human diseases that reproduce a model of disease in vitro and summary the pathological phenotypes and the etiopathology of the diseases. So, their use allow the development of innovative therapies, drug screening and toxicological testing[1-3].
For some genetic diseases no therapeutic treatment is available and the animal model does not always fully possess the variability of the disease. In addition, the understanding of the pathogenetic mechanism at the base of the disease are slows.
The ultimate goal of reprogramming is the transplantation of progenitor cell, genetically corrected in vitro before transplantation, derived from a patient-specific human induced pluripotent stem cells (hiPSCs). These cells will not trigger an immune response, avoid tumour formation and recover the target-damaged tissue.
In 2007, iPSCs were obtained from human fibroblasts by manipulation and expression of genes involved in dedifferentiation and in the maintenance of “stemness”[4,5]. Reprogramming somatic cells using the defined OCT4, SOX2, KLF4, and c-MYC (OSKM) factors led Yamanaka S and Gurdon JB to win the Nobel Prize in Physiology or Medicine in 2012. Thomson’s group follow-up research produced iPSCs using NANOG and LIN28 instead of KLF4 and c-MYC[5] and later, many others researchers developed alternative methods of reprogramming[6].
The most commonly used method is the use of viral transduction of defined factors to somatic cells. Lentiviral-based systems, for example, are the most efficient and reproducible, driving the integration of the reprogramming factors. Unfortunately, viral-based disease models still bear the risks of oncogene reactivation, insertional mutagenesis, immunogenicity, reactivation of reprogramming genes or their uncontrollable silencing, making them unacceptable for human clinical applications. In terms of the aspect of safety of reprogramming, various alternative approaches of gene delivery have been developed. Instead of integrating vectors[1], plasmids[7], Cre/loxP system[8], piggy Bac vectors[9], and minicircle vectors have been investigated[10], in order to partially prevent transgene integrations and in the same time to simplify the methods to obtain cell reprogramming[6].
Current studies have successfully reported the generation of transgene-free iPSCs using different approach, such as: Protein transduction[11], non-integrating viral vectors such as: The Sendai virus[12], episomal vectors[13], transfection of modified mRNA transcripts[14], and chemicals[15]. Nevertheless, when using protein as inducing factor for reprogramming the efficiency is lower (approximately 0.001).
A modified mRNA-based strategy is currently being explored to produce transgene-free iPSCs[16,17]. Other methods dealing with small molecules have also been reported to enhance the performance of iPSCs derivation[18-25]. Similarly human telomerase reverse transcriptase (hTERT), P53 siRNA and Simian Vacuolating Virus 40 large T (SV40LT) Antigen successfully stimulate the reprogramming kinetics[26,27]. Some others, like Estrogen related receptor β (Esrrb), Utf1, Lin28, and developmental pluripotency-associated 2 (Dppa2) generate iPSCs without of OSKM factors with single-cell level identification of reprogramming events[28].
The typical yields of iPSCs production by the methods aforesaid range from 0.01%-5%, depending on the target cell and reprogramming system. Rais et al[29] reported the reprogramming efficiency of methyl-binding protein 3 deletion that reached up to nearly 100% within a few days, supporting that iPSCs reprogramming represents a deterministic process.
One factor strongly influencing the efficiency of reprogramming is the type of cell used as target. This choice can depend on the amount of DNA methylation, gene expression and stability of the pluripotent phenotype, as well as the epigenetic memory of the cell type. In addition, the gene delivery methods, and culture conditions as well as the transcription factors combination might also be the reason of the differences between the various iPS cell populations created. At least, several uncontrollable stochastic events can influence the success of the reprogramming[30].
For all these reasons, researchers have studied the “best candidate parental population” to create iPSCs for in vitro investigations and eventual clinical trials[31]. Fibroblasts are the main source of iPSCs, although other sources for iPSCs have been reported, like hepatocytes and mature B cells[32]. To better understand developments in personalized medicine, we will focus our review on the production and application of human iPSCs derived from foetal tissues, highlighting their higher responsiveness during the reprogramming.
Stem cells have been identified in several foetal tissues and from amniotic fluid, umbilical cord blood, and placenta at term[33-35]. These cells have mesenchymal origin and they are capable of self-renewal and differentiation into multiple tissue types[36-39]. According to their tissue derivation and to the gestational age, the heterogeneity of foetal stem cell populations is emblematic, in agreement to their phenotypic characteristic, properties and cell marker expression.
Trophoblast cells form the foetal part of the placenta. The placenta is an organ indispensable for the growth and survival of the developing embryo. The placenta is constituted by different trophoblast cell types aimed for embryo implantation, for vascular connection to the maternal circulation and nutrition of the fetus and immunological adaptation. Trophoblast cells derive from the trophoectoderm, which gives rise to both attachment and implantation of the embryo. The trophoectoderm is composed of floating and anchored villi and their specialized cell types, the syncytium and the cytotrophoblast. For the first 3 wk of pregnancy, it represents a continuously renewing epithelium[40]. During gestation, the trophoblast changes morphologically and functionally. In particular, the cytotrophoblast, a self-renewing population located in the proliferation zone, divides continuously and fuses to form syncytiotrophoblasts, in which some authors report the presence of possible stem cells important for its renewal[41,42]. Spitalieri et al[39] has isolated and characterized a subpopulation of multipotent cells, named human cytotrophoblastic-derived multipotent cells (hCTMCs) obtained from human chorionic villus sampling (hCVSs) with characteristics that are “intermediate” between mesenchymal and pluripotent stem cells. These cells express stem cell markers, such as ALP, SSEA4, OCT-4, CD117, NANOG, and SOX2. Also, these cells are capable of generate in vitro cells belonging to ectoderm, mesoderm and endoderm layers, but, if inoculated into Nod/SCID mice, they are unable to form teratomas. If injected into mouse blastocysts, hCTMCs are integrated and could be tracked into various tissues of the adult chimeric mice. These cells may be also a promising target for gene editing approaches, such as small fragment homologous recombination, as we report for an in vitro genetic modification of SMN gene in a fetus affected by spinal muscular atrophy (SMA). They can be genetically edited with high performance, allowing an innovative therapeutic approach to cure genetic defects[39,43,44].
During amniocentesis, at the sixteenth week of gestation, small amount of fluid carry hAFSCs. Stem cells are present also in amniotic fluid at term (from routine caesarean deliveries). Kaviani et al[45] collected up to 20000 cells from 2 mL of amniotic fluid, 80% of which are having the ability to grow. Membrane receptor c-kit (CD117), marker of stemness, is expressed on ESCs[46], primordial germ cells and many others somatic stem cells, including a sort multipotent subpopulation of human amniotic fluid stem cells (hAFCs) (round 1%)[47]. Moreover, hAFSCs show a high telomerase activity with a self-renewal capacity, and display normal G1 and G2 cell cycle checkpoints. On top, at late passages they maintain a normal karyotype. Like ESCs, hAFSCs are capable of differentiating into all three germ layers, their efficiency depends on the gestational age[33,48-50]. But, hAFSCs are unable to form teratoma, when inoculated into immunodeficient mice in vivo. It is possible, also, to use hAFSCs in a technique involving retrovirally tagged cells, because they have a high clonal capacity.
From a single term amnion membrane it is possible isolate a multipotent epithelial cell population and obtain approximately 120 million viable epithelial cells[51-53]. hAECs possess ability of multipotent differentiation[54], low immunogenicity[55] and anti-inflammatory functions[56].
Therefore, as reported in the literature, both hCVSs and hAFCs are heterogeneous populations composed by several stem/progenitor lineages[57]. In response to external stimuli, they modulate gene and protein expression for their high plasticity, as already published[58]. Moreover foetal stem cells have an higher proliferative capacity if compared to adult cells. Again, congenital malformations or genetic diseases in newborns could be treated thanks to the capacity to separate pluripotent autogenic progenitor cells during pregnancy[59]. Only recently have these cells, traditionally used for prenatal diagnosis, been explored for their stemness and for reprogramming efficiency[3,60].
When compared with embryonic cells, iPSCs differentiate less efficiently into specialized cell lines, due to their “molecular identity”[61]. Moreover, some iPSCs have a greater capacity to silence some genes demonstrated to be required for foetal development and differentiation[62]. These differences constitute an active area of research that still requires a direct comparison of the pluripotency of hiPSCs vs hESCs. An explanatory situation is represented by the FMR1 gene, involved in Fragile X syndrome (#300624), a genetic condition characterized by learning disabilities and cognitive impairment. The protein product is necessary for normal brain development. In Fragile X syndrome the FMR1 gene acquires a silencing mutation. While this gene functions normally in human embryonic cells and becomes silenced as the cells differentiate, in hiPSCs remains inactive[63].
Several studies suggest that cells undergoing reprogramming go through an intermediary state via resetting of the epigenetic landscape, whereby c-MYC and KLF4 are initially required to prime the cells that are then driven towards pluripotency by OCT4 and SOX2. These observations open interesting scenarios for further investigations focused to discover methods to directly create progenitor therapeutic cell types from somatic cells[6], bypassing the pluripotency step.
Thus, iPSCs that conserve genomic stability and free from any integrated agents represent an important aim for therapeutic uses. Recent studies suggest the use of a high-resolution method, such as the Affymetrix Cytoscan HD array (Affymetrix, Santa Clara, CA, United States), for monitoring genomic alterations throughout iPSC preparation to preserve clinical applications[64].
Given the high level of manipulation and the lack of knowledge about their role in vivo, the use of hiPSCs in human trials is complex. Whether hiPSCs will prove an useful substitute for hESCs has yet to be determined, hESCs are still considered the gold standard for embryonic cell lines[65].
