Published online Sep 26, 2015. doi: 10.4252/wjsc.v7.i8.1078
Peer-review started: January 24, 2015
First decision: March 6, 2015
Revised: June 27, 2015
Accepted: August 20, 2015
Article in press: August 21, 2015
Published online: September 26, 2015
Processing time: 244 Days and 15.5 Hours
A small subset of cancer cells that act as tumor initiating cells or cancer stem cells (CSCs) maintain self-renewal and growth promoting capabilities of cancer and are responsible for drug/treatment resistance, tumor recurrence and metastasis. Due to their potential clinical importance, many researchers have put their efforts over decades to unravel the molecular mechanisms that regulate CSCs functions. MicroRNAs (miRNAs) which are 21-23 nucleotide long, endogenous non-coding RNAs, regulate gene expression through gene silencing at post-transcriptional level by binding to the 3’-untranslated regions or the open reading frames of target genes, thereby result in target mRNA degradation or its translational repression and serve important role in several cellular, physiological and developmental processes. Aberrant miRNAs expression and their implication in CSCs regulation by controlling asymmetric cell division, drug/treatment resistance and metastasis make miRNAs a tool of great therapeutic potential against cancer. Recent advancements on the biological complexities of CSCs, modulation in CSCs properties by miRNA network and development of miRNA based treatment strategies specifically targeting the CSCs as an attractive therapeutic targets for clinical application are being critically analysed.
Core tip: Cancer stem cells (CSCs) which are believed to be the prerequisite for metastasis and tumor recurrence, are endowed with ability to undergo symmetric cell division, capacity for self-renewability, long term proliferation and resistance to anti-neoplasic therapeutic drugs. Regulatory characteristics of microRNAs (miRNAs) which are the clusters of non-coding RNA molecules, include widespread changes in gene expression through gene silencing at post-transcriptional level and are dysregulated in human cancer. Over the past two decades, miRNAs have gained widespread attention due to their involvement in acquisition of stem cell-like properties, regulation and reprogramming by cancer cells during cancer progression. Many studies are coming up which document miRNAs as novel therapeutic tool in targeting CSCs functions, sensitizing them to apoptotic effects of anti-cancer drugs, and reducing tumor burden with no relapse in current clinical settings.
- Citation: Garg M. Emerging role of microRNAs in cancer stem cells: Implications in cancer therapy. World J Stem Cells 2015; 7(8): 1078-1089
- URL: https://www.wjgnet.com/1948-0210/full/v7/i8/1078.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i8.1078
Despite high response rates to initial treatments including chemotherapy, radiotherapy or sometimes even after combinational chemotherapies, cancer remains the most lethal disease. Recent technological advancements and animal studies have validated the presence of samll subset of cancer cells among the heterogenous population of tumor cells and are known as cancer stem cells (CSCs) or tumor initiating cells. They have different biological properties, increased tumorgenic potential and are supposed to serve important role in drug resistance, tumor development and its recurrence. Anti-cancer therapeies are being employed on the basis of their ability to shrink tumor cells, but CSCs survive and proliferate owning to an increased intrinsic (de novo) or acquired resistance to anti-cancer drugs. This makes the clinical therapy ineffective, leads to tumor recurrence and results in high mortality rates in cancer patients. Decreased experssion of energy dependent transporters to retain drugs inside the cancer cells and increased efficacy of drug response by sensitizing these subset of cancer cells to drug induced apoptosis could be key mechanisms towards improved treatment of cancer. Therefore, there is an urgent need to understand the molecular mechanisms controlling CSCs population and functions to develop effective therapies to eradicate recurrence which is a real threat to complete cancer cure. Emerging evidences have suggested the critial involvement of protein-coding genes as well as non-coding RNAs (ncRNAs) including microRNAs (miRNAs) in various types of cancers and serve important role in CSCs functions. Recent findings on the miRNAs, responsible for the maintenance and regulation of CSCs properties like tumorigenicity and drug resistance, are briefly summarized in the later sections. Gaining further insight into the potential and therapeutic approaches by targeting CSCs using miRNAs as a tool would be helpful for deciphering and designing future strategies for human cancer treatment with better outcome by preventing metastatic progression and tumor recurrence.
CSCs with tumor intiating potential constitute only a fraction of neoplastic cells and were first reported by Lapidot et al[1]. These cells are endowed with the capacity of long term proliferation, ability to undergo symmetric cell division and seed new tumors by virtue of their intrinsic self-protection and self-renewal capabilities. Capacity for self-renewal, propagation of differentiated progenitors and the expression of specific stem cell genes are some of the biological properties that are shared by both normal and CSCs. Accumulation of genetic and epigenetic alterations degregulate the basic stem cell biology and distinguish CSCs from the normal stem cells in their chemoresistance, enhanced tumorigenic and metastatic activities[2]. Recent experimental studies have identified the activation of oncogenes and inactivation of tumor suppressor genes to be responsible for self-renewal ability and pluripotency of CSCs[3]. Concept of heterogeneity among CSCs as stationary and migrating CSCs came into existence with the discovery of process of epitheilal-to-mesencymal transition (EMT). EMT is a crucial event which allows the stationary CSCs at the primary site to loose their epithelial character, morphological transition, and loss of cell-cell adherence. This transition imparts high motility to the stationary CSCs, which is a necessary requirement by migrating CSCs for local invasion and metastatic dissemination. The concept of migrating CSCs and especially EMT as a widespread mechanism of stem cell generation validates the dynamic nature of CSCs and strongly supports the current CSC theory[4,5].
Over the past few years, number of studies have reported the isolation and charecterization of CSCs for hematological malignancies including acute lymphoblastic leukemia, chronic myeloid leukemia, acute myelogenous leukemia, multiple myeloma and also for solid tumors of breast, colon, stomach, brain, lung, liver, skin, prostate, testis, ovary and pancreas[6]. The subpopulation of CSCs are identified, isolated and charecterized on the basis of capacity to form sphere-clusters with a high clonogenic efficiency, a stronger self-renewal ability and much higher levels of cell cycle markers which account for G1-S/G2-M phases progression, side population profile and stemness gene expression.
Stem cell maintenance, survival, self-renewal and differentiation of CSCs are known to be regulated by several signaling pathways and molecules. Notch, Wnt/catenin, sonic hedgehog, bone morphogenetic protein, receptor tyrosine kinase and TGF-β signaling pathways are studied to be aberrantly active in CSCs and regulate self-renewal activity, thereby play significant role in tumor initiation and development in various malignancies including breast, colorectal, prostate, pancreatic, glioma, leukemia and colon cancers[7-11].
CSCs are not only the source of initial tumor formation, postoperative recurrence and metastasis but also contribute to chemo/radioresistance. High invasiveness and frequent recurrence due to the presence of inefficient DNA repair mechanisms, which render CSCs highly resistant to chemotherapeutic and ionizing radiations, are the main reasons for treatment failure and recurrent disease[12,13]. Targeting the pathways and mechanisms behind the development of chemo/radioresistance, modifying the niche of cancer cells, regulating the cellular response to damage by modulating apoptosis, cell cycle proliferation, DNA repair, invasion and differentiation functions might help in eliminating CSCs subpopulation and developing therapeutic strategies for complete cancer treatment with no relapse.
MiRNAs have gained widespread attention in modulating the CSCs functions through affecting the expression level of target genes and proteins that are involved in signaling pathways including cell proliferation and cell death. Recent advancements on the role of miRNAs in regulation and maintenance of tumor cells with stem cell properties are discussed in later sections.
The most important advancement over the past recent years is the discovery of non-coding RNA families which are actively transcribed from the genome of many organisms and generate widespread changes by regulating vital biological processes. Among them, miRNAs are the novel small non-protein coding, evolutionarily conserved, 21-23 nucleotide long single-stranded RNA molecules. Many important diverse functions of miRNAs include the regulation of cellular differentiation, proliferation and apoptosis by regulating the stability or translational efficiency of target messenger RNAs which are expressed in tissue-specific or developmental stage-specific manner.
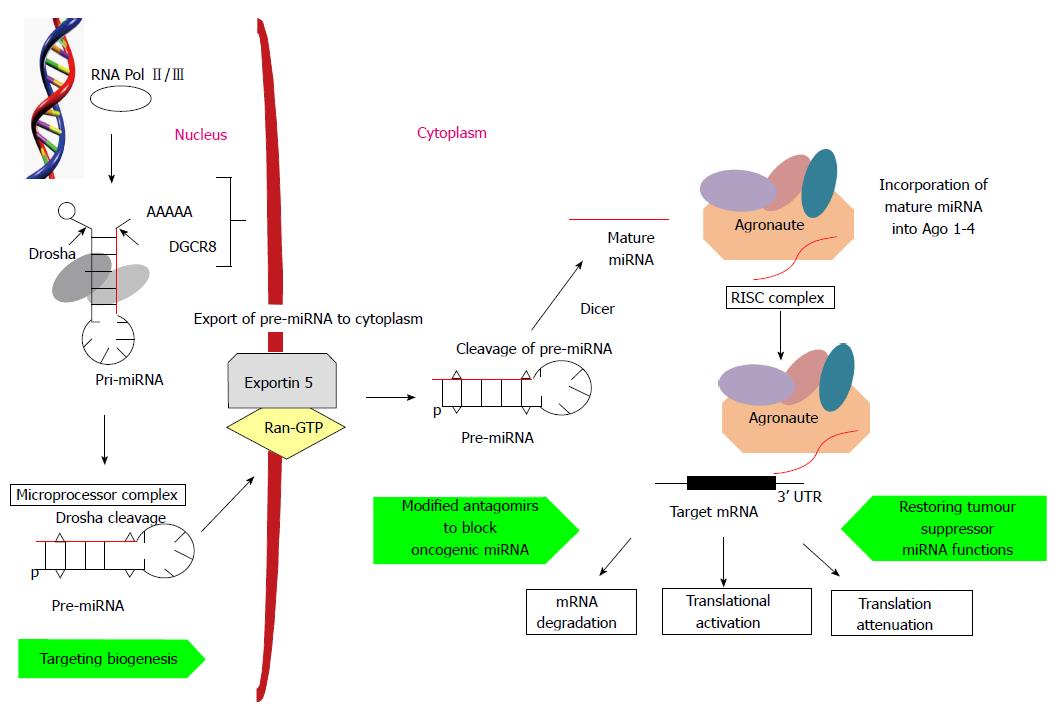
MiRNA-mRNA target recognition due to sequence complementarity of the lin-4 miRNAs to multiple conserved sites within the lin-14 3’ untranslated region (3’UTR) leads to the discovery of miRNAs way back in 1993 during genome study in Caenorhabditis elegans development[13,14]. MiRNAs are randomly located in mammalian genome and the complex process of biogenesis begins in nucleus with the transcription of polycistronic primary-miRNA (pri-miRNA) transcript by RNA polymerases II and III[15,16]. Hundreds or thousands of nucleotides long pri-miRNA transcripts with one or many stem loops, 5’ capping and 3’ polyadenylation, undergo further cleavage by nuclear microprocessor complex containing enzymes Drosha, RNA III endonuclease and double stranded RNA binding protein DiGeorge syndrome critical region 8 (DGCR8)[17,18]. The resulting 70 nucleotide long pre-miRNAs are exported to cytoplasm by exportin 5 and Ran-GTP[19]. Another enzyme, RNA III endonuclease known as Dicer, cleaves hairpin-like pre-miRNA into two complementary fragments and one of which is mature miRNA[20]. Mature miRNA strand is then incorporated into the members of Agronaute protein family, which constitutes the catalytic portion of the multi-protein RNA-induced silencing complex (RISC). MiRNAs then direct RISC to target mRNAs which share sequence complementation in seed region that consists of nucleotides at position 2-8 of 5’ end of mature miRNA. Complementation between the seed sequence and 3’UTR of target mRNA results in mRNA transcript degradation while imperfect complementation results translational repression[21]. Nuclear pri-miRNA and cytoplasmic pre-miRNA cleavage steps and mature miRNA-target mRNA recognition sites are the potential therapeutic points against cancer by regulating miRNA processing, its biogenesis and miRNA functions (Figure 1).
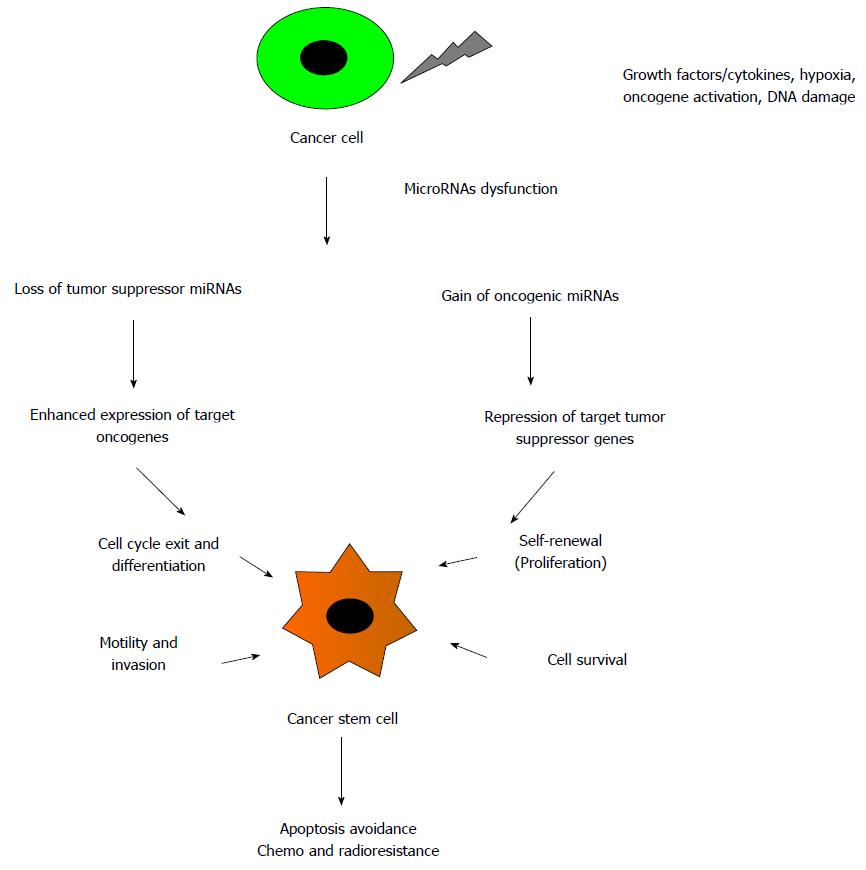
Statsitical methods and profiling studies validate the presence of miRNAs in close proximity to chromosomal breakpoints, cancer associated genomic regions and fragile sites where mutations/deletions can occur. Loss of tumor suppressive miRNAs and increased expression of oncogenic miRNAs enhance the expression of target oncogenes and represse the target tumor suppressor genes respectively. Deregulation of novel miRNAs expression leads to induction of anti-apoptotoic activity, tumor invasion, drug resistance and metastasis and has been correlated with the pathogenesis of cancer. MiRNAs are present in complex regulatory circuits to regulate stem cells function and are being examined to play important role in generation of CSCs, maintenance of enhanced self-renewal capacities of CSCs, pluripotency and their neoplastic transformation into tumors (Figure 2).
Many important functions of miRNAs in embryonic development and stem cells regulation in mammals are investigated. Dynamic expression profile of miRNAs in CSCs validates its significant role in controlling the self-renewal ability, pluripotency, diffentiation of progenitor cells, prosurvival and antistress mechanisms (Table 1).
| MicroRNAs | Transcript target | Relevance to cancer progression and its biological functions | Ref. |
| Upregulated microRNAs | |||
| miR-1246 | CCNG2 | Pancreatic cancer: Induces chemoresistance and CSC-like properties | [53] |
| miR-495 | E-cadherin and REDD1 | Breast cancer: Promotes oncogenesis and hypoxia resistance | [54] |
| miR-371-373 cluster | Wnt/β-catenin, DKK1 | Many cancers: Promotes cell growth and invasive activity | [55,56] |
| Myc | Liver cancer: Regulates the properties of CSCs | ||
| miR-216a/217 | PTEN and SMAD7 | Hepatocellular carcinoma: Increased proliferation, migration and metastatic ability | [57] |
| miR-210 | Nanog, Oct4 and EZH2 | Pancreatic cancer: Increased cell migration and invasion | [58] |
| miR-191 | BASP1, Wnt/β-catenin | Lung cancer: Increased migratory potential and neoplastic properties | [59] |
| miR-130b | P53-induced nuclear protein 1 | Acute myeloid leukemia: Regulates hematopoietic stem cells | [60] |
| miR-29a | P53-induced nuclear protein 1 | Acute myeloid leukemia: Regulates hematopoietic stem cells | [61] |
| miR-21 | Nanog, Oct4 and EZH2 | Pancreatic cancer: Increased cell migration and invasion | [58] |
| miR-18 | DLL4, inhibitor of Notch signaling | Glioma: Promotes tumorigenic potential of GSCs | [62] |
| Downregulated microRNAs | |||
| Let-7 | Lin-28 | Colon adenocarcinomas: Promotes cell migration, invasion and transforms immortalized colonic epithelial cells | [24,63] |
| Pancreatic cancer: Increased pluripotency | |||
| miR-487b | SUZ12, BMI1, WNT5A, MYC, and KRAS | Lung cancer: Increased proliferation and invasion | [64] |
| miR-451 | SMAD 3 and 4 | GBM: Controls GBM stem cells differentiation | [41] |
| miR-326 | Hh smoothened signal transducer | Chronic myeloid leukemia: Increased cell proliferation and decreased apoptosis | [65] |
| miR-204 | Sox4 and Ephrin receptor EphB2 | Glioma: Involved in GSCs self-renewal and invasion | [66] |
| miR-200 family | VEGFR1, VEGFR2 and EMT-related transcription factors | Pancreatic cancer: Regulates CSCs properties | [63] |
| miR-200a | ZEB1, ZEB2, SNAIL and SLUG N-cadherin, ZEB1,vimentin, | Pancreatic cancer: Increased cell migration and invasion | [67] |
| E-cadherin | |||
| miR-181 | ATM | Breast cancer: Regulates the properties of CSCs | [68] |
| miR-150 | MYb | Acute myeloid leukemia: Blocking of myeloid differentiation | [69] |
| miR-143/145 cluster | KRAS2 and its downstream effector RREB1 | Pancreatic cancer: Regulates CSCs survival | [70] |
| miR-145 | Oct4, Sox2, Nanog, Klf4 as well as Kras and Rreb1 | Pancreatic cancer: Increased pluripotency | [63] |
| miR-128 | Histone methylation [H3K27me(3)], Akt phosphorylation, p21(CIP1) Bmi-1 | Glioma: Increased self-renewal and proliferation | [71] |
| miR-107 | Nanog, Oct3/4, and Sox2 | Head and neck squamous cell carcinoma: Increased CSC proliferation | [72] |
| miR-100/let-7a-2/miR-125b-1 cluster | Myc | Liver cancer: Regulates the properties of CSCs | [56] |
| miR-34 family | Notch and Bcl-2 | Pancreatic cancer: Involved in self-renewal of CSCs | [28,29] |
| miR-29b-1 | CD133, N-Myc, CCND2, E2F1 and E2F2, Bcl-2, IAP-2, Oct3/4, Sox2 and Nanog | Osteosarcoma: Increased proliferation, self-renewal and chemoresistance | [73] |
| miR-27a | 14-3-3theta;, Bax and Bad | Acute leukemia: Regulate apoptosis | [74] |
| miR-23b | Cell cycle arrest | Glioma: Inhibits proliferation | [75] |
MiRNAs playing role in differentiation processes can either directly suppress the self-renewal state by suppressing the markers of pluripotency including Nanog, POU class 5 homoebox 1 (POU5f1) also known as Oct4, Kruppel like factor 4 and sex-determining region Y-box containing gene 2 [Sox2, a crucial transcription factor for the maintenance of embryonic stem cell (ESC) pluripotency and the determination of cell fate], or stabilize the differentiated cell fate by targeting the transcripts that are regulated by the pluripotency transcription factors including Oct4, Sox2, Nanog and Tcf3[22]. The transfection of let-7c, member of Let-7 family has been shown to rescue the differentiation defects in DGCR8-/- cells by downregulating the stemness genes including Oct4, Sox2 and Nanog[22]. Negative feedback regulation between Lin-28, a marker of undifferentiated ESCs and Let-7 family members has been reported in mouse differentiated cells and embryonic carcinoma cells[23]. Reduced patient survival and tumor relapse in human colon adenocarcinomas has been correlated with increased expression of Lin-28[24]. Studies on inhibition of Sox2 and placenta-specific 1 gene by miR-126 has been reported in gastric carcinogenesis[25].
First oncomiR reported as miR-17-92 polycistron has been examined to regulate c-Myc expression and accelerate tumor development in stomach, prostate, pancreatic, colon, lung and breast cancers. Novel stem cell specific miRNAs including miR-290, miR-302/367 and miR-371 clusters are identified in human ESCs and exhibit altered cell cycle profile and inhibit ESCs transition from self-renewal to differentiated state. Members of the miR-302 family has been shown to reprogram human skin carcinoma cells ino pluripotent ESC-like state[26].
Inhibition of tumor sphere growth in vitro and tumor formation in vivo upon restoration of miR-34 has been concluded to be associated with the suppression of growth of CSCs with biomarkers of CD44+ and CD133+ in human pancreatic cancer cells. These CSCs are deficient in miR-34 and show higher expression of Bcl-2 and Notch, which are the target genes for tumor suppressor like p53, and are involved in survival and self-renewal of CSCs. MiR-34 has been shown to inhibt the reprogramming through repressing pluripotency genes like Nanog, Sox2 and N-Myc[27]. Further research has confirmed that restoration of miR-34 directly regulates Bcl-2 and Notch target genes, activates caspase-3, induces apoptosis, thereby could increase chemotherapeutic and radiotherapeutic sensitivities in pancreatic cancer cells[28,29]. Suppression of tumorigenesis in vitro has been found to be regulated by miR-134 by downregulating the Notch target proteins and affecting G2/M pahse of cells in human endometrial CSCs[30]. Independent study by Park et al[31], 2013 revealed the differential expression of 43 miRNAs and their putative target genes such as p53, ErbB1, Notch, Wnt, and TGF-β signaling pathways which are the key regulators for stem cells and are mainly involved in cell death, cellular development, cellular growth, proliferation and maintenance of cancer and stem cell in glioblastoma multiforme (GBM) which is the most aggressive primary brain tumor, and notorious for resistance to chemo/radiotherapy.
Owning to high degree of biochemical specificity and potency shown by miRNAs in regulating the multiple vital pathways that can significantly affect the cancer progression, development of miRNAs as therapeutic molecule in association with anti-cancer drugs provides immense opportunities to counteract chemo/radioresistance and improve treatment outcome in various human cancers. Recent advances in the mechanisms, co-delivery of therapeutic molecules using nano-carriers and various advantages of combinational therapy in elimination of CSCs population and in complete cure of human cancer with no recurrence or metastasis are discussed in the subsequent section.
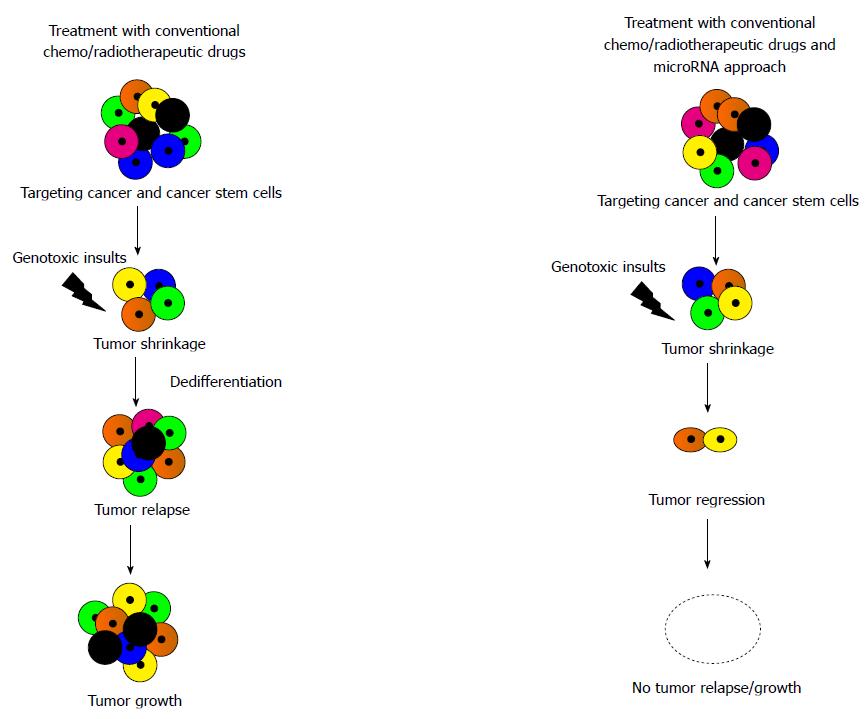
MiRNAs involved in acquisition of stem cell-like properties, regulation and reprogramming by cancer cells during cancer progression are not only exploited as molecular biomarkers to predict the risk of metastasis, systemic treatment resistance, and disease relapse of patients with cancer but are also an important novel therapeutic tools to improve patient-tailored treatments based on the unique signatures of a patient disease. MiRNAs can function as tumor suppressors or oncogenes and thereby either silence or express hundreds of genes and at the same time, where one gene can be targeted by multiple miRNAs. Local administration of miRNA-sponges which are the transcripts having multiple, tandem binding sites to the miRNAs of interest; antagomiRNAs/anti-miRNAs (highly chemically modified miRNA passenger strand) which include strong RNA binding analog locked nucleic acid in mixers with DNA, charged neutral anti-miRNA oligonucleotides like peptide nucleic acid, phosphorodiamidate morpholino oligonucleotides, 2’-modified anti-miRNA oligonucleotides such as 2’-O-methyl, 2’-O-methoxyethyl (MOE), 2’-fluoro/2’-methoxyethyl mixers (2’F/MOE) enhance binding affinity for their RNA targets, steric-block potency and biostability and induce inhibition of endogenous miRNA functions following miRNA- anti-miRNA oligonucleotide binding in a chemistry dependent manner[32,33]. Expression levels of related miRNA families that share the same seed sequence can be effectively reduced by using synthetic mRNAs, which contain multiple binding sites for endogenous miRNAs[34]. MiRNA replacement therapy is another important therapeutic intervention which is based on the systematic delivery of miRNA mimics into the cytoplasm of target cells to gain tumor suppressor functions. Inducing miRNA-mediated biological processes or blocking specific targets either could reverse the development of cancer or delay tumor growth[35]. Besides this, based on the relationship between expression of miRNA-regulated target genes and their reaction to drug therapies, some other miRNAs are explored which can mediate molecular target drugs and regulate chemosensitivities and chemoresistances of cancer. Experimental evidences document that the co-administration of CSCs with either antagomiRNAs/anti-miRNAs or miRNA mimics along with potential anti-cancer drugs can better sensitize cancer cells to promote apoptosis and autophagy, downregulate efflux transporters, revert EMT and inhibit tumor angiogenesis and could be employed as a more popular therapeutic approach for better clinical outcome in the treatment of human malignancies (Figure 3).
Viral vectors like lentiviral/adenovirus vectors are efficient delivery systems for miRNAs but toxicity and immunogenicity are the major hurdles for their clinical use. MiRNAs enclosed into non-viral particles, nanoparticles (NP) to form micelle-like structures for their effective delivery and enhanced stability in circulation. Some of the commonly used NPs for miRNAs delivery include polycationic-liposome hyaluronic acid, neutral lipid 1,2 dioleoyl-sn-glycero-3 phosphatidylcholine, polyethyleneimine, dendrimers, protamine, atelocollagen, poly(lactide-co-glycolide) particles, gold- or silica-based inorganic NPs. Binding of tumor specific ligands to NPs or conjugation with different compounds that have specific affinity with tumor cells ensure tissue-specific targeted delivery of miRNAs[36]. Some of the significant recent work being conducted in cancer cell lines and or animal models to understand the cellular processes that regulate CSCs functions and investigate the therapeutic effects of miRNAs and add new dimensions in cancer treatment, are being discussed here (Table 2).
| Co-delivery of microRNAs/anti-cancer drug | Biological functions | Cancer type | Ref. |
| Oncogenic microRNAs | |||
| miR-9/temozolomide | Inhibit the expression of drug efflux transporter, P-glycoprotein | Glioblastoma multiforme | [39] |
| miR-21/metformin, 5-Fluorouracil and Oxaliplatin | Target Wnt/β-catenin | Colon cancer | [38] |
| miR-125b/temozolomide | Target PIAS3, which contributes to reduced STAT3 transcriptional activity and subsequent decreased expression of MMP-2 and -9 | Glioblastoma | [40] |
| miR-125b/temozolomide | Target pro-apoptotic Bcl-2 antagonist killer 1 and sensitize CSCs to temozolomide induced apoptosis | Glioblastoma | [76] |
| Tumor suppressor microRNAs | |||
| miR-612/cisplatin, 5-Fluorouracil | Target Wnt/β-catenin, regulate EMT and inhibit cell proliferation, migration, invasion, and metastasis | Liver cancer | [48] |
| miR-205/gemcitabine | Decreased tumor cell population and increased apoptosis | Pancreatic cancer | [42] |
| miR-200c/docetaxel | Reduced TUBB3 level, and reversed EMT | Gastric cancer | [43] |
| miR-146a/cetuximab | Target Numb to stabilize β-catenin, regulate EMT, direct ACD-to-SCD switch | Colorectal cancer | [77] |
| miR-145/metformin, 5-Fluorouracil and Oxaliplatin | Target Wnt/β-catenin | Colon cancer | [38] |
| miR-34 family/5-Fluorouracil | Repression of c-Kit by p53 | Colorectal cancer | [45] |
| miR-34a/doxorubicin | Target Notch1 and reduce cancer stem cell properties | Breast cancer | [44] |
The effects of knockdown of miR-21 expression include induced apoptosis, reduced cell proliferation, invasion, and colony formation ability of colon tumor cells, inhibited G1/S cell cycle transition and increased sensitivity of cancer cells to 5-Fluorouracil (5-FU) chemotherapy. Direct targeting of human mut S homolog 2 and indirect regulation of the expression of thymidine phosphorylase and dihydropyrimidine dehydrogenase by miR-21 confirms its important role in enhancing the sensitivity of 5-FU resistance of colon cancer cells. Re-emergence of chemotherapy-resistant CSCs could be one of the possible reasons for recurrence in colon cancer[37]. The synergistic effect of increased miR-145 and reduced miR-21 expression which target Wnt/β-catenin signaling pathway along with co-delivery of metformin, in combination with 5-FU and oxaliplatin, has been examined inducing cell death in chemoresistant colon cancer cells, inhibiting colonospheres formation and enhancing colonospheres disintegration in the treatment of recurring colon cancer[38]. The delivery of synthetic Cy5-tagged anti-miR-9 to the resistant GBM cells has been observed to reverse the expression of the drug efflux transporter, P-glycoprotein and stimulate the sensitization of GBM cells to temozolomide, as shown by increased cell death and caspase activity[39]. Experimental studies by Shi et al[40], 2014 demonstrate the therapeutic effects of miR-125b inhibitor in enhancing the invasion-prevention activity of temozolomide in glioblastoma stem cells through targeting PIAS3 [protein inhibitor of activated signal transducer and activator of transcription (STAT)], which contributes to reduced STAT3 transcriptional activity and subsequent decreased expression of matrix metalloproteinase-2 (MMP-2) and MMP-9.
The cooperative effect of miR-451 (having a target site for SMAD in its promoter region) in combination with imatinib mesylate treatment in dispersal of GBM neurospheres and reduced tumorigenicity in glioblastoma validates this co-treatment as a new potential drug against the stem-cell like characteristics of glioblastoma[41]. The delivery of combinational formulations of gemcitabine and miR-205 conjugated copolymers in pancreatic cancer cells effectively reversed chemoresistance, invasion and migration by inducing apoptosis and inhibiting the growth and proliferation of CSCs[42]. In an another study, synergetic effects of targeted delivery of miR-200c and docetaxel (DOC) by gelatinases-stimuli NPs on inhibition of CSCs and non-CSC cancer cells in gastric cancer has been verified. The treatment of cancer cells with miR-200c/DOC NPs has been shown to significantly enhance the cytotoxicity of DOC, possibly by decreasing tubulin beta 3 Class III (TUBB3) level and reversing EMT, thereby affect tumor cell viability, migration and invasion[43]. Ectopic expression of another tumor suppressor, miR34a has been studied to reduce CSCs properties and increase sensitivity to doxorubicin treatment by directly targeting Notch1 in chemoresistant breast cancer cells[44]. c-Kit has been established as a new direct target of miR-34 where p53-induced miR-34 microRNA family mediated repression of c-Kit via a conserved seed-matching sequence in the c-Kit 3’-UTR, result reduced chemoresistance to 5-FU, reduced migration/invasion and stemness, further proves the hypothesis that this regulation interferes with several c-Kit-mediated effects in colorectal cancer cells[45]. Recent preliminary studies document the tumor growth inhibition in human non-small cell lung cancer xenografts and KRAS-G12D transgenic mouse model or the elimination of self-renewing breast CSCs upon therapeutic delivery of Let-7 mimics[46,47]. Up-regulation of miR-30e with tumor suppressor functions by downregulating BCR-ABL (Breakpoint cluster region-Abelson murine leukemia viral oncogene homolog 1) expression in chronic myeloid leukemic cells has been examined to render therapeutic efficacy against this disease. Owing to presence of putative target site for miR-30e in the 3’UTR of the ABL gene, enforced expression of miR-30e in K562 cells has been shown to suppress proliferation and induce apoptosis of these cells and sensitize them to imatinib treatment[48]. Attenuation of EMT by retaining epithelial cell morphology, reducing the levels of α-smooth muscle actin and increasing the levels of E-cadherin in human kidney proximal tubular epithelial cells has been observed after enforced expression of all the three members of miR-106b-25 cluster (miR-106b, miR-93 and miR-25) in salvianolic acid B[49].
Protective functions of decreased expression of oncogenic miR-21 and increased expression oftumor suppressor miR-138 regulated by α-solanine has been shown in prostate cancer. These therapeutic effects have been experimentally examined to be mediated by suppressing extracellular signal-regulated kinases and phosphatidylinositol-3 kinase - serine/threonine kinase signaling pathways, inhibiting EMT, reducing proliferation and inducing apoptosis[50].
Systemic administration of cationic lipid NP/pre-miR-107 significantly has been shown to retard tumor growth by 45.2% compared to NP/pre-miR-control by decreasing the expression of miR-107 targets including protein kinase Cε, cyclin-dependent kinase 6, and hypoxia-inducible factor 1-β. NP/pre-miR-107 further has been shown to reduce the cancer-initiating cell population and dampen the expression of the core embryonic stem cell transcription factors, Nanog, Oct3/4, and Sox2 and inhibit the clonogenic survival, cell invasion, and cell migration of head and neck squamous cell carcinoma cells[51].
Overexpression of miR-612 has been shown to suppress the stemness of hepatocellular carcinoma by reducing the number and size of tumorospheres, clone formation in soft agar and relieve the cancer cells from drug resistance to cisplatin and 5-FU by targeting Wnt/β-catenin signaling, thereby control EMT-associated stem cell-like traits[52].
Diverse cellular pathways that modulate CSCs functions act in concert with various molecular events and promote tumor initiation, progression and metastasis. Over the past recent years, emergence of miRNAs and molecular-targeted therapies has intensified research in the development of miRNA based therapeutic agents for cancer treatment with improved clinical outcome. Preclinical therapeutic data on mechanistic regulation of metastasis by miRNAs from in vitro studies need to be reevaluated in genetically engineered animal models which could further help us to provide new insights into clinical applications of miRNAs. Understanding the complex network of biological properties of CSCs, their control by multifunctional miRNAs and potential involvement of miRNAs in the development of anti-cancer drug resistance, reversion of cancer stemness and thereby rendering advanced cancers more susceptible to long term control are some of the open challenges to design novel strategies that can be used in preventive and treatment settings as an adjuvant to current cancer therapeutics.
CSCs which reside as a subset in many cancers are capable of self-renewal, tumor initiation, recurrence, metastasis, conferring resistance to anti-neoplastic drugs are still considered as huge obstacles on the way to cure cancer. This highlights the need to design strategies and therapeutics that specifically target and kill CSCs to eliminate cancer. Over the past few years, small ncRNAs, called miRNAs, have gained widespread attention in molecular biology due to their involvement in DNA translational control, their impression on mRNA and protein expression levels, and their ability to reprogram molecular signaling pathways in cancer. Differential miRNA profile in CSCs make them as potential biomarkers for aggressive tumor biology and therapeutic resistance and their biological specificity in targeting the various properties of CSCs make them strong targets for improving the response to anti-cancer drugs by sensitizing these cells to enhanced apoptotic effects of drugs. Novel anticancer therapies are based on the manipulation of oncogenic or tumor suppressor miRNAs by reducing or increasing their expression levels respectively. Success of using miRNAs as specific drug target in clinically relevant animal models as well as in preclinical development has been tested and determined by normal organ morphology, blood chemistries, serum cytokine levels, significant tumor regression and prolonged survival.
P- Reviewer: Jun Y, Kiselev SL, Papaccio G S- Editor: Gong XM L- Editor: A E- Editor: Jiao XK
| 1. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3316] [Cited by in RCA: 3384] [Article Influence: 109.2] [Reference Citation Analysis (0)] |
| 2. | Colmont CS, Harding KG, Piguet V, Patel GK. Human skin cancer stem cells: a tale of mice and men. Exp Dermatol. 2012;21:576-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 3. | Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198:281-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 284] [Cited by in RCA: 303] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 4. | Hatina J. The dynamics of cancer stem cells. Neoplasma. 2012;59:700-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Garg M. Epithelial-mesenchymal transition - activating transcription factors - multifunctional regulators in cancer. World J Stem Cells. 2013;5:188-195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 168] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 6. | Garg M. Mismatch repair system: Therapeutic approaches to cancer stem cells. Singh SR, editor. India: Research Signpost 2010; 271-291. |
| 7. | Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657-667. [PubMed] |
| 8. | Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, Clouthier SG, Wicha MS. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;7:e1000121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 393] [Cited by in RCA: 419] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 9. | Yan K, Yang K, Rich JN. The evolving landscape of glioblastoma stem cells. Curr Opin Neurol. 2013;26:701-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 10. | Takahashi RU, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Front Genet. 2014;4:295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 11. | So JY, Suh N. Targeting cancer stem cells in solid tumors by vitamin D. J Steroid Biochem Mol Biol. 2015;148:79-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Garg M. Potential therapeutic applications of microRNAs in response to DNA damage in cancer stem cells. J Stem Cells. 2011;6:51-65. [PubMed] |
| 13. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 14. | Garg M. MicroRNAs, stem cells and cancer stem cells. World J Stem Cells. 2012;4:62-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051-4060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2902] [Cited by in RCA: 2996] [Article Influence: 142.7] [Reference Citation Analysis (0)] |
| 16. | Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 938] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 17. | Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1290] [Cited by in RCA: 1281] [Article Influence: 61.0] [Reference Citation Analysis (0)] |
| 18. | Gregory RI, Chendrimada TP, Shiekhattar R. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol. 2006;342:33-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 19. | Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 946] [Cited by in RCA: 1002] [Article Influence: 47.7] [Reference Citation Analysis (0)] |
| 20. | Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834-838. [PubMed] |
| 21. | Pratt AJ, MacRae IJ. The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem. 2009;284:17897-17901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 368] [Cited by in RCA: 408] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 22. | Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 523] [Cited by in RCA: 536] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 23. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7589] [Cited by in RCA: 7242] [Article Influence: 402.3] [Reference Citation Analysis (0)] |
| 24. | King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS, Rustgi AK. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71:4260-4268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 205] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 25. | Otsubo T, Akiyama Y, Hashimoto Y, Shimada S, Goto K, Yuasa Y. MicroRNA-126 inhibits SOX2 expression and contributes to gastric carcinogenesis. PLoS One. 2011;6:e16617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 138] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 26. | Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115-2124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 336] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 27. | Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353-1360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 328] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 28. | Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433-8438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 500] [Cited by in RCA: 535] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 29. | Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 532] [Cited by in RCA: 542] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 30. | Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS Lett. 2015;589:207-214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 31. | Park EC, Kim G, Jung J, Wang K, Lee S, Jeon SS, Lee ZW, Kim SI, Kim S, Oh YT. Differential expression of MicroRNAs in patients with glioblastoma after concomitant chemoradiotherapy. OMICS. 2013;17:259-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 32. | Garg M. Targeting microRNAs in epithelial-to-mesenchymal transition-induced cancer stem cells: therapeutic approaches in cancer. Expert Opin Ther Targets. 2015;19:285-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 33. | Torres AG, Fabani MM, Vigorito E, Gait MJ. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA. 2011;17:933-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 34. | Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 504] [Cited by in RCA: 532] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 35. | Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027-7030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 489] [Cited by in RCA: 485] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 36. | Rothschild SI. microRNA therapies in cancer. Mol Cell Ther. 2014;2:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 37. | Deng J, Lei W, Fu JC, Zhang L, Li JH, Xiong JP. Targeting miR-21 enhances the sensitivity of human colon cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys Res Commun. 2014;443:789-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 38. | Nangia-Makker P, Yu Y, Vasudevan A, Farhana L, Rajendra SG, Levi E, Majumdar AP. Metformin: a potential therapeutic agent for recurrent colon cancer. PLoS One. 2014;9:e84369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 39. | Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol Ther Nucleic Acids. 2013;2:e126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 340] [Cited by in RCA: 406] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 40. | Shi L, Wan Y, Sun G, Zhang S, Wang Z, Zeng Y. miR-125b inhibitor may enhance the invasion-prevention activity of temozolomide in glioblastoma stem cells by targeting PIAS3. BioDrugs. 2014;28:41-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 41. | Gal H, Pandi G, Kanner AA, Ram Z, Lithwick-Yanai G, Amariglio N, Rechavi G, Givol D. MIR-451 and Imatinib mesylate inhibit tumor growth of Glioblastoma stem cells. Biochem Biophys Res Commun. 2008;376:86-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 177] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 42. | Mittal A, Chitkara D, Behrman SW, Mahato RI. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014;35:7077-7087. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 43. | Liu Q, Li RT, Qian HQ, Wei J, Xie L, Shen J, Yang M, Qian XP, Yu LX, Jiang XQ. Targeted delivery of miR-200c/DOC to inhibit cancer stem cells and cancer cells by the gelatinases-stimuli nanoparticles. Biomaterials. 2013;34:7191-7203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 44. | Park EY, Chang E, Lee EJ, Lee HW, Kang HG, Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H. Targeting of miR34a-NOTCH1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014;74:7573-7582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 165] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 45. | Siemens H, Jackstadt R, Kaller M, Hermeking H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget. 2013;4:1399-1415. [PubMed] |
| 46. | Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, Homer R, Brown D, Bader AG, Weidhaas JB. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580-1587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 360] [Cited by in RCA: 376] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 47. | Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759-764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 448] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 48. | Hershkovitz-Rokah O, Modai S, Pasmanik-Chor M, Toren A, Shomron N, Raanani P, Shpilberg O, Granot G. MiR-30e induces apoptosis and sensitizes K562 cells to imatinib treatment via regulation of the BCR-ABL protein. Cancer Lett. 2015;356:597-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 49. | Tang Q, Zhong H, Xie F, Xie J, Chen H, Yao G. Expression of miR-106b-25 induced by salvianolic acid B inhibits epithelial-to-mesenchymal transition in HK-2 cells. Eur J Pharmacol. 2014;741:97-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 50. | Shen KH, Liao AC, Hung JH, Lee WJ, Hu KC, Lin PT, Liao RF, Chen PS. α-Solanine inhibits invasion of human prostate cancer cell by suppressing epithelial-mesenchymal transition and MMPs expression. Molecules. 2014;19:11896-11914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 51. | Piao L, Zhang M, Datta J, Xie X, Su T, Li H, Teknos TN, Pan Q. Lipid-based nanoparticle delivery of Pre-miR-107 inhibits the tumorigenicity of head and neck squamous cell carcinoma. Mol Ther. 2012;20:1261-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 52. | Tang J, Tao ZH, Wen D, Wan JL, Liu DL, Zhang S, Cui JF, Sun HC, Wang L, Zhou J. MiR-612 suppresses the stemness of liver cancer via Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2014;447:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 53. | Hasegawa S, Eguchi H, Nagano H, Konno M, Tomimaru Y, Wada H, Hama N, Kawamoto K, Kobayashi S, Nishida N. MicroRNA-1246 expression associated with CCNG2-mediated chemoresistance and stemness in pancreatic cancer. Br J Cancer. 2014;111:1572-1580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 54. | Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463-2474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 55. | Zhou AD, Diao LT, Xu H, Xiao ZD, Li JH, Zhou H, Qu LH. β-Catenin/LEF1 transactivates the microRNA-371-373 cluster that modulates the Wnt/β-catenin-signaling pathway. Oncogene. 2012;31:2968-2978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 56. | Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci USA. 2010;107:20471-20476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 57. | Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 323] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 58. | Bao B, Ali S, Ahmad A, Azmi AS, Li Y, Banerjee S, Kong D, Sethi S, Aboukameel A, Padhye SB. Hypoxia-induced aggressiveness of pancreatic cancer cells is due to increased expression of VEGF, IL-6 and miR-21, which can be attenuated by CDF treatment. PLoS One. 2012;7:e50165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 59. | Xu W, Ji J, Xu Y, Liu Y, Shi L, Liu Y, Lu X, Zhao Y, Luo F, Wang B. MicroRNA-191, by promoting the EMT and increasing CSC-like properties, is involved in neoplastic and metastatic properties of transformed human bronchial epithelial cells. Mol Carcinog. 2015;54 Suppl 1:E148-E161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 60. | Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, Man K, Wong N, To KF. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 322] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 61. | Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475-489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 228] [Cited by in RCA: 240] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 62. | Turchi L, Debruyne DN, Almairac F, Virolle V, Fareh M, Neirijnck Y, Burel-Vandenbos F, Paquis P, Junier MP, Van Obberghen-Schilling E. Tumorigenic potential of miR-18A* in glioma initiating cells requires NOTCH-1 signaling. Stem Cells. 2013;31:1252-1265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 63. | Sureban SM, May R, Qu D, Weygant N, Chandrakesan P, Ali N, Lightfoot SA, Pantazis P, Rao CV, Postier RG. DCLK1 regulates pluripotency and angiogenic factors via microRNA-dependent mechanisms in pancreatic cancer. PLoS One. 2013;8:e73940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 123] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 64. | Xi S, Xu H, Shan J, Tao Y, Hong JA, Inchauste S, Zhang M, Kunst TF, Mercedes L, Schrump DS. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J Clin Invest. 2013;123:1241-1261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 65. | Babashah S, Sadeghizadeh M, Hajifathali A, Tavirani MR, Zomorod MS, Ghadiani M, Soleimani M. Targeting of the signal transducer Smo links microRNA-326 to the oncogenic Hedgehog pathway in CD34+ CML stem/progenitor cells. Int J Cancer. 2013;133:579-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 66. | Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H, Li W, Hu B, Cheng SY, Li M. Loss of miR-204 expression enhances glioma migration and stem cell-like phenotype. Cancer Res. 2013;73:990-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 133] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 67. | Lu Y, Lu J, Li X, Zhu H, Fan X, Zhu S, Wang Y, Guo Q, Wang L, Huang Y. MiR-200a inhibits epithelial-mesenchymal transition of pancreatic cancer stem cell. BMC Cancer. 2014;14:85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 68. | Wang Y, Yu Y, Tsuyada A, Ren X, Wu X, Stubblefield K, Rankin-Gee EK, Wang SE. Transforming growth factor-β regulates the sphere-initiating stem cell-like feature in breast cancer through miRNA-181 and ATM. Oncogene. 2011;30:1470-1480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 230] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 69. | Morris VA, Zhang A, Yang T, Stirewalt DL, Ramamurthy R, Meshinchi S, Oehler VG. MicroRNA-150 expression induces myeloid differentiation of human acute leukemia cells and normal hematopoietic progenitors. PLoS One. 2013;8:e75815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 70. | Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470-1480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 239] [Cited by in RCA: 234] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 71. | Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125-9130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 529] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 72. | Collet G, Skrzypek K, Grillon C, Matejuk A, El Hafni-Rahbi B, Lamerant-Fayel N, Kieda C. Hypoxia control to normalize pathologic angiogenesis: potential role for endothelial precursor cells and miRNAs regulation. Vascul Pharmacol. 2012;56:252-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 73. | Di Fiore R, Drago-Ferrante R, Pentimalli F, Di Marzo D, Forte IM, D’Anneo A, Carlisi D, De Blasio A, Giuliano M, Tesoriere G. MicroRNA-29b-1 impairs in vitro cell proliferation, self-renewal and chemoresistance of human osteosarcoma 3AB-OS cancer stem cells. Int J Oncol. 2014;45:2013-2023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 74. | Scheibner KA, Teaboldt B, Hauer MC, Chen X, Cherukuri S, Guo Y, Kelley SM, Liu Z, Baer MR, Heimfeld S. MiR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3theta;. PLoS One. 2012;7:e50895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 75. | Geng J, Luo H, Pu Y, Zhou Z, Wu X, Xu W, Yang Z. Methylation mediated silencing of miR-23b expression and its role in glioma stem cells. Neurosci Lett. 2012;528:185-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 76. | Chen J, Fu X, Wan Y, Wang Z, Jiang D, Shi L. miR-125b inhibitor enhance the chemosensitivity of glioblastoma stem cells to temozolomide by targeting Bak1. Tumour Biol. 2014;35:6293-6302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 77. | Hwang WL, Jiang JK, Yang SH, Huang TS, Lan HY, Teng HW, Yang CY, Tsai YP, Lin CH, Wang HW. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol. 2014;16:268-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 223] [Article Influence: 20.3] [Reference Citation Analysis (0)] |