Published online May 26, 2015. doi: 10.4252/wjsc.v7.i4.700
Peer-review started: October 22, 2014
First decision: November 27, 2014
Revised: January 30, 2015
Accepted: March 18, 2015
Article in press: March 20, 2015
Published online: May 26, 2015
Processing time: 221 Days and 13.2 Hours
In the adult mouse brain, the subventricular zone lining the lateral ventricles and the subgranular zone in the dentate gyrus of the hippocampus are two zones that contain neural stem cells (NSCs) with the capacity to give rise to neurons and glia during the entire life of the animal. Spatial and temporal regulation of gene expression in the NSCs population is established and maintained by the coordinated interaction between transcription factors and epigenetic regulators which control stem cell fate. Epigenetic mechanisms are heritable alterations in genome function that do not involve changes in DNA sequence itself but that modulate gene expression, acting as mediators between the environment and the genome. At the molecular level, those epigenetic mechanisms comprise chemical modifications of DNA such as methylation, hydroxymethylation and histone modifications needed for the maintenance of NSC identity. Genomic imprinting is another normal epigenetic process leading to parental-specific expression of a gene, known to be implicated in the control of gene dosage in the neurogenic niches. The generation of induced pluripotent stem cells from NSCs by expression of defined transcription factors, provide key insights into fundamental principles of stem cell biology. Epigenetic modifications can also occur during reprogramming of NSCs to pluripotency and a better understanding of this process will help to elucidate the mechanisms required for stem cell maintenance. This review takes advantage of recent studies from the epigenetic field to report knowledge regarding the mechanisms of stemness maintenance of neural stem cells in the neurogenic niches.
Core tip: Neural stem cells (NSCs) are capable of extensive self-renewal while preserving the ability to generate cell progeny that can differentiate into different cell types from the nervous system. Intrinsic mediators as well as extrinsic cues provided by the neurogenic niche (microenvironment where NSCs reside in vivo) are important for stem cell self-renewal and differentiation. Epigenetic changes, including alterations in DNA methylation, histone modifications and imprinting alter the way a gene interacts with the cell transcribing machinery, turning genes “on” or “off”. These heritable changes must be reversible and context-dependent being responsible of stem cell plasticity.
- Citation: Montalbán-Loro R, Domingo-Muelas A, Bizy A, Ferrón SR. Epigenetic regulation of stemness maintenance in the neurogenic niches. World J Stem Cells 2015; 7(4): 700-710
- URL: https://www.wjgnet.com/1948-0210/full/v7/i4/700.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i4.700
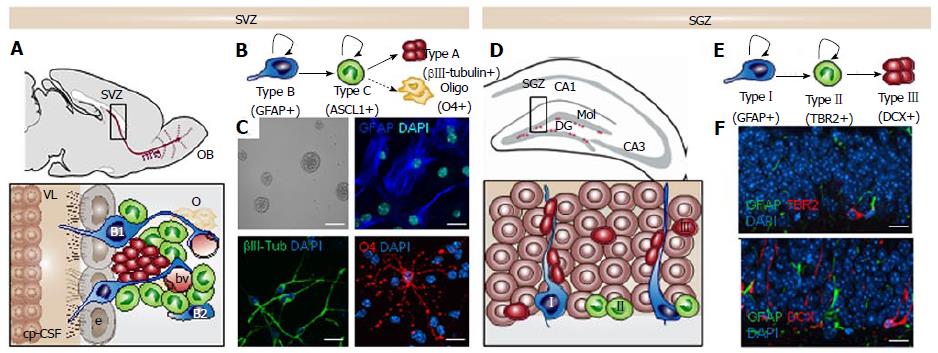
Adult stem cells have the ability to divide, self-renew and generate functional differentiated cells that replace lost cells throughout an organism’s lifetime. The existence of adult stem cells was first described in tissues with high proliferation rates, such as the hematopoietic system and the intestine. Since then, stem cells have been found in almost all adult tissues including the nervous system[1]. In the adult mouse brain two main regions continue to generate new neurons throughout adulthood: the subventricular zone (SVZ) in the walls of the lateral ventricles[2] (Figure 1A-C) and the subgranular zone (SGZ) in the dentate gyrus (DG) of the hippocampus[3] (Figure 1D-F). Adult neurogenesis is supported by multipotent neural stem cells (NSCs) deriving from embryonic radial-glia and thus expressing astroglial characteristics[4,5]. Astrocytic-like stem cells are relatively quiescent and can be identified by the expression of the glial fibrillary acidic protein (GFAP), the stemness-related transcription factor Sox2 [Sex determining region Y (SRY)-box 2], and the neural progenitor marker Nestin[2,6,7]. Moreover, their slow division rate can be detected by the label retention of thymidine analogs incorporated during DNA replication[6,8,9]. NSCs can also be isolated from their natural niche and cultured in vitro in the presence of the epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) mitogens. In culture, NSCs form free-floating aggregates called “neurospheres” (Figure 1C). Self-renewal and multipotency characteristics of NSCs are assessed in vitro by clonal analysis in which single cells give rise to neurospheres[10,11] (Figure 1C).
The SVZ is located lining the walls of the lateral ventricles and constitutes a complex microenvironment or niche in which proliferation and self-renewal of NSCs are strongly regulated by multiple extracellular factors such as EGF, bFGF, bone morphogenetic protein and pigment epithelium derived factor[12-14]. This significant extrinsic signaling is possible because of the special cytoarchitecture of the niche that allows NSCs to be in direct contact with the cerebrospinal fluid (CSF) produced by the choroid plexus in the ventricles, with the vasculature and with other cells from the niche like astrocytes or microglia[15,16]. Subventricular NSCs (also known as type B1 cells) present a radial glia-like morphology, with an apical primary cilium contacting the ventricular lumen and a basal process reaching the basal lamina and the vascular structures[17,18] (Figure 1A). The walls of the lateral ventricles show a typical organization where the small apical process of one or more type B1 cells are surrounded by a rosette of epithelial ependymal cells that form structures known as pinwheels at the surface[19]. There is another astrocyte-like type B cell that is more frequently located close to the underlying striatal parenchyma known as type B2 cells[20]. When activated, these slowly dividing NSCs give rise to fast cycling cells called transit-amplifying progenitors (TAP or type C cells). TAP cells contribute to reducing the number of cell division rounds that NSCs have to undergo to preserve their genome integrity. Mash1-positive type C cells generate neuroblasts (type A cells) that migrate along the rostral migratory stream (RMS) into the olfactory bulb (OB) where they differentiate and integrate into interneurons (Figure 1B). These chains of polysialylated neural cell adhesion molecule (PSA-NCAM) and DCX (doublecortin) positive neuroblasts reach the core of the OB, where they detach from the RMS and migrate radially into the granular and periglomerular layers[21-23]. These immature neurons then integrate and differentiate into inhibitory interneurons, playing an important role in rodent olfaction. In addition of being a neurogenic region, the SVZ can serve as a niche of oligodendrocytes although generated in much lower numbers than neuroblasts. Thereby, Olig2-positive transit amplifying cells give rise to oligodendroblasts that migrate to the corpus callosum and striatum while tightly associated with blood vessels[24], where they differentiate into myelinating and nonmyelinating oligodendrocytes[25].
Along with the SVZ, the subgranular zone in the dentate gyrus of the hippocampus constitutes the other main neurogenic niche in the adult mouse brain[26-28]. The SGZ is also a complex microenvironment in which the vasculature plays an important role. Dividing stem cells in the SGZ are in close proximity to an extensive network of interconnected blood vessels and parenchymal astrocytes that can regulate their proliferation and differentiation via paracrine signaling[29]. The SGZ is located between the granular layer and the hilus of the DG and the SGZ NSCs constitute a subpopulation of GFAP-positive cells that are analogous to subventricular type B1 cells[30]. In this region, two types of neural progenitors can be identified according to different expression of molecular markers and their morphologies[23] (Figure 1D and E). Type I progenitors exhibit a radial process spanning the granule cell layer and arborizing profusely in the molecular layer[27]. These cells express nestin, GFAP, and Sox2[31]. Type II hippocampal progenitors have short processes and contrary to type I cells, express TRB2 but not GFAP (Figure 1F). There is evidence suggesting that type II cells may derive from type I cells but a lineage relationship study is still lacking[31]. In the adult SGZ, precursors give rise by asymmetrical divisions to intermediate neuronal lineage-restricted progenitor cells and in a minor number, to glial lineage-restricted progenitor cells (both of them are GFAP-negative cells). Compared to the SVZ, few oligodendrocytes are generated in the SGZ. Type II cells generate in turn type III cells, which are neuronal precursors that express markers of immature migrating neurons, such DCX and PSA-NCAM (Figure 1F). These differentiated cells integrate neuronal circuits into the hippocampal CA3 region forming dendrites and spreading their axons[22]. In addition to the production of granular neurons, a low percentage of activated NSCs divide asymmetrically to give rise to astrocytes. The latter migrate into the hilus and the molecular layer where they lose their stem cell identity and cause the depletion of the pool of NSCs[32,33] thus explaining the possible decrease in hippocampal neurogenesis associated with ageing.
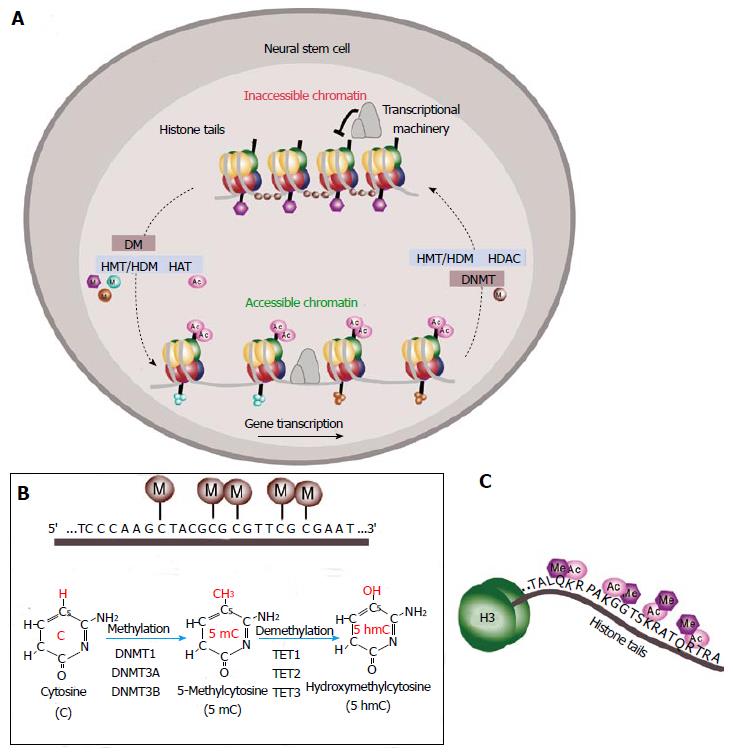
Epigenetic is defined as the study of heritable alterations in genome function that do not involve changes in DNA sequence itself[34,35]. These epigenetic marks modulate gene expression either by directly altering the chromatin structure or by creating bindings sites for chromatin and transcription regulatory subunits. Two general classes of epigenetic regulation can be defined: covalent modifications to DNA and post-translational covalent modifications to the histones (H) around which the DNA is bound, influencing whether DNA is accessible or not for gene transcription[36,37] (Figure 2A). Moreover, the three-dimensional structure and arrangement of chromatin within the nucleus are both regulated by and contribute to the establishment and maintenance of epigenetic states[34]. These different classes of epigenetic modifications are intimately related, resulting in multiple layers of control allowing cells to maintain their identity over time[34,38]. Dysregulation of these mechanisms leads to new cellular phenotypes by causing altered gene expression without a change in genotype. In the neurogenic niches, epigenetic regulators and the associated transcription factors play an important role in the control and maintenance of NSC stemness.
DNA methylation involves the addition of a methyl group to the fifth carbon in the cytosine pyrimidine ring (Figure 2B). In most mammalian genes, CpG dinucleotides are methylated and concentrated in clusters called “CpG islands” which often have regulatory functions and tend to be found in the promoter and first exon regions of genes[39] where it promotes a closed chromatin structure and aids the prevention of expression[34,40]. DNA methylation marks repress gene expression either by attracting DNA methyl-binding domain proteins (MBDs) such as methyl-CpG binding protein 2 (MeCP2) which recruit repressors and chromatin remodeling molecules to generate an inactive chromatin environment or by directly inhibiting transcription factor binding[41-43]. MBD proteins have been suggested to play a role in neurogenesis. For example, mice deficient in MBD1 show decreased neurogenesis and hippocampus-related behaviour defects. Indeed, Mbd1-deficient NSCs generate a reduced number of neurons when compared to wild-type cells, suggesting a role for MBD1 in neuronal fate commitment[44].
There are two types of methylation reactions both mediated by DNA methyltransferases (DNMTs). One is de novo methylation catalyzed by DNMT3a and DNMT3b, important for normal embryogenesis and development and responsible for the establishment of methylation patterns. The other type is maintenance methylation mediated by DNMT1 that effectively maintains CpG methylation upon DNA replication and provides the heritable “memory” of the methylation state of the parent cell[45,46]. DNMT1 is highly expressed in the embryonic, perinatal and adult CNS in both dividing neural progenitors and mature neurons where it maintains DNA methylation[47-49]. A lack of DNMT1 alters neuronal excitability and increases apoptosis in post-mitotic cortical neurons[50]. In support of this, mice deficient for Dnmt1 specifically in neural progenitors at embryonic stages exhibit deficits in neuronal function and die postnatally, suggesting a requirement for methylation in brain development[51]. DNMT3a and DNMT3b are highly expressed in postnatal NSCs and are required for neurogenesis and neuronal maturation[48,49,52,53]. Loss of DNMT3a results in gene silencing[53] and depletion of DNMT3b leads to deficient neuronal differentiation in vitro[54].
DNA is hypomethylated in neural progenitor cells and methylation is progressively increased during lineage commitment[55]. The suppression of astrogliogenesis during neuronal specification is also associated with changes in DNA methylation[56,57]. This silencing is attenuated later in development resulting in the generation of astrocytes which correlates with the suppression of neurogenesis. Demethylation and expression of the genes coding for the astrocytic markers Gfap and the calcium binding protein S100β during astrocytic maturation, correlates with methylation and downregulation of neurogenic genes such as Neurogenin 1[58-60]. Activation of the Gfap promoter requires binding of the signal transducer and activator of transcription 3 (STAT3) to a consensus sequence. Early progenitors are refractory to astrocyte differentiation due to methylation of the STAT3 binding site. At later development stages, loss of STAT3-binding element methylation is associated with Gfap promoter activation[60,61]. A similar alteration in methylation pattern occurs at another STAT3 binding site in the S100β promoter[58].
DNA methylation marks are reversible through both passive replication-dependent demethylation and active demethylation which probably involve the recently characterized 5-hydroxymethyl (5hmC) intermediate[62] (Figure 2B). In mammals, three members of the ten-eleven translocation (TET) family of enzymes have been identified: TET1, TET2 and TET3[63,64]. TET hydroxylases may catalyze active DNA demethylation by oxidation of 5mC to 5hmC[65-67] (Figure 2B). 5hmC is relatively abundant in mouse embryonic stem cells (ESCs), the early embryo and in adult brain[68,69]. In the brain, 5hmC is enriched at active genes, associated with the strong depletion of 5mC from these regions[70]. It has been proposed that TET enzymes in the blastocyst and ESCs are involved in pluripotency by maintaining the hypomethylated state of key regulatory regions[69,71]. Recent studies have also shown that TET1 is involved in the epigenetic regulation of neural progenitor cell proliferation in the adult hippocampus[72-74]. Mice lacking Tet1 exhibit impaired hippocampal neurogenesis accompanied by poor learning and memory[72-74]. However, the full role and importance of hydroxymethylation in the brain remains to be elucidated.
In eukaryotic cells, a histone octamer including two H2A-H2B dimers and a H3-H4 tetramer acts as a scaffold around which DNA is wrapped to form a nucleosome[75,76]. The interaction between histones and DNA is mediated by an N-terminal tail of histone proteins available for post-translational modifications that control the chromatin structure[75] (Figure 2C). These covalent modifications in the histone tails alter the interaction between adjacent nucleosomes and/or between histones and the DNA, changing the three-dimensional chromatin structure. Modifications in the body of histones have also been shown to alter chromatin structure influencing gene expression[77]. Histone modifications are divided into repressive and active marks according to how they correlate with levels of transcriptional activity. For example, histone acetylation of lysine residues of histones, catalyzed by histone acetyltransferases enhances the recruitment and activation of the transcriptional machinery and is generally associated with areas of active gene transcription[78]. However, histone deacetylases (HDACs) remove acetyl groups promoting the condensation of chromatin[79] (Figure 2A). HDAC1 is expressed by GFAP-positive cells within the SVZ whereas HDAC2 is found in migrating neuroblasts and in TAP cells within the SVZ[80]. Deletion of HDAC2 in the SVZ results in a defective neurogenesis to the OB[81] and neurospheres treated with class I and II HDAC inhibitors promotes neuronal differentiation[82] suggesting a role for this enzyme in neuronal fate determination. Furthermore, oligodendrocyte fate commitment is accompanied by a decrease in histone deacetylation at transcriptional repressors of oligodendrocytic differentiation such as Sox11[83] and at neuronal genes such as Sox2[84].
Histone methylation is associated with both active and silent chromatin and is catalyzed by histone methyltransferases (HMTs). Trimethylation of lysine (K)-27 and lysine 9 of histone H3 (H3K27me3 and H3K9me3) tends to associate with regions of inactive gene transcription, whereas H3K4, H3K36 and H3K79 methylations are associated with active transcription[85]. Histone demethylases (HDMs) also have a key role in regulating neural development[86]. During neural stem cell commitment, H3K27 methylation decreases in key developmental genes following downregulation of the HMT Enhancer of Zest homolog 2 (EZH2) and upregulation of the HDM Jumonji domain containing-3 (JMJD3). Indeed, deletion of Ezh2 in SGZ progenitor cells results in cell proliferation restriction leading to a reduced number of neurons that ultimately leads to impairment in spatial learning and memory[87]. Additionally, JMJD3 is upregulated in neuroblasts, and Jmjd3 deletion targeted to SVZ NSCs in both developing and adult mice impairs neuronal differentiation. JMJD3 regulates neurogenic gene expression via interaction at not only promoter regions but also neurogenic enhancer elements such as Dlx2[88]. Moreover, H3K9me3 is enriched in the adult murine SVZ and it has been recently shown that its repression in undifferentiated cells is engaged in the maintenance of cell type integrity in this neurogenic niche[89]. MLL1, another HMT that methylates H3K4, has been associated with the trithorax group of transcription factors. In mice where Mll1 is knocked out in NSCs, neurogenesis is impaired. Mll1 is associated with the promoter of the homeobox transcription factor Dlx2 and although loss of Mll1 does not affect the methylation of H3K4, it does increase H3K27me3 on the promoter indicating that Mll1 is recruiting a H3K27 demethylase[90]. In summary, the above studies indicate that different chromatin modifiers have a critical role in adult neurogenesis[91].
Imprinted genes are expressed predominantly from one chromosome in a parental-origin dependent manner. While most genes are expressed from both alleles, imprinted genes are functionally monoallelic and are expressed from either the maternally or the paternally inherited chromosome[92]. In mammals, this affects around 100 genes that are found in clusters. Imprinting control regions (ICRs) regulate the parental allele-specific pattern of gene expression and have differentially methylated regions (DMRs) on the two parental chromosomes. ICRs can be divided into those which are methylated on the paternally inherited copy and those with maternally inherited methylation[93]. DMRs are also characterized by the asymmetrical accumulation of different histone modifications on the two parental chromosomes and the recent identification of a “tri-mark”, comprising the trimethylation of H3K4 and H3K9 and the trimethylation of H4K20 at all known ICRs[94]. The majority of imprinted genes are expressed in the brain and several exhibit brain-specific imprinting. Their monoallelic expression makes these loci very vulnerable as mutation of the expressed allele can compromise expression and lead to severe developmental defects. For example, human congenital imprinting syndromes including Angelman syndrome and Prader-Willi syndrome are all characterized by neurological and behavioral impairments and learning difficulties[95]. Evidence is suggesting that selective regulation of imprinting is a normal mechanism of modulating gene dosage and is associated with the control of stem cell potential in the neurogenic niche. For instance, relaxation of imprinting of the gene for the atypical NOTCH ligand delta-like homologue 1 (Dlk1) usually expressed from the paternally inherited chromosome has been shown in the neural stem cells and niche astrocytes within the SVZ[96]. Notably, this selective absence of Dlk1 imprinting is associated with acquisition of DNA methylation at the germline-derived imprinting control region[96]. Igf2 is also an imprinted gene expressed only from the paternally-inherited allele although it is specifically biallelically expressed in postnatal human and mouse choroid plexus epithelium and leptomeninges[97,98]. Thus, CSF produced from the choroid plexus and blood vessels is a biallelic source of neurogenesis-promoting IGF2[99].
Epigenetic reprogramming consists in the transition from one cell type to another, permitted by the loss of the molecular characteristics of the cell of origin and the acquisition of an entirely new molecular identity without changing the genomic sequence[100]. Reprogramming involves changes in the transcriptome and chromatin state of the reprogrammed cell type to that of a pluripotent cell[101-103]. This implicates different levels of changes in DNA factor binding, transcription and chromatin state[103]. Since the discovery by Takahashi and Yamanaka in 2006 that the introduction of four transcription factors, Oct3/4, Klf4, c-Myc, Sox2 (known as OKMS) could reprogram mouse embryonic and adult fibroblasts into induced pluripotent stem cells (iPSCs)[104], the field of reprogramming has considerably evolved and several studies have reported the use of sets of these transcription factors in various combinations to reprogram mouse and human somatic cells[105-108]. More recently, murine B lymphocytes, liver, stomach and pancreatic β-cells were showed to reprogram into iPSCs using the combination of factors OKMS[109-111]. In 2008, Eminli et al[112] reported the generation of iPSCs from murine NSCs by retroviral infection of the same combination of factors. Since neurosphere cultures express Sox2 and c-myc, a considerable advance consisted in showing that they could be reprogrammed only with Oct4 and Klf4 at similar efficiency to the reprogramming rate of murine fibroblasts with the original four factors[112-114]. Finally, the forced expression of Oct4 alone was shown sufficient to reprogram murine NSCs, albeit with a ten-fold lower efficiency than with two factors[113]. Because NSCs are originally closer to the pluripotency state than somatic cells and require fewer factors to be reprogrammed, they constitute a more simple and attractive system to study epigenetic mechanisms occurring during the acquisition of pluripotency. Importantly, iPSCs derived from human and murine NSCs exhibited markers of ESCs, showed demethylation of pluripotency genes, formed teratomas, and contributed to viable chimeras[112-114].
Reprogramming of somatic cells is a stochastic event[115]. However, in NSCs, Oct4 only seems sufficient to repress genes responsible for NSCs molecular identity and activate the pluripotency genes, suggesting that epigenetic of NSCs renders them easier to reprogram and that the combination of factors necessary for reprogramming is dependent on cellular context[112]. iPSCs have lower levels of methylation than somatic cells, suggesting that demethylation is an important chromatin feature to achieve pluripotency[116]. During reprogramming, it is stipulated that reprogramming factors interfere with methylation of the newly synthesized DNA by binding to specific promoters or enhancer regions leading to demethylation and activation of the pluripotency genes. In addition, active DNA demethylation mechanisms could be required for the reactivation of pluripotency genes[117]. Recent studies in NSCs have shown the importance of methylation level in the context of reprogramming. Undifferentiated neurospheres highly express DNMT1 and contain methylated chromatin suggesting the role of methylation for the maintenance of the quiescent or undifferentiated state of NSCs[118]. It is then probable that NSC chromatin is dynamically remodelled and that DNA methylation modification is essential for reprogramming to a pluripotent state. For instance, histone methyltransferase G9a is responsible for the downregulation of Oct4 during NSC differentiation and its inhibition results in iPSC formation after overexpression of exogenous Klf4 and c-myc only[119]. In addition, interference with DNMT1 promotes iPSC formation, also supporting that DNA methylation is a feature limiting reprogramming to pluripotency[101]. All reprogramming techniques involve demethylation of the genome thus appearing as a crucial process for successfully achieving pluripotency[120,121].
During reprogramming, NSCs downregulate specific genes like Nestin and progressively express the markers of pluripotency Oct4, Nanog, Fgf4, Zfp42[113,114]. In addition, efficiency and timing of reprogramming highly depends on the differentiation state of the initial cell type. Importantly, comparative studies with ESCs reported that efficiently reprogrammed iPSCs show transcriptional pattern and epigenetic marks highly similar to ESCs. For instance, Oct4 and Nanog promoters are demethylated and histones H3 lysine 4 (K4) and lysine 27 mostly exhibit patterns of trimethylation[101,106,122]. However, reprogramming of NSCs into iPSCs is often incomplete and leaves epigenetic marks including DNA methylation, chromatin modification and transcriptional regulation in the resulting iPSC genome[123,124] known as epigenetic “memory”. Partially reprogrammed cell lines are characterized by an absence of complete downregulation of the exogenous reprogramming factors and partial demethylation and reactivation of pluripotency genes[101,104]. During reprogramming, somatic markers get progressively downregulated demonstrating the importance of silencing its differentiation program as a step towards pluripotency. Treatment of partially reprogrammed iPSCs with inhibitors of ERK1/2 and GSK3b signaling[125], induced genome demethylation of 30% explained by decreased levels of DNMT3a/b and their targeting factor DNMT3L[126-128]. The two inhibitors repress DNMT3A/B expression inducing demethylation of certain genomic regions in ESCs. Thus, DNA demethylation of the reprogrammed cell type as a way to remove epigenetic marks is important for complete reprogramming into iPSCs. Reprogrammed iPSCs often present the limitation of not being fully reprogrammed thus keeping epigenetic traces of the tissue of origin. Future generation of iPSCs without epigenetic memory is an important challenge in the field to ensure that differentiation decisions are not affected by events from the past[116].
Determining the mechanisms by which neural stem cells maintain self-renewal capacity and at the same time generate differentiated progeny is a central challenge in stem cell biology. Several recent studies have demonstrated that epigenetic gene regulation plays a crucial role in the control of stem cell behaviour. Epigenetic mechanisms include changes in chromatin structure that provides a way for coordinately activating or repressing genes during proliferation and differentiation. Extracellular signaling from the microenvironment or niche in which NSCs reside in vivo interacts with these diverse epigenetic mechanisms, thus regulating transcription factors and intracellular pathways. These changes in gene expression are often heritable and reversible, features that support stem cell plasticity such as the ability to dedifferentiate or become reprogrammed under certain conditions. Finally, aberrant epigenetic mechanisms are known to be involved in the development of many neurological diseases. Characterizing epigenetic changes associated with a particular neural pathology may be used as biomarkers of disease and the manipulation of those epigenetic mechanisms holds great promise as a potential therapeutic strategy.
P- Reviewer: Markic D, Santulli G, Wagner KD S- Editor: Ji FF L- Editor: A E- Editor: Wu HL
| 1. | Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 854] [Cited by in RCA: 815] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 2. | Doetsch F, García-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046-5061. [PubMed] |
| 3. | Gage FH, Kempermann G, Palmer TD, Peterson DA, Ray J. Multipotent progenitor cells in the adult dentate gyrus. J Neurobiol. 1998;36:249-266. [PubMed] |
| 4. | Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003;6:1127-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 555] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 5. | Merkle FT, Tramontin AD, García-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci USA. 2004;101:17528-17532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 646] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 6. | Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805-3819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 490] [Cited by in RCA: 561] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 7. | Morshead CM, Reynolds BA, Craig CG, McBurney MW, Staines WA, Morassutti D, Weiss S, van der Kooy D. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071-1082. [PubMed] |
| 8. | Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 213] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 9. | Ponti G, Obernier K, Guinto C, Jose L, Bonfanti L, Alvarez-Buylla A. Cell cycle and lineage progression of neural progenitors in the ventricular-subventricular zones of adult mice. Proc Natl Acad Sci USA. 2013;110:E1045-E1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 192] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 10. | Ferron SR, Andreu-Agullo C, Mira H, Sanchez P, Marques-Torrejon MA, Farinas I. A combined ex/in vivo assay to detect effects of exogenously added factors in neural stem cells. Nat Protoc. 2007;2:849-859. [PubMed] |
| 11. | Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707-1710. [PubMed] |
| 12. | Faigle R, Song H. Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochim Biophys Acta. 2013;1830:2435-2448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 255] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 13. | Porlan E, Perez-Villalba A, Delgado AC, Ferrón SR. Paracrine regulation of neural stem cells in the subependymal zone. Arch Biochem Biophys. 2013;534:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Ramírez-Castillejo C, Sánchez-Sánchez F, Andreu-Agulló C, Ferrón SR, Aroca-Aguilar JD, Sánchez P, Mira H, Escribano J, Fariñas I. Pigment epithelium-derived factor is a niche signal for neural stem cell renewal. Nat Neurosci. 2006;9:331-339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 332] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 15. | Shen Q, Goderie SK, Jin L, Karanth N, Sun Y, Abramova N, Vincent P, Pumiglia K, Temple S. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1137] [Cited by in RCA: 1143] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 16. | Silva-Vargas V, Crouch EE, Doetsch F. Adult neural stem cells and their niche: a dynamic duo during homeostasis, regeneration, and aging. Curr Opin Neurobiol. 2013;23:935-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 17. | Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703-716. [PubMed] |
| 18. | Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 845] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 19. | Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265-278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 823] [Cited by in RCA: 828] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 20. | Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674-686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 288] [Cited by in RCA: 282] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 21. | Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145-1148. [PubMed] |
| 22. | Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687-702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2119] [Cited by in RCA: 1973] [Article Influence: 140.9] [Reference Citation Analysis (0)] |
| 23. | Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2152] [Cited by in RCA: 2331] [Article Influence: 137.1] [Reference Citation Analysis (0)] |
| 24. | Cayre M, Courtès S, Martineau F, Giordano M, Arnaud K, Zamaron A, Durbec P. Netrin 1 contributes to vascular remodeling in the subventricular zone and promotes progenitor emigration after demyelination. Development. 2013;140:3107-3117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 25. | Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907-7918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 664] [Cited by in RCA: 771] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 26. | Bonaguidi MA, Song J, Ming GL, Song H. A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Curr Opin Neurobiol. 2012;22:754-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 27. | Encinas JM, Sierra A, Valcárcel-Martín R, Martín-Suárez S. A developmental perspective on adult hippocampal neurogenesis. Int J Dev Neurosci. 2013;31:640-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | Palmer TD, Willhoite AR, Gage FH. Vascular niche for adult hippocampal neurogenesis. J Comp Neurol. 2000;425:479-494. [PubMed] |
| 30. | Seri B, García-Verdugo JM, McEwen BS, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21:7153-7160. [PubMed] |
| 31. | Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515-528. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 656] [Cited by in RCA: 651] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 32. | Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142-1155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 731] [Cited by in RCA: 669] [Article Influence: 47.8] [Reference Citation Analysis (0)] |
| 33. | Encinas JM, Michurina TV, Peunova N, Park JH, Tordo J, Peterson DA, Fishell G, Koulakov A, Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 753] [Cited by in RCA: 691] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 34. | Bird A. Perceptions of epigenetics. Nature. 2007;447:396-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1909] [Cited by in RCA: 1833] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 35. | Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4367] [Cited by in RCA: 4245] [Article Influence: 193.0] [Reference Citation Analysis (0)] |
| 36. | Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693-705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7446] [Cited by in RCA: 7672] [Article Influence: 426.2] [Reference Citation Analysis (0)] |
| 37. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6156] [Cited by in RCA: 6154] [Article Influence: 246.2] [Reference Citation Analysis (0)] |
| 38. | Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409-418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 535] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 39. | Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007;90:314-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 40. | Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023-2036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 251] [Cited by in RCA: 274] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 41. | Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5074] [Cited by in RCA: 4879] [Article Influence: 212.1] [Reference Citation Analysis (0)] |
| 42. | Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19:269-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 296] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 43. | Tost J. DNA methylation: an introduction to the biology and the disease-associated changes of a promising biomarker. Methods Mol Biol. 2009;507:3-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 44. | Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci USA. 2003;100:6777-6782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 288] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 45. | Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611-2617. [PubMed] |
| 46. | Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002-33010. [PubMed] |
| 47. | Brooks PJ, Marietta C, Goldman D. DNA mismatch repair and DNA methylation in adult brain neurons. J Neurosci. 1996;16:939-945. [PubMed] |
| 48. | Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 861] [Cited by in RCA: 774] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 49. | Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39-44. [PubMed] |
| 50. | Hutnick LK, Golshani P, Namihira M, Xue Z, Matynia A, Yang XW, Silva AJ, Schweizer FE, Fan G. DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet. 2009;18:2875-2888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 145] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 51. | Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, Biniszkiewicz D, Bates B, Lee PP, Kuhn R. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. J Neurosci. 2001;21:788-797. [PubMed] |
| 52. | Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247-257. [PubMed] |
| 53. | Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 525] [Cited by in RCA: 482] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 54. | Bai S, Ghoshal K, Datta J, Majumder S, Yoon SO, Jacob ST. DNA methyltransferase 3b regulates nerve growth factor-induced differentiation of PC12 cells by recruiting histone deacetylase 2. Mol Cell Biol. 2005;25:751-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 80] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 55. | Sikorska M, Sandhu JK, Deb-Rinker P, Jezierski A, Leblanc J, Charlebois C, Ribecco-Lutkiewicz M, Bani-Yaghoub M, Walker PR. Epigenetic modifications of SOX2 enhancers, SRR1 and SRR2, correlate with in vitro neural differentiation. J Neurosci Res. 2008;86:1680-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 56. | Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645-657. [PubMed] |
| 57. | Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 375] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 58. | Namihira M, Nakashima K, Taga T. Developmental stage dependent regulation of DNA methylation and chromatin modification in a immature astrocyte specific gene promoter. FEBS Lett. 2004;572:184-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 80] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 59. | Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365-376. [PubMed] |
| 60. | Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749-758. [PubMed] |
| 61. | Fan G, Martinowich K, Chin MH, He F, Fouse SD, Hutnick L, Hattori D, Ge W, Shen Y, Wu H. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345-3356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 315] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 62. | Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci. 2013;368:20110330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 319] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 63. | Ito T. Role of histone modification in chromatin dynamics. J Biochem. 2007;141:609-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 64. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4860] [Cited by in RCA: 4389] [Article Influence: 274.3] [Reference Citation Analysis (0)] |
| 65. | Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67-79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 667] [Cited by in RCA: 620] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 66. | He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1977] [Cited by in RCA: 2110] [Article Influence: 150.7] [Reference Citation Analysis (0)] |
| 67. | Loenarz C, Schofield CJ. Oxygenase catalyzed 5-methylcytosine hydroxylation. Chem Biol. 2009;16:580-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 68. | Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA. 2011;108:3642-3647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 526] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 69. | Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129-1133. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2151] [Cited by in RCA: 1978] [Article Influence: 131.9] [Reference Citation Analysis (0)] |
| 70. | Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417-1430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 733] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 71. | Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200-213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 657] [Cited by in RCA: 625] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 72. | Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 319] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 73. | Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 340] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 74. | Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 283] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 75. | Luger K, Richmond TJ. DNA binding within the nucleosome core. Curr Opin Struct Biol. 1998;8:33-40. [PubMed] |
| 76. | Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol. 2003;4:809-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 167] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 77. | Tropberger P, Schneider R. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat Struct Mol Biol. 2013;20:657-661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 78. | Marmorstein R, Trievel RC. Histone modifying enzymes: structures, mechanisms, and specificities. Biochim Biophys Acta. 2009;1789:58-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 232] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 79. | Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3359] [Cited by in RCA: 4040] [Article Influence: 288.6] [Reference Citation Analysis (0)] |
| 80. | Foti SB, Chou A, Moll AD, Roskams AJ. HDAC inhibitors dysregulate neural stem cell activity in the postnatal mouse brain. Int J Dev Neurosci. 2013;31:434-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 81. | Jawerka M, Colak D, Dimou L, Spiller C, Lagger S, Montgomery RL, Olson EN, Wurst W, Göttlicher M, Götz M. The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 2010;6:93-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 82. | Siebzehnrubl FA, Buslei R, Eyupoglu IY, Seufert S, Hahnen E, Blumcke I. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Exp