At the same time, hiPSCs represent an ideal autologous cellular model for the study and the treatment of diseases, reaching the goal of personalized medicine. The field of personalized medicine is based on the idea that life is variable and that individuals behave differently from each other under disease conditions. By taking into account individual clinical, genetic and environmental information, personalized medicine optimizes medical care and outcomes for customized disease prevention, detection and treatment.
hiPSCs coincide perfectly with the concept of personalized medicine for disease modelling and further clinical application. In fact, hiPSCs bypass the limitation of immune rejection, being patient-specific cells, united to a “rejuvenation” of telomere length during reprogramming[66], epigenetic memory and functional properties, offering enormous clinical potential.
The tumorigenic risk of hiPSCs arising from the use of integrating vectors for their derivation supports the use of integrating vectors that can be subsequently removed from the genome. Sommer et al[67], for example, reports the use of the human STEMCCA excisable polycistronic lentiviral vector[3]. The delivery vectors are designed so that the ectopic genes are flanked by loxP sites, thus enabling their removal by transient Cre protein expression[68,69]. This approach generates hiPSCs free of transgenic sequences that can improve and increase the safety of derivation methods.
Due to ethical and technical challenges, human embryonic stem cells (hESCs) can represent unsuitable candidates for disease modelling. Therefore, hiPSCs, closely resembling the key features of hESCs such as self-renewal and pluripotent potentials, can be extensively exploited to study various inherited disorders.
Dimos et al[70] (2008) and Park et al[71] (2008) reported for the first time disease-specific iPSC lines in 2008, mimicking human disease. This new strategy consists of screening patients for genetic mutations, isolating cell lines, returning them to iPSCs, and finally differentiating the iPSCs into one or more cell types phenotypically developing the disease.
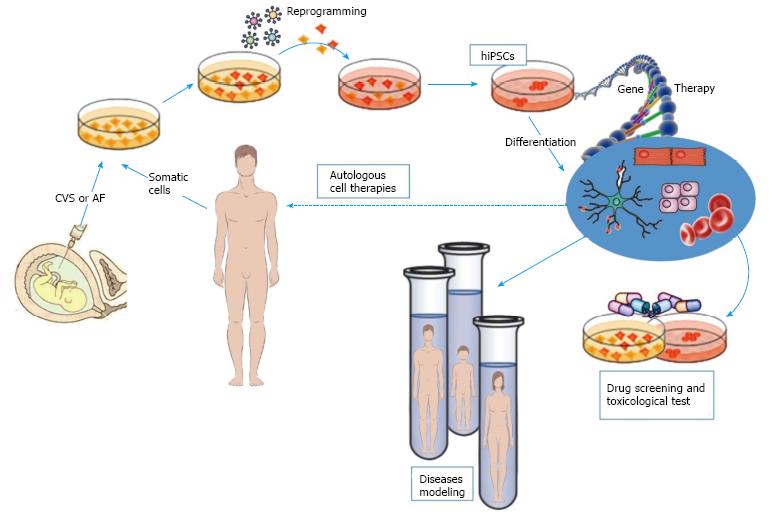
The possibility of dedifferentiating patient-specific cells back to stem cells and again to differentiate them into cells representative of the disease organ or tissue, allows one to faithfully replicate the key aspects of the disease in a ‘‘petri dish’’ and to quantify disease phenotype in different tissues (Figure 1). Each of these points is difficult to obtain; however, many disorders have been successfully recapitulated in vitro contributing to research in the field of disease modelling. Most of these studies have focused on monogenic disorders that exhibit strong phenotypes in vitro. Moreover, obtaining pluripotent cells from patients with developmental or degenerative disorders also allows for new opportunities for drug discovery[72]. When differentiated in vitro into relevant somatic lineages, hiPSCs are used both for assessing personalized pharmacological therapy and for in vivo cell therapy.
Therefore, iPSCs-disease models mimic human pathological development rather than trials utilizing conventional rodent and cell lines[73]. Human cell culture assists the research using animal models of disease. Murine models of human inherited and acquired diseases are helpful systems but human pathophysiology cannot always be faithfully reproduced. When murine and human physiology are different, disease-specific pluripotent cells able of differentiation into the several cells affected establish disease pathophysiology, allowing in vitro investigation in a human tool monitored and proving a large number of genetically-modifiable cells in a specific manner for each genetic defects.
The main advantages of iPSCs-based model systems are: (1) iPSCs can be obtained from several sources (adult somatic cells, embryonic/foetal cells, adult stem cells and cancer cells); (2) iPSCs naturally maintain the genetic background; (3) iPSCs have the ability to differentiate into any desired cell types in vitro; (4) iPSCs can self-renew and maintain their undifferentiated state and pluripotent capacity; and (5) iPSCs resume early human embryo development during differentiation in vitro.
For all these reasons, iPSCs represent an available model system for studying the pathogenetic mechanisms of various diseases, particularly in those cases where animal models do not exactly reproduce human phenotype or when disease-target cells types are not available for research.
This review provides an overview of the current state of modelling monogenic disease by using hiPSCs and the ultimate applications of these in vitro models for cell therapy. We consider and summarize some peculiar aspects such as the type of parental cell used for reprogramming, the methods currently used to induce expression of the reprogramming factors, and the type of iPSC-derived differentiated cells, relating them to the genetic basis of diseases and to their inheritance model (Tables 1,2 and 3).
| Diseases | Genetic defects | Target cells for reprogramming | Delivery methods | Disease relevant cells | Ref. |
| Huntington’s disease (OMIM #143100) | Expanded CAG repeat in HTT gene | HF | Lentiviral vectors (O, S, K, M, NANOG and LIN28) | Neurons | [79] |
| HF | Retroviral vectors (O, S, K, M) | Astrocytes | [80] | ||
| HF | Lentiviral vectors (O, S, K, M) | Neurons | [81] | ||
| HF | Retroviral vectors (O, S, K, M) | Neurons | [82] | ||
| HF | Retroviral vectors (O, S, K, M) | GABAergic neurons | [83] | ||
| HF | Retroviral vectors (O, S, K, M) | Striatal neurons | [84] | ||
| Marfan syndrome (OMIM #154700) | FBN1 mutations | HF | Retroviral vectors (O, S, K, M) | Osteogenic cells chondrogenic cells | [87] |
| Myotonic dystrophy type 1 (OMIM #160900) | Expanded CTG repeat in DMPK gene | HF | Retroviral vectors (O, S, K, M) | NS | [92] |
| HF | Retroviral vectors (O, S, K, M) | NSC | [94] | ||
| Achondroplasia (OMIM #100800) | FGFR3 mutations | HF | Episomal plasmid vectors (O, S, K, M, LIN28 and p53 shRNA) | Chondrocytes | [97] |
| Familial hypercholesterolemia (OMIM #143890) | LDLR mutations | HF | Retroviral vectors (O, S, K, M) | HLC | [99] |
| HF | Lentiviral vectors (O, S, NANOG, LIN28) | Hepatocytes | [100] | ||
| HF | Retroviral vectors (O, S, K, M) | HLC | [101] | ||
| Timothy syndrome (OMIM #601005) | CACNA1C mutations | HF | Retroviral vectors (O, S, K, M) | Cardiac myocytes | [102] |
| HF | Retroviral vectors (O, S, K, M) | Cortical neuronal precursor cells and neurons | [103] | ||
| HF | Retroviral vectors (O, S, K, M) | Cortical neuronal precursor cells and neurons | [104] |
| Diseases | Genetic defects | Target cells for reprogramming | Delivery methods | Disease relevant cells | Ref. |
| Spinal muscular atrophy (OMIM #253300) | SMN1 mutations | HF | Lentiviral vectors (O, S, NANOG, LIN28) | Neurons/astrocytes/motor neurons | [109] |
| HF | Retroviral vectors (O, S, K, M) | Motor neurons | [110] | ||
| HF | Lentiviral vectors (O, S, NANOG, LIN28) | Motor neurons | [112] | ||
| HF | Retroviral vectors (O,S, K, M) | GABAergic neurons | [113] | ||
| β-thalassaemia (OMIM #613985) | Point mutations or deletions in the β-globin (HBB) gene | HF/AF/CVS | Retroviral vectors (O, S, K, M) | Hematopoietic cells | [116] |
| AF | Lentiviral vectors (O, S, K, M) | Hematopoietic cells | [118] | ||
| HF/MSCs | Lentiviral vectors (O, S, K, M) | Erythroid cells | [119] | ||
| HF | PiggyBac transposon | Hematopoietic cells | [120] | ||
| Cystic fibrosis (OMIM #219700) | CFTR mutations | HF | Lentiviral vectors (O, S, K, M) | Mature airway epithelial cells | [126] |
| HF | Modified RNAs (RiPSC) | Mature airway epithelial cells | [127] | ||
| HF | Retroviral vectors (O, S, K, M) | Mature airway epithelial cells | [128] | ||
| HF | Retroviral vectors (O, S, K, M) | CLCs | [130] | ||
| Sickle cell disease (OMIM #603903) | HBB mutations | HF | Retroviral vectors (O, S, K, M) | Erythrocytes | [135] |
| HF | Lentiviral vectors (O, S, K, M) | None | [136] | ||
| Hutchinson-gilford progeria syndrome (OMIM #176670) | LMNA mutations | HF | Retroviral vectors (O, S, K, M) | Neural progenitors, endothelial cells, fibroblasts, VSMCs, and MSCs | [137] |
| HF | Retroviral vectors (O, S, K, M) | Vascular SMCs | [138] | ||
| HF | Retroviral vectors (O, S, K, M) | MSCs and osteogenic cells | [139] | ||
| Niemann–pick disease type C1 (OMIM #257220) | NPC1 mutations | HF | Retroviral vectors (O, S, K, M) | Neurons | [140] |
| HF | Lentiviral vectors (O, S, K, M) | Neurons | [141] | ||
| HF | SeV vectors (O, S, K, M) | HLCs and neural progenitors | [142] |
| Diseases | Genetic defects | Target cells for reprogramming | Delivery methods | Disease relevant cells | Ref. |
| Fragile X syndrome (OMIM #300624) | FMR1 silencing | HF | Retroviral vectors (O, S, K, M) | Neurons | [146] |
| HF | Retroviral vectors (O, S, K, M) | Forebrain Neurons | [147] | ||
| HF | Sendai virus | NPC | [148] | ||
| Duchenne muscular dystrophy (OMIM #310200) | Dystrophin gene mutations | HF | Retroviral vectors (O, S, K, M) | Myogenic cells | [154] |
| HF | Retroviral vectors (O, S, K, M) | CMs | [155] | ||
| HF | Retroviral vectors (O, S, K, M) | Neurons | [156] | ||
| Wiskott-aldrich syndrome (OMIM #301000) | WASP mutations | HF | Retroviral vectors/sendai virus vectors (O, S, K, M) | Megakaryocytes | [157] |
| Rett syndrome (OMIM #312750) | MeCP2/CDKL5 mutations | HF | Retroviral vectors (O, S, K, M) | Neurons | [158] |
| HF | Retroviral vectors (O, S, K, M) | NPCs/mature neurons | [159] | ||
| HF | Retroviral vectors (O, S, K, M) | NPCs/mature neurons | [160] | ||
| Hemophilia A (OMIM #306700) | Deficiency of factor VIII | Urine cells | Episomal vectors (O, S, K, SV40LT) | Hepatocytes | [161] |
Huntington’s disease (HD; OMIM #143100) is an autosomal dominant neurodegenerative disorder that shows up later in life. It is caused by an expansion of the “CAG” triplets repeated in exon 1 of the HTT gene. The protein encoded by HTT gene is expressed in many tissues and organs, especially in the brain and testis[74-77]. The normal function of HTT is still not fully identified as it differs from other known proteins.
HD is known as a neurological disease; however, peripheral HD-associated pathologies, such as cardiac defeat and skeletal muscle malfunction, have also been described. The generation of HD-iPSC lines can facilitate the match of the affected phenotypes in “dish” with clinical discoveries in sick individual of their families, highlighting the genetic basis and molecular mechanism leading to the development of the HD disease[78]. For this purpose, the HD-iPSC consortium recently promoted and defined an in vitro model of HD based on the creation of iPSCs and established by multiple lines, clones and repeat lengths, exhibiting that the clear association between the extension of the CAG repeats and the clinical pathology severity observed in HD patients could also be reproduced in vitro by HD-iPSC differentiated into neurons[79].
Juopperi et al[80] generated an in vitro system deriving patient-specific iPSCs to study HD pathogenesis. Thanks to this model, they were able to describe a specific vacuolation phenotype in iPSCs-derived astrocytes. The same characteristics were previously observed in primary lymphocytes derived from HD patients[80]. This study opens up new potential investigations using human iPSCs for model HD and for therapeutic drug screening. iPSC lines obtained from two homozygous individuals bringing 42/44 and 39/42 CAG repeats and from one heterozygote having 17/45 CAG repeats show a lysosomal activity increased when cultured in vitro and when differentiated in neurons[81]. The sizes of the CAG repeats persisted constant during the period of culture. On the other hand, another HD-iPSC line carrying 72 CAG repeats had no phenotype when cultured in vitro both in undifferentiated state and in neural precursors. However, the HD-iPSCs display manifestation of HD disorder under condition of oxidative stress[82], or proteasome inhibitors or if injected into neonatal brains for 33 wk[83]. Increased caspase activation in HD-iPSCs is also observed in iPSCs-derived neurons[84]. Thus, HD-iPSCs recapitulate the disease phenotype and represent an available tool to study HD and to develop novel therapeutics.
Marfan syndrome (MFS; OMIM #154700) is an autosomal dominant hereditary disorder of connective tissue strongly involving skeletal, ocular, and cardiovascular systems[85,86]. The mutated gene responsible for MFS is located on chromosome 15 and encodes for fibrillin-1 (FBN1).
Originally, the fibrous connective tissue disorders of MFS were attributed to structural weakness of the fibrillin-rich extracellular matrix. Increased bioavailability of TGFβ is associated with the pathological signs, indicating the resemblance to MFS-related disorders. In fact, mesenchymal cells, derived from both MFS iPSCs and ESCs differentiate spontaneously into chondrogenic cells contrary to wild type iPSC/ESC-derived mesenchymal cells that need exogenous TGFβ chondrocytes, demonstrating an alteration of TGFβ signaling in MFS cells[87]. This model is in agreement with the skeletal manifestations of MFS and increases the knowledge of molecular mechanisms underlying the pathogenesis of abnormal skeletogenesis in human diseases caused by mutations in FBN1.
The use of the iPS technology permits the reprogramming of MFS adult fibroblasts containing different FBN1 mutations, allowing the clearance of the mechanisms underlying the pathological variability, and shows the benefit of personalized therapeutic interventions.
Myotonic dystrophy type 1 (DM1; or Steinert’s disease, OMIM #160900) is the most common muscular dystrophy in adults[88]. DM segregates as an autosomal dominant pathology and is caused by the expansion of a CTG repeat located within the 3′-untranslated region of the dystrophia myotonica protein kinase (DMPK) gene on chromosome 19q13.3[89]. In the classic form, the major features include myotonia, muscle weakness and wasting, cardiomyopathy with conduction defects, insulin-resistance, frontal balding, cataracts and disease-specific serological abnormalities[90]. hiPSCs offer the possibility to study unstable repeat expansions by generating a model disease and disease-impaired cells in culture. These aspects are helpful in order to investigate unstable repeat pathologies[91].
In particular, DM1 patient-derived iPSCs could be an ideal model to study triplet-repeat instability. Du et al[92] first generated iPSCs from DM1 patient fibroblasts and detected CTG.CAG triplet repeats in each iPSC clone. Homogeneous lengths of CTG.CAG triplet repeats in each iPSC clone allows for the study of the mechanisms of repeat expansion, and offers knowledge of a general mechanism of triplet-repeat expansion in iPSCs[92].
Recently, Xia et al[93,94] reported neural stem cells (NSCs) derived from iPSCs of DM1 patients, a helpful device for the study of DM1-NSCs neuropathogenesis. Both DM1 iPSCs and iPS-derived NSCs show the presence of nuclear RNA foci, representing a molecular hallmark of disease, allowing them to be used as cellular models to understand the dynamic changes of RNA foci during the cell cycles[93,94].
Achondroplasia (ACH; OMIM #100800) is the most common skeletal dysplasia, with disproportionate short-limb dwarfism. The mutated gene encodes for fibroblast growth factor receptor 3 (FGFR3)[95,96]. The study of skeletal dysplasia, as well as many other diseases, exploits the development of iPSCs technology.
Yamashita et al[97] demonstrated that the chondrogenically differentiated ACH-hiPSCs adequately recapitulate the primary abnormalities found in FGFR3-related disease patients. These cells manifested lower proliferation and higher apoptosis when differentiated into chondrocytes[97]. Thus, hiPSCs technology is instrumental in investigating the effects of several therapeutic molecules, including statins, on ACH iPSCs-derived chondrocytes[98] (Table 1).
SMA (OMIM #253300) is an autosomal recessive neurodegenerative disorder. SMA is caused by mutation or deletion of the survival motor neuron-1 (SMN1) gene[105,106]. The clinical phenotype is typically characterized by the degeneration of α-motor neurons in the spinal cord, leading to muscle weakness, atrophy and premature death[107,108].
All SMA patients have also a highly homologous gene copy (SMN2) in different copy number. SMN2 is not able to produce sufficient levels of SMN protein, due to its defective splicing pattern. However SMN2 copy number is inversely correlated with the severity of the SMA phenotype.
One study describes human iPSCs derived from skin fibroblasts to model SMA[109]. The main characteristic is the degeneration of motor neurons caused by a loss of SMN1 protein in all cells of the body. The use of SMA-iPSCs-derived motor neurons may help to elucidate the role of SMN1 in disease initiation and progression, but also to screen new drug useful in future pharmacological therapies for SMA.
Later studies reported the establishment of five iPSC lines from type 1 SMA fibroblasts. iPSCs-derived neurons with a decreased ability to generate motor neurons and an altered neurite outgrowth. Exogenously induced expression of SMN in these iPSC lines determined a normal motor neuron differentiation and rescued the aberrant neurite outgrowth, confirming the role of the SMN defect in the disease[110]. Successively, several reports have been published using these cells to test novel compounds for efficacy prior to administration to patients, increasing the possibility of success in the treatment of this serious disorder[111-113].
β-thalassemia (β-Thal; OMIM #613985) is an inherited autosomal recessive blood disorder, caused by either point mutations or deletions of nucleotides in the β-globin gene, provoking a reduced/abnormal or absent synthesis of β-globin chains that make up hemoglobin. Affected patients have severe anemia and an shortened life span[114].
The generation of patient-specific iPSCs and the subsequent editing of the disease-causing mutations provide an ideal therapeutic solution to β-thalassemia and other haemoglobinopathies[115]. Disease-specific autologous iPSCs have been generated from somatic cells and differentiated into haematopoietic cells, both in vitro and in vivo in SCID mice[116,117]. Fan et al[118] used cultured β-thalassemia-amniotic fluid cells as target cells for an efficient reprogramming by using a single polycistronic lentiviral vector. iPSCs producing insufficient amounts of β-globin can be induced to increase β-globin product by infecting them with a viral vector carrying an exogenous copy of the β-globin gene and successively to differentiate into “restored” erythrocytes[119]. Recently, other approaches have been used for targeting the HBB gene in β-thalassemia-derived iPSCs, demonstrating how TALENs was able to mediate a higher homologous recombination efficiency than that obtained by CRISPR/Cas9[120]. The development of innovative gene editing protocols opens new promising prospects for the use of iPSCs as a target of gene therapy for monogenic diseases.
Cystic fibrosis (CF; OMIM #219700) is an autosomal recessive disorder. The primary defect is the regulation of epithelial chloride transport by a chloride channel protein, encoded by the CF transmembrane conductance regulator (CFTR) gene[121]. CF is a multisystem disorder characterized by loss of function in the CFTR in organs with secretory function[122,123]. Recurrent pulmonary infections are responsible for 80%-90% of the deaths in CF patients[124].
Fibroblasts from patients with CF can be reprogrammed to iPSCs and differentiated into lung airway epithelium[125]. From the point of view of translational medicine, patient-specific iPSC-derived airway epithelial cells open the way to personalize therapeutic interventions for the treatment of serious lung diseases. Different groups report the generation of iPSCs from CF patients and their differentiation into pulmonary cells, creating a platform for dissecting human lung disease[126-129]. Unfortunately, the development of iPSCs-based models of human lung disease is hampered by the inability to differentiate hiPSCs into lung progenitors and subsequently into mature pulmonary epithelial cell types.
Moreover mutations in the CFTR gene are also responsible for CF-associated pathologies, such as cholangiopathy, resulting in reduced intraluminal chloride secretion, increased bile viscosity and focal biliary cirrhosis. Sampaziotis et al[130] generated hiPSCs from skin fibroblasts of a CF patient and differentiated them into cholangiocytes (CLCs), showing that CF-hiPSC-derived CLCs (CF-CLCs) represent a good model CF biliary disease in vitro. In fact, their use for treatment with the experimental CF drug VX809 has demonstrated the in vitro rescue of the disease phenotype (phase 2a clinical trials)[131,132]. The use of gene targeting specific nucleases to correct CFTR gene sequences has been reported[129,133,134].
Crane et al[124] designed zinc-finger nucleases to target endogenous CFTR for editing the inherited genetic mutation in patient-derived iPSCs via homology-directed repair (HDR). When induced to differentiate in vitro, modified hiPSCs demonstrated a restored expression of the CFTR gene, recovering the expression of the mature CFTR glycoprotein and of the chloride channel functions[124] (Table 2).
Fragile X syndrome (FXS, OMIM #300624) is an inherited disorders due to CGG triplet expansion located within the 5’ untranslated region of the Fragile X mental retardation gene (FMR1). The expansion causes the epigenetic silencing and the consequent loss of the Fragile X mental retardation protein (FMRP), a cytoplasmic mRNA transport factor[143-145]. Species-specific differences in molecular and neurodevelopmental aspects of FXS require a human FXS model and hiPSCs enable disease modelling.
It has been reported that FXS hESCs and hiPSCs differ in the epigenetic state of the FMR1 gene. In fact, the FMR1 gene is unmethylated and expressed in hESCs, presenting full-mutation repeats, converting to methylated and silenced in the differentiated state[64]. In contrast, FXS hiPSCs do not return to the naive epigenetic state because the FMR1 gene remains methylated and silenced during reprogramming[64,146]. Although FX-hiPSCs do not reproduce the methylation state of the FMR1 gene, they represent a useful model for studying the role of FMR1 in neural cells. The distinction between FX-hES and FX-hiPSCs at the FMR1 locus suggests a more general epigenetic phenomenon in human pluripotent stem cells, which highlights the need for more studies to clarify the similarity and differences between ESCs and iPSCs.
Different hiPSCs cell lines have been generated from multiple patients with FXS and successively induced to differentiate into post-mitotic neurons and glia[146]. In these cells, an aberrant neuronal differentiation of FXS hiPSCs is observed, directly associated to the epigenetic modification of the FMR1 gene and to a loss of FMR protein expression, evidencing a key role for the FMR protein early in human neurodevelopment prior to synaptogenesis. iPSCs-derived neurons represent disease-associated cellular phenotypes, very useful for discovering novel therapies for FXS and other diseases sharing common pathophysiology.
Another paper reports the reprogramming of hiPSC lines from FXS fibroblasts. FXS forebrain neurons have been differentiated from these iPSCs, displaying both defective neurite initiation and extension[147]. iPSCs constitute a platform to examine potential neuronal deficits caused by FXS and develop assays for drug discovery[148].
Because the main consequence of the lack of FMRP in FXS is the synaptic defect, another group has reported a cellular model focused on neuronal cells that well represent the disease and express a transcriptional and proteomic pattern similar to that present in the neurons of the brain[148]. This aspect is important for identification of the target that modulates FMRP expression because its study in other cellular models could be erroneous. Current therapy for FXS is only at the behavioral level.
Duchenne muscular dystrophy (DMD; OMIM #310200) is the most prevalent congenital pediatric muscular dystrophy. It is an X-linked genetic degenerative myopathy and multisystem disease characterized by disease-specific serological abnormalities, dilated cardiomyopathy, cataracts, insulin-resistance, cardiac conduction defect, myotonia and muscular dystrophy, which can lead to the loss of motor function in puberty[149].
The disease is a myopathy that affects in approximately 1 in 5000 male births and is caused by mutations within the dystrophin gene (locus Xp21.2)[150,151]. The disease is characterized by a reduction in dystrophin, a protein assembles with the dystrophin glycoprotein complex (DGC), associating the cytoskeleton to the extracellular matrix in skeletal and cardiac muscles[152]. Consequences of DGC inefficiency are severe muscle wasting, contraction-induced damage, necrosis and inflammation[153].
Cell transplantation and hiPSCs offers an encouraging way for cell-based therapy, in fact myogenic cells derived from hiPSCs are an unlimited source for cell-based therapy of DMD. Goudenege et al[154] have established the usefulness of genetically corrected human multipotent cells for muscle repair. Transplantation studies in hiPSCs highlight the advantages to correct the patient own cells, avoiding the immune response against the donor myoblast or mesoangioblast. Lin et al[155] have utilized DMD iPSCs to replicate and analyze the major phenotypes of dilated cardiomyopathy (CMs) found in DMD-affected individuals, and thus to reveal the disease mechanism. Their study has identified a pathway determining increased apoptosis in DMD-CMs that can be regulated by drug therapy. Thus, these cells might represent an in vitro system for preclinical testing of future therapy[155] (Table 3).
The opportunity to derive patient-specific iPSCs in combination with the current development of gene modification protocols surely represents a good opportunity for cell therapy of several inherited genetic diseases.
Different gene editing methods have been demonstrated to modify defective genes in hiPSCs. Their choice depends on the gene correction approach and on the mutation type.
In the last few years, substantial progress has been made by using BACs (bacterial artificial chromosomes)[162-164], viral vectors[165-167], and other relatively new methods, such as zinc finger nucleases (ZFNs)[168-171], transcription activator-like effector nucleases (TALENs)[172,173] and especially the clusters of regularly interspaced palindromic repeats (CRISPR) /Cas-derived RNA-guided endonucleases[174,175].
BAC-based targeting vectors can obtain a high efficiency of homologous recombination in iPSCs of different genetic backgrounds, but this approach has the difficulty of confirming the homologous recombination event. Adenoviral and retroviral vector-mediated gene targeting appear to be effective considering the efficiency of transduction and the homologous recombination. The preparation of these viral vectors needs expertise and an enough biosafety facility. Moreover, their use is confined to insertional mutagenesis, although, self-inactivating (SIN) lentiviral vectors have now become almost safe clinical strategies[167,176-178].
The correction of disease mutations by nucleases in iPSCs has been described for different diseases. These nucleases cleave chromosomal DNA, generating DNA double strand breaks (DSBs), whose repair via endogenous mechanisms, such as homologous recombination (HR) or non-homologous end-joining (NHEJ), leads to targeted mutagenesis and chromosomal rearrangements[179-181]. Hence, before delivery to patients, gene-corrected-iPSCs are differentiated into the appropriate somatic cells, to evaluate the expression of the corrected gene and to avoid teratoma formation in patients. In particular, different research groups have created nucleases for genome engineering in hiPSCs by linking the cleavage domain of the Fokl restriction enzyme to a designed zinc finger protein (ZFP). The ZFN, which works as a dimer, is mediated by its linked ZFP domain. In particular, the ZFNs are designed to bind to a genomic sequence of sufficient length (18-36 bp)[134,137,170,173,182,183]. The ZFN method is rapid and suitable but has a poor targeting density and lack of targetable sites for genome editing in small DNA sequences.
A recent and interesting approach for engineering DNA binding specificities is based upon TALEs from Xanthomonas plant pathogens. TALEs are a different way to the creation of site-specific nucleases[184-187]. TALEs are transcription factors that specifically bind and regulate plant genes during pathogenesis[188,189]. The DNA binding domain of TALEs is composed of multiple 34 amino acid units (TALE repeats) that are organized in tandem. Their sequence is almost equivalent, but presents two highly variable amino acids that set up the base recognition specificity for each unit. Each single domain allows the specificity of binding to one of the four possible nucleotides in the TALE recognition sequence, so any desired genomic sequence can be recognized as a DNA-binding domain. An example of TALENs application to correct a defective gene in iPSCs is the correction of the Niemann-Pick type C (NPC1) mutations in iPSCs-derived hepatic and neuronal cells of patients affected by NPC disease, a lipid storage disorder causing severe neurodegeneration and liver dysfunction. This approach allows for the rescue of the phenotype, including the dysfunctional autophagic flux directly linked to loss of NPC1 protein function[190]. It is important to keep in mind that the DNA methylation and histone acetylation in inactive chromatin could influence the efficiency of genome editing via TALENs.
The latest and the greatest mechanism for gene editing is represented by the bacterial cluster and regularly interspaced short palindromic repeats/CRISPR-associate nuclease 9 (CRISPR-Cas9). CRISPR is yet another example for how scientists have learned from nature’s inventions helping them to discover new gene functions with high sensitivity and precision. This technology was created from type II CRISPR-Cas systems, by which bacteria degrade targeted nucleic acids. CRISPRs are components of the genomes of most bacteriophage-resistant Bacteria and Archaea. Cas9, a CRISPR-associated endonuclease, can be confined to specific DNA loci to induce double-strand breaks beneath the guidance of the trans-activating CRISPR RNA (tracrRNA): CRISPR RNA (crRNA) duplex. The dual tracrRNA:crRNA was additionally evolved as a single guide RNA (sgRNA) for genome engineering and consists of the 5’ end 20-nucleotide sequence disposing the DNA target site conforming to Watson-Crick base pairing and 3’ end double-stranded structure binding Cas9. The sgRNA could lead CRISPR-Cas 9 to any target DNA sequence with a protospacer-adjacent motif (PAM) by modifying the guide RNA sequences[174,175]. In a genome editing system, the right choice of delivery system is crucial for the targeted cells.
Different delivery protocols have been adopted to vehicle in vitro plasmid DNA encoding Cas9-gRNA complexes through cell membranes in cell culture including electroporation, nucleofection and lipofectamine-mediated transfection[177,191,192].
An important characteristic of the CRISPR/gRNA is its easy design and preparation. In addition, the system recognizes approximately 23 bp of the target site, which is a relatively shorter sequence than that recognized by TALENs. Recent papers have also corroborated that the RNA-mediated CRISPR/CAS9 system has a high tolerance to a few base pair mismatches towards the 5’half of the target site and a high potential for off-target risk in human cell lines.
A lot of studies have successfully modified genes in monogenic disorders using patient-specific iPSCs.
An interesting use of CRISPR-Cas9 is the correction of the HBB gene in human iPSCs generated from patients carrying a homozygous missense point mutation in the HBB gene. Using a specific guide RNA and Cas9, Huang et al[193] corrected, without off-targeting mutations, one allele of the HBB gene by homologous recombination with a donor DNA template containing the wild-type HBB DNA. Erythrocytes were then generated by differentiating hiPSCs in which normal expression of the β-globin protein (due to CRISPR-Cas9 modified HBB allele) was detected[193].
Recently, another group has demonstrated the restoration of the dystrophin protein in DMD patient-derived iPSCs by TALEN or CRISPR-Cas9. They used three correction methods (exon skipping, frameshifting and exon knock-in) to restore protein levels. After investigating the genome integrity, they differentiated the corrected iPSCs towards skeletal muscle cells and detected the expression of the full-length dystrophin protein[194].
Haemophilia A is an X-linked genetic disorder caused by mutations in the F8 gene encoding the blood coagulation factor VIII. The CRISPR-Cas9 protocol has been used by Park et al[195] to repair two large inverted regions back to the normal orientation in Haemophilia A patient-derived iPSCs. They demonstrated the expression of the F8 gene in vitro in endothelial cells after their differentiation. Targeted deep sequencing and whole genome sequencing analyses showed no off-target effects, as they chose a target sequence that differs from any other site in the human genome by three nucleotides[195].
The CRISPR protocol has been recently adopted for correcting the ∆F508 mutation in CFTR- mutated hiPSCs. An excisable selection system is used to improve the efficiency of the correction. The possibility of any genomic footprint at the target site allowed Firth et al[196] to edit a point mutation in the genome, while leaving no genomic scar in patient cells. Modified hiPSCs were then differentiated to mature airway epithelial cells in which the functional recovery of the CFTR channel protein was demonstrated. Half of the CRISPR-corrected epithelial cells stably responded to stimulation by the whole-cell patch clamp method[196].
The number of disease-specific hiPSC lines is increasing and the technology of gene editing using engineered nucleases holds considerable promise for progressing science and enhancing human health with the potential to create a variety of novel therapeutics for a range of diseases, many of which are untreatable today. Through the use of guiding RNA and nuclease activity, genes can be located and modified in a less expensive way. These technologies allow researchers to make changes to human DNA (human germline editing) in the nuclei of cells, in eggs, in sperm or in human embryos. In this way replacing or eliminating disease-causing genes could reverse disease symptoms.
However, these technologies also induce several social considerations and have not yet carried out an ethical experimental protocol useful for current medicinal practice. As a result, the National Academy of Sciences and the National Academy of Medicine are promoting an important initiative to lead decision making about research activities involving human gene editing. Many researchers report problems linked to mutations in other genes or off-target effects of the Cas9 nuclease whose consequence is largely unknown. These problems are most likely caused by the high level of functionality of the Cas9 protein, posing a dilemma in research goals requiring both powerful methods and efficacy. Several methods may minimize these problems, selecting for example unique target sequences and changing the structure of the crRNA or sgRNA, strengthening nuclease expression levels, making use of truncated sgRNAs or recombinant proteins instead of plasmids.
Nonetheless, more detailed and comprehensive studies will be needed to determine the relative merits of these technologies in different experimental procedures to open the next era of gene therapy.
Human iPSCs are central in modelling and studying monogenic diseases. They represent an affordable and applicable tool to investigate the pathogenesis and the progression of human disease and, at the same time, can be used for the in vitro screening of new therapeutic compounds.
The advantages of induced pluripotent stem cells are represented by the generation of patient-derived cells and the maintenance of a versatile differentiation potential, opening new perspectives for the development of cell-based therapy and personalized medicine.
The possibility of obtaining a wide variety of disease relevant cells would allow investigators to design an efficient test system that permits large-scale screening of drugs for the targeted treatment of specific human diseases in vitro.
Moreover, this technology offers the opportunity to develop cell therapy protocols for several serious diseases requiring the restoration of cells or organs damaged in the pathological process. The possibility of using powerful methods for genome engineering, such as ZFNs, TALENs, and the CRISPR/Cas9 system, allows the correction of mutated genes in vitro and thus safe transplantation back to the patients to assess their therapeutic efficacy.
When concerning iPSC-derived models, it is necessary to examine the inherited genetic and epigenetic variations among patients in the phenotypic analysis. Regardless their potential, iPSCs-based clinical trials cannot be used except if all obstacles discussed in this review are overcame. For example, the residual pluripotent stem cells form teratomas after cells transplantation. Any aberrations regarding tumour formation or malfunctions of epigenetic memory, as well as the genomic instability, the delivery and expression of the reprogramming factors, and the growth of cells in culture, should also be closely monitored.
However, promising results have already been obtained in pre-clinical studies in different disease models, and also in the first clinical study currently on-going in Japan[197,198]. In summary, only 8 years after their first report, iPS cell technologies provide a promising prospect for clinical uses.
We acknowledge Bonelli G for the technical assistance.
P- Reviewer: Liu SH, Wakao H, Zou ZM S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Jang J, Quan Z, Yum YJ, Song HS, Paek S, Kang HC. Induced pluripotent stem cells for modeling of pediatric neurological disorders. Biotechnol J. 2014;9:871-881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Trounson A, Shepard KA, DeWitt ND. Human disease modeling with induced pluripotent stem cells. Curr Opin Genet Dev. 2012;22:509-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Spitalieri P, Talarico RV, Botta A, Murdocca M, D’Apice MR, Orlandi A, Giardina E, Santoro M, Brancati F, Novelli G. Generation of Human Induced Pluripotent Stem Cells from Extraembryonic Tissues of Fetuses Affected by Monogenic Diseases. Cell Reprogram. 2015;17:275-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] |
| 5. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [PubMed] |
| 6. | Hussein SM, Nagy AA. Progress made in the reprogramming field: new factors, new strategies and a new outlook. Curr Opin Genet Dev. 2012;22:435-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1459] [Cited by in RCA: 1328] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 8. | Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 493] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 9. | Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1423] [Cited by in RCA: 1263] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 10. | Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 575] [Cited by in RCA: 508] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 11. | Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1410] [Cited by in RCA: 1226] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 12. | Ando M, Nishimura T, Yamazaki S, Yamaguchi T, Kawana-Tachikawa A, Hayama T, Nakauchi Y, Ando J, Ota Y, Takahashi S. A Safeguard System for Induced Pluripotent Stem Cell-Derived Rejuvenated T Cell Therapy. Stem Cell Reports. 2015;5:597-608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 13. | Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1382] [Cited by in RCA: 1541] [Article Influence: 110.1] [Reference Citation Analysis (0)] |
| 14. | Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618-630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2155] [Cited by in RCA: 1917] [Article Influence: 127.8] [Reference Citation Analysis (0)] |
| 15. | Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 938] [Cited by in RCA: 1024] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 16. | Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun. 2010;394:189-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 164] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 17. | Rosa A, Brivanlou AH. Synthetic mRNAs: powerful tools for reprogramming and differentiation of human cells. Cell Stem Cell. 2010;7:549-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1205] [Cited by in RCA: 1112] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 19. | Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA. 2010;107:9222-9227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 645] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 20. | Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 994] [Cited by in RCA: 949] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 21. | Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 739] [Cited by in RCA: 752] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 22. | Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713-720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 357] [Cited by in RCA: 325] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 23. | Moriguchi H, Chung RT, Mihara M, Sato C. Generation of human induced pluripotent stem cells from liver progenitor cells by only small molecules. Hepatology. 2010;52:1169; author reply 1169-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, Long Y, Zhao J, Pei G. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 167] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 25. | Wang Q, Xu X, Li J, Liu J, Gu H, Zhang R, Chen J, Kuang Y, Fei J, Jiang C. Lithium, an anti-psychotic drug, greatly enhances the generation of induced pluripotent stem cells. Cell Res. 2011;21:1424-1435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 26. | Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 352] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 27. | Mathew R, Jia W, Sharma A, Zhao Y, Clarke LE, Cheng X, Wang H, Salli U, Vrana KE, Robertson GP. Robust activation of the human but not mouse telomerase gene during the induction of pluripotency. FASEB J. 2010;24:2702-2715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209-1222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 670] [Cited by in RCA: 654] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 29. | Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, Mansour AA, Caspi I, Krupalnik V, Zerbib M. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 409] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 30. | Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 527] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 31. | Spinelli V, Guillot PV, De Coppi P. Induced pluripotent stem (iPS) cells from human fetal stem cells (hFSCs). Organogenesis. 2013;9:101-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 32. | Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22:1987-1997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 245] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 33. | De Coppi P, Bartsch G, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25:100-106. [PubMed] |
| 34. | Miki T, Lehmann T, Cai H, Stolz DB, Strom SC. Stem cell characteristics of amniotic epithelial cells. Stem Cells. 2005;23:1549-1559. [PubMed] |
| 35. | Prusa AR, Hengstschlager M. Amniotic fluid cells and human stem cell research: a new connection. Med Sci Monit. 2002;8:RA253-RA257. [PubMed] |
| 36. | Li CD, Zhang WY, Li HL, Jiang XX, Zhang Y, Tang P, Mao N. WITHDRAWN: Isolation and Identification of a Multilineage Potential Mesenchymal Cell from Human Placenta. Placenta. 2005;Sep 19; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 37. | Sakuragawa N, Kakinuma K, Kikuchi A, Okano H, Uchida S, Kamo I, Kobayashi M, Yokoyama Y. Human amnion mesenchyme cells express phenotypes of neuroglial progenitor cells. J Neurosci Res. 2004;78:208-214. [PubMed] |
| 38. | Tsai MS, Lee JL, Chang YJ, Hwang SM. Isolation of human multipotent mesenchymal stem cells from second-trimester amniotic fluid using a novel two-stage culture protocol. Hum Reprod. 2004;19:1450-1456. [PubMed] |
| 39. | Spitalieri P, Cortese G, Pietropolli A, Filareto A, Dolci S, Klinger FG, Giardina E, Di Cesare S, Bernardini L, Lauro D. Identification of multipotent cytotrophoblast cells from human first trimester chorionic villi. Cloning Stem Cells. 2009;11:535-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Demir R, Kosanke G, Kohnen G, Kertschanska S, Kaufmann P. Classification of human placental stem villi: review of structural and functional aspects. Microsc Res Tech. 1997;38:29-41. [PubMed] |
| 41. | James JL, Stone PR, Chamley LW. The isolation and characterization of a population of extravillous trophoblast progenitors from first trimester human placenta. Hum Reprod. 2007;22:2111-2119. [PubMed] |
| 42. | Pidoux G, Gerbaud P, Laurendeau I, Guibourdenche J, Bertin G, Vidaud M, Evain-Brion D, Frendo JL. Large variability of trophoblast gene expression within and between human normal term placentae. Placenta. 2004;25:469-473. [PubMed] |
| 43. | d’Ercole C, Shojai R, Desbriere R, Chau C, Bretelle F, Piéchon L, Boubli L. Prenatal screening: invasive diagnostic approaches. Childs Nerv Syst. 2003;19:444-447. [PubMed] |
| 44. | Sangiuolo F, Filareto A, Spitalieri P, Scaldaferri ML, Mango R, Bruscia E, Citro G, Brunetti E, De Felici M, Novelli G. In vitro restoration of functional SMN protein in human trophoblast cells affected by spinal muscular atrophy by small fragment homologous replacement. Hum Gene Ther. 2005;16:869-880. [PubMed] |
| 45. | Kaviani A, Perry TE, Dzakovic A, Jennings RW, Ziegler MM, Fauza DO. The amniotic fluid as a source of cells for fetal tissue engineering. J Pediatr Surg. 2001;36:1662-1665. [PubMed] |
| 46. | Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat Biotechnol. 2005;23:699-708. [PubMed] |
| 47. | Murphy SV, Atala A. Amniotic fluid and placental membranes: unexpected sources of highly multipotent cells. Semin Reprod Med. 2013;31:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 48. | Fauza D. Amniotic fluid and placental stem cells. Best Pract Res Clin Obstet Gynaecol. 2004;18:877-891. [PubMed] |
| 49. | Guillot PV, Gotherstrom C, Chan J, Kurata H, Fisk NM. Human first-trimester fetal MSC express pluripotency markers and grow faster and have longer telomeres than adult MSC. Stem Cells. 2007;25:646-654. [PubMed] |
| 50. | O’Donoghue K, Fisk NM. Fetal stem cells. Best Pract Res Clin Obstet Gynaecol. 2004;18:853-875. [PubMed] |
| 51. | Murphy S, Rosli S, Acharya R, Mathias L, Lim R, Wallace E, Jenkin G. Amnion epithelial cell isolation and characterization for clinical use. Curr Protoc Stem Cell Biol. 2010;Chapter 1:Unit 1E.6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 52. | Parolini O, Alviano F, Bagnara GP, Bilic G, Bühring HJ, Evangelista M, Hennerbichler S, Liu B, Magatti M, Mao N. Concise review: isolation and characterization of cells from human term placenta: outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells. 2008;26:300-311. [PubMed] |
| 53. | Miki T, Marongiu F, Ellis E, C Strom S. Isolation of amniotic epithelial stem cells. Curr Protoc Stem Cell Biol. 2007;Chapter 1:Unit 1E.3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 54. | Ilancheran S, Michalska A, Peh G, Wallace EM, Pera M, Manuelpillai U. Stem cells derived from human fetal membranes display multilineage differentiation potential. Biol Reprod. 2007;77:577-588. [PubMed] |
| 55. | Bailo M, Soncini M, Vertua E, Signoroni PB, Sanzone S, Lombardi G, Arienti D, Calamani F, Zatti D, Paul P. Engraftment potential of human amnion and chorion cells derived from term placenta. Transplantation. 2004;78:1439-1448. [PubMed] |
| 56. | Li H, Niederkorn JY, Neelam S, Mayhew E, Word RA, McCulley JP, Alizadeh H. Immunosuppressive factors secreted by human amniotic epithelial cells. Invest Ophthalmol Vis Sci. 2005;46:900-907. [PubMed] |
| 57. | Rosner M, Schipany K, Shanmugasundaram B, Lubec G, Hengstschläger M. Amniotic fluid stem cells: future perspectives. Stem Cells Int. 2012;2012:741810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 58. | Thomas MG, Stone L, Evill L, Ong S, Ziman M, Hool L. Bone marrow stromal cells as replacement cells for Parkinson’s disease: generation of an anatomical but not functional neuronal phenotype. Transl Res. 2011;157:56-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 59. | Weiss ML, Troyer DL. Stem cells in the umbilical cord. Stem Cell Rev. 2006;2:155-162. [PubMed] |
| 60. | Amendola D, Nardella M, Guglielmi L, Cerquetti L, Carico E, Alesi V, Porru M, Leonetti C, Bearzi C, Rizzi R. Human placenta-derived neurospheres are susceptible to transformation after extensive in vitro expansion. Stem Cell Res Ther. 2014;5:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 61. | Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285-290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1842] [Cited by in RCA: 1685] [Article Influence: 112.3] [Reference Citation Analysis (0)] |
| 62. | Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 655] [Cited by in RCA: 608] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 63. | Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 333] [Cited by in RCA: 294] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 64. | Kang X, Yu Q, Huang Y, Song B, Chen Y, Gao X, He W, Sun X, Fan Y. Effects of Integrating and Non-Integrating Reprogramming Methods on Copy Number Variation and Genomic Stability of Human Induced Pluripotent Stem Cells. PLoS One. 2015;10:e0131128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 65. | Hyun I, Li W, Ding S. Scientific and ethical reasons why iPS cell research must proceed with human embryonic stem cell research. Stanford J Law Sci Policy. 2010;3:43-48. |
| 66. | Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 377] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 67. | Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, Alt FW, Murphy GJ, Kotton DN, Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 187] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 68. | Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771-775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 976] [Cited by in RCA: 855] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 69. | Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964-977. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1202] [Cited by in RCA: 1071] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 70. | Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 457] [Cited by in RCA: 332] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 71. | Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877-886. [PubMed] |
| 72. | Cai S, Chan YS, Shum DK. Induced pluripotent stem cells and neurological disease models. Sheng Li Xue Bao. 2014;66:55-66. [PubMed] |
| 73. | Kim C. Disease modeling and cell based therapy with iPSC: future therapeutic option with fast and safe application. Blood Res. 2014;49:7-14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 74. | The Huntington Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971-983. [PubMed] |
| 75. | Li SH, Schilling G, Young WS, Li XJ, Margolis RL, Stine OC, Wagster MV, Abbott MH, Franz ML, Ranen NG. Huntington’s disease gene (IT15) is widely expressed in human and rat tissues. Neuron. 1993;11:985-993. [PubMed] |
| 76. | Strong TV, Tagle DA, Valdes JM, Elmer LW, Boehm K, Swaroop M, Kaatz KW, Collins FS, Albin RL. Widespread expression of the human and rat Huntington’s disease gene in brain and nonneural tissues. Nat Genet. 1993;5:259-265. [PubMed] |
| 77. | Sharp AH, Loev SJ, Schilling G, Li SH, Li XJ, Bao J, Wagster MV, Kotzuk JA, Steiner JP, Lo A. Widespread expression of Huntington’s disease gene (IT15) protein product. Neuron. 1995;14:1065-1074. [PubMed] |
| 78. | Jacquet L, Neueder A, Földes G, Karagiannis P, Hobbs C, Jolinon N, Mioulane M, Sakai T, Harding SE, Ilic D. Three Huntington’s Disease Specific Mutation-Carrying Human Embryonic Stem Cell Lines Have Stable Number of CAG Repeats upon In Vitro Differentiation into Cardiomyocytes. PLoS One. 2015;10:e0126860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 79. | The HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 395] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 80. | Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, Ming GL, Song H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 2012;5:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 183] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 81. | Camnasio S, Delli Carri A, Lombardo A, Grad I, Mariotti C, Castucci A, Rozell B, Lo Riso P, Castiglioni V, Zuccato C. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 2012;46:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 82. | Chae JI, Kim DW, Lee N, Jeon YJ, Jeon I, Kwon J, Kim J, Soh Y, Lee DS, Seo KS. Quantitative proteomic analysis of induced pluripotent stem cells derived from a human Huntington’s disease patient. Biochem J. 2012;446:359-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 83. | Jeon I, Lee N, Li JY, Park IH, Park KS, Moon J, Shim SH, Choi C, Chang DJ, Kwon J. Neuronal properties, in vivo effects, and pathology of a Huntington’s disease patient-derived induced pluripotent stem cells. Stem Cells. 2012;30:2054-2062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 84. | Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2010;2:RRN1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 193] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 85. | Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337-339. [PubMed] |
| 86. | Pereira L, Andrikopoulos K, Tian J, Lee SY, Keene DR, Ono R, Reinhardt DP, Sakai LY, Biery NJ, Bunton T. Targetting of the gene encoding fibrillin-1 recapitulates the vascular aspect of Marfan syndrome. Nat Genet. 1997;17:218-222. [PubMed] |
| 87. | Quarto N, Leonard B, Li S, Marchand M, Anderson E, Behr B, Francke U, Reijo-Pera R, Chiao E, Longaker MT. Skeletogenic phenotype of human Marfan embryonic stem cells faithfully phenocopied by patient-specific induced-pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:215-220. [PubMed] |
| 88. | Harper PS. Myotonic Dystrophy. 3rd ed. London: WB Saunders 2001; 37. |
| 89. | Fu YH, Pizzuti A, Fenwick RG, King J, Rajnarayan S, Dunne PW, Dubel J, Nasser GA, Ashizawa T, de Jong P. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256-1258. [PubMed] |
| 90. | Moxley RT, Corbett AJ, Minaker KL, Rowe JW. Whole body insulin resistance in myotonic dystrophy. Ann Neurol. 1984;15:157-162. [PubMed] |
| 91. | Yanovsky-Dagan S, Mor-Shaked H, Eiges R. Modeling diseases of noncoding unstable repeat expansions using mutant pluripotent stem cells. World J Stem Cells. 2015;7:823-838. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 92. | Du J, Campau E, Soragni E, Jespersen C, Gottesfeld JM. Length-dependent CTG·CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells. Hum Mol Genet. 2013;22:5276-5287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 93. | Xia G, Santostefano KE, Goodwin M, Liu J, Subramony SH, Swanson MS, Terada N, Ashizawa T. Generation of neural cells from DM1 induced pluripotent stem cells as cellular model for the study of central nervous system neuropathogenesis. Cell Reprogram. 2013;15:166-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 94. | Xia G, Ashizawa T. Dynamic changes of nuclear RNA foci in proliferating DM1 cells. Histochem Cell Biol. 2015;143:557-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 95. | Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252-254. [PubMed] |
| 96. | Shiang R, Thompson LM, Zhu YZ, Church DM, Fielder TJ, Bocian M, Winokur ST, Wasmuth JJ. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335-342. [PubMed] |
| 97. | Yamashita A, Morioka M, Kishi H, Kimura T, Yahara Y, Okada M, Fujita K, Sawai H, Ikegawa S, Tsumaki N. Statin treatment rescues FGFR3 skeletal dysplasia phenotypes. Nature. 2014;513:507-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 156] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 98. | Karagiannis P, Tsumaki N. Cell reprogramming for skeletal dysplasia drug repositioning. Cell Cycle. 2014;13:3791-3792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 99. | Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127-3136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 498] [Cited by in RCA: 441] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 100. | Cayo MA, Cai J, DeLaForest A, Noto FK, Nagaoka M, Clark BS, Collery RF, Si-Tayeb K, Duncan SA. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012;56:2163-2171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 101. | Fattahi F, Asgari S, Pournasr B, Seifinejad A, Totonchi M, Taei A, Aghdami N, Salekdeh GH, Baharvand H. Disease-corrected hepatocyte-like cells from familial hypercholesterolemia-induced pluripotent stem cells. Mol Biotechnol. 2013;54:863-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 102. | Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 927] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 103. | Paşca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Paşca AM, Cord B, Palmer TD, Chikahisa S, Nishino S. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657-1662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 465] [Cited by in RCA: 443] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 104. | Tian Y, Voineagu I, Paşca SP, Won H, Chandran V, Horvath S, Dolmetsch RE, Geschwind DH. Alteration in basal and depolarization induced transcriptional network in iPSC derived neurons from Timothy syndrome. Genome Med. 2014;6:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 105. | Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, Rudnik-Schöneborn S, Wienker T, Zerres K. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet. 1999;64:1340-1356. [PubMed] |
| 106. | Munsat TL, Davies KE. International SMA consortium meeting. (26-28 June 1992, Bonn, Germany). Neuromuscul Disord. 1992;2:423-428. [PubMed] |
| 107. | Iannaccone ST, Smith SA, Simard LR. Spinal muscular atrophy. Curr Neurol Neurosci Rep. 2004;4:74-80. [PubMed] |
| 108. | Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat. 2000;15:228-237. [PubMed] |
| 109. | Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1169] [Cited by in RCA: 1039] [Article Influence: 64.9] [Reference Citation Analysis (0)] |
| 110. | Chang T, Zheng W, Tsark W, Bates S, Huang H, Lin RJ, Yee JK. Brief report: phenotypic rescue of induced pluripotent stem cell-derived motoneurons of a spinal muscular atrophy patient. Stem Cells. 2011;29:2090-2093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 111. | Ebert AD, Svendsen CN. Human stem cells and drug screening: opportunities and challenges. Nat Rev Drug Discov. 2010;9:367-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 123] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 112. | Sareen D, Ebert AD, Heins BM, McGivern JV, Ornelas L, Svendsen CN. Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS One. 2012;7:e39113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 115] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 113. | Garbes L, Heesen L, Hölker I, Bauer T, Schreml J, Zimmermann K, Thoenes M, Walter M, Dimos J, Peitz M. VPA response in SMA is suppressed by the fatty acid translocase CD36. Hum Mol Genet. 2013;22:398-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 114. | Cao A, Galanello R. Beta-thalassemia. Genet Med. 2010;12:61-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 506] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 115. | Finotti A, Breda L, Lederer CW, Bianchi N, Zuccato C, Kleanthous M, Rivella S, Gambari R. Recent trends in the gene therapy of β-thalassemia. J Blood Med. 2015;6:69-85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 116. | Ye L, Chang JC, Lin C, Sun X, Yu J, Kan YW. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci USA. 2009;106:9826-9830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 149] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 117. | Wang Y, Jiang Y, Liu S, Sun X, Gao S. Generation of induced pluripotent stem cells from human beta-thalassemia fibroblast cells. Cell Res. 2009;19:1120-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 118. | Fan Y, Luo Y, Chen X, Li Q, Sun X. Generation of human β-thalassemia induced pluripotent stem cells from amniotic fluid cells using a single excisable lentiviral stem cell cassette. J Reprod Dev. 2012;58:404-409. [PubMed] |
| 119. | Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol. 2011;29:73-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 231] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 120. | Xu P, Tong Y, Liu XZ, Wang TT, Cheng L, Wang BY, Lv X, Huang Y, Liu DP. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2-654 (C & gt; T) mutation in β-thalassemia-derived iPSCs. Sci Rep. 2015;5:12065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 121. | Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073-1080. [PubMed] |
| 122. | Grody WW. Cystic fibrosis: molecular diagnosis, population screening, and public policy. Arch Pathol Lab Med. 1999;123:1041-1046. [PubMed] |
| 123. | Sangiuolo F, D’Apice MR, Bruscia E, Lucidi V, Novelli G. Towards the pharmacogenomics of cystic fibrosis. Pharmacogenomics. 2002;3:75-87. [PubMed] |
| 124. | Crane AM, Kramer P, Bui JH, Chung WJ, Li XS, Gonzalez-Garay ML, Hawkins F, Liao W, Mora D, Choi S. Targeted correction and restored function of the CFTR gene in cystic fibrosis induced pluripotent stem cells. Stem Cell Reports. 2015;4:569-577. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 157] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 125. | Moodley Y, Thompson P, Warburton D. Stem cells: a recapitulation of development. Respirology. 2013;18:1167-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 126. | Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, Ying L, Sommer AG, Jean JM, Smith BW. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728-1740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 325] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 127. | Mou H, Zhao R, Sherwood R, Ahfeldt T, Lapey A, Wain J, Sicilian L, Izvolsky K, Musunuru K, Cowan C. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell. 2012;10:385-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 242] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 128. | Wong AP, Bear CE, Chin S, Pasceri P, Thompson TO, Huan LJ, Ratjen F, Ellis J, Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol. 2012;30:876-882. [PubMed] |
| 129. | Sargent RG, Suzuki S, Gruenert DC. Nuclease-mediated double-strand break (DSB) enhancement of small fragment homologous recombination (SFHR) gene modification in human-induced pluripotent stem cells (hiPSCs). Methods Mol Biol. 2014;1114:279-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 130. | Sampaziotis F, Cardoso de Brito M, Madrigal P, Bertero A, Saeb-Parsy K, Soares FA, Schrumpf E, Melum E, Karlsen TH, Bradley JA. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015;33:845-852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 293] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 131. | Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, Decker CJ, Miller M, McCartney J, Olson ER. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc Natl Acad Sci USA. 2011;108:18843-18848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 788] [Cited by in RCA: 854] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 132. | Clancy JP, Rowe SM, Accurso FJ, Aitken ML, Amin RS, Ashlock MA, Ballmann M, Boyle MP, Bronsveld I, Campbell PW. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax. 2012;67:12-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 402] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 133. | Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 609] [Cited by in RCA: 520] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 134. | Lee CM, Flynn R, Hollywood JA, Scallan MF, Harrison PT. Correction of the ΔF508 Mutation in the Cystic Fibrosis Transmembrane Conductance Regulator Gene by Zinc-Finger Nuclease Homology-Directed Repair. Biores Open Access. 2012;1:99-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 135. | Zou J, Mali P, Huang X, Dowey SN, Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599-4608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 240] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 136. | Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, Goodwin MJ, Hawkins JS, Ramirez CL, Batista LF. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 246] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 137. | Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse HF. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8:31-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 138. | Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, Panopoulos AD, Suzuki K, Kurian L, Walsh C. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472:221-225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 486] [Cited by in RCA: 442] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 139. | Blondel S, Jaskowiak AL, Egesipe AL, Le Corf A, Navarro C, Cordette V, Martinat C, Laabi Y, Djabali K, de Sandre-Giovannoli A. Induced pluripotent stem cells reveal functional differences between drugs currently investigated in patients with hutchinson-gilford progeria syndrome. Stem Cells Transl Med. 2014;3:510-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 140. | Lee H, Lee JK, Park MH, Hong YR, Marti HH, Kim H, Okada Y, Otsu M, Seo EJ, Park JH. Pathological roles of the VEGF/SphK pathway in Niemann-Pick type C neurons. Nat Commun. 2014;5:5514. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 141. | Efthymiou AG, Steiner J, Pavan WJ, Wincovitch S, Larson DM, Porter FD, Rao MS, Malik N. Rescue of an in vitro neuron phenotype identified in Niemann-Pick disease, type C1 induced pluripotent stem cell-derived neurons by modulating the WNT pathway and calcium signaling. Stem Cells Transl Med. 2015;4:230-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 142. | Soga M, Ishitsuka Y, Hamasaki M, Yoneda K, Furuya H, Matsuo M, Ihn H, Fusaki N, Nakamura K, Nakagata N. HPGCD outperforms HPBCD as a potential treatment for Niemann-Pick disease type C during disease modeling with iPS cells. Stem Cells. 2015;33:1075-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 143. | Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817-822. [PubMed] |
| 144. | Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905-914. [PubMed] |
| 145. | Hagerman RJ, Hagerman PJ. Fragile X Syndrome: Diagnosis, Treatment, and Research. The John Hopkins University Press, Baltimore. Heredity. 2003;90:419-420. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 146. | Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 237] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 147. | Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang SC, Abbeduto L, Bhattacharyya A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 148. | Kaufmann M, Schuffenhauer A, Fruh I, Klein J, Thiemeyer A, Rigo P, Gomez-Mancilla B, Heidinger-Millot V, Bouwmeester T, Schopfer U. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen. 2015;20:1101-1111. [PubMed] |
| 149. | Mercuri E, Muntoni F. Muscular dystrophy: new challenges and review of the current clinical trials. Curr Opin Pediatr. 2013;25:701-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 150. | Emery AE. Muscular dystrophy into the new millennium. Neuromuscul Disord. 2002;12:343-349. [PubMed] |
| 151. | Mendell JR, Rodino-Klapac L, Sahenk Z, Malik V, Kaspar BK, Walker CM, Clark KR. Gene therapy for muscular dystrophy: lessons learned and path forward. Neurosci Lett. 2012;527:90-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 152. | Braun R, Wang Z, Mack DL, Childers MK. Gene therapy for inherited muscle diseases: where genetics meets rehabilitation medicine. Am J Phys Med Rehabil. 2014;93:S97-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 153. | Lapidos KA, Kakkar R, McNally EM. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ Res. 2004;94:1023-1031. [PubMed] |
| 154. | Goudenege S, Lebel C, Huot NB, Dufour C, Fujii I, Gekas J, Rousseau J, Tremblay JP. Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol Ther. 2012;20:2153-2167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 127] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 155. | Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S, Bett GC, Rasmusson RL, Denning C, Yang L. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy. Dis Model Mech. 2015;8:457-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 156. | Luo Y, Fan Y, Chen X, Yue L, Yu B, Li Q, Chen Y, Sun X. Modeling induced pluripotent stem cells from fibroblasts of Duchenne muscular dystrophy patients. Int J Neurosci. 2014;124:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 157. | Ingrungruanglert P, Amarinthnukrowh P, Rungsiwiwut R, Maneesri-le Grand S, Sosothikul D, Suphapeetiporn K, Israsena N, Shotelersuk V. Wiskott-Aldrich syndrome iPS cells produce megakaryocytes with defects in cytoskeletal rearrangement and proplatelet formation. Thromb Haemost. 2015;113:792-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 158. | Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527-539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1082] [Cited by in RCA: 998] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 159. | Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci USA. 2011;108:14169-14174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 164] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 160. | Livide G, Patriarchi T, Amenduni M, Amabile S, Yasui D, Calcagno E, Lo Rizzo C, De Falco G, Ulivieri C, Ariani F. GluD1 is a common altered player in neuronal differentiation from both MECP2-mutated and CDKL5-mutated iPS cells. Eur J Hum Genet. 2015;23:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 161. | Jia B, Chen S, Zhao Z, Liu P, Cai J, Qin D, Du J, Wu C, Chen Q, Cai X. Modeling of hemophilia A using patient-specific induced pluripotent stem cells derived from urine cells. Life Sci. 2014;108:22-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 162. | Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319-321. [PubMed] |
| 163. | Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat Biotechnol. 2007;25:1477-1482. [PubMed] |
| 164. | Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 165. | Suzuki K, Mitsui K, Aizawa E, Hasegawa K, Kawase E, Yamagishi T, Shimizu Y, Suemori H, Nakatsuji N, Mitani K. Highly efficient transient gene expression and gene targeting in primate embryonic stem cells with helper-dependent adenoviral vectors. Proc Natl Acad Sci USA. 2008;105:13781-13786. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 86] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 166. | Aizawa E, Hirabayashi Y, Iwanaga Y, Suzuki K, Sakurai K, Shimoji M, Aiba K, Wada T, Tooi N, Kawase E. Efficient and accurate homologous recombination in hESCs and hiPSCs using helper-dependent adenoviral vectors. Mol Ther. 2012;20:424-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 167. | Aiuti A, Roncarolo MG. Ten years of gene therapy for primary immune deficiencies. Hematology Am Soc Hematol Educ Program. 2009;682-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 168. | Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. [PubMed] |
| 169. | Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. [PubMed] |
| 170. | Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646-651. [PubMed] |
| 171. | Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279-1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 353] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 172. | Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731-734. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 996] [Cited by in RCA: 958] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 173. | Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 387] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 174. | Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10336] [Cited by in RCA: 11145] [Article Influence: 928.8] [Reference Citation Analysis (0)] |
| 175. | Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6555] [Cited by in RCA: 6996] [Article Influence: 583.0] [Reference Citation Analysis (0)] |
| 176. | Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 916] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 177. | Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 814] [Article Influence: 67.8] [Reference Citation Analysis (0)] |
| 178. | Sauer AV, Di Lorenzo B, Carriglio N, Aiuti A. Progress in gene therapy for primary immunodeficiencies using lentiviral vectors. Curr Opin Allergy Clin Immunol. 2014;14:527-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 179. | Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 215] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 180. | Kim S, Lee HJ, Kim E, Kim JS. Analysis of targeted chromosomal deletions induced by zinc finger nucleases. Cold Spring Harb Protoc. 2010;2010:pdb.prot5477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 181. | Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22:539-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 182. | Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169-1175. [PubMed] |
| 183. | Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93:1156-1160. [PubMed] |
| 184. | Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1811] [Article Influence: 113.2] [Reference Citation Analysis (0)] |
| 185. | Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1451] [Cited by in RCA: 1370] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 186. | Osborn MJ, Starker CG, McElroy AN, Webber BR, Riddle MJ, Xia L, DeFeo AP, Gabriel R, Schmidt M, von Kalle C. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21:1151-1159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 206] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 187. | Park CY, Kim J, Kweon J, Son JS, Lee JS, Yoo JE, Cho SR, Kim JH, Kim JS, Kim DW. Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc Natl Acad Sci USA. 2014;111:9253-9258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 188. | Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645-648. [PubMed] |
| 189. | Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648-651. [PubMed] |
| 190. | Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, Gao Q, Mitalipova M, Jaenisch R. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Reports. 2014;2:866-880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 164] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 191. | Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 402] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 192. | Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2216] [Cited by in RCA: 2422] [Article Influence: 201.8] [Reference Citation Analysis (0)] |
| 193. | Huang X, Wang Y, Yan W, Smith C, Ye Z, Wang J, Gao Y, Mendelsohn L, Cheng L. Production of Gene-Corrected Adult Beta Globin Protein in Human Erythrocytes Differentiated from Patient iPSCs After Genome Editing of the Sickle Point Mutation. Stem Cells. 2015;33:1470-1479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 194. | Li HL, Fujimoto N, Sasakawa N, Shirai S, Ohkame T, Sakuma T, Tanaka M, Amano N, Watanabe A, Sakurai H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 390] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 195. | Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS. Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell. 2015;17:213-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 225] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 196. | Firth AL, Menon T, Parker GS, Qualls SJ, Lewis BM, Ke E, Dargitz CT, Wright R, Khanna A, Gage FH. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep. 2015;12:1385-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 241] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 197. | Cyranoski D. Stem cells cruise to clinic. Nature. 2013;494:413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 198. | Song P, Inagaki Y, Sugawara Y, Kokudo N. Perspectives on human clinical trials of therapies using iPS cells in Japan: reaching the forefront of stem-cell therapies. Biosci Trends. 2013;7:157-158. [PubMed] |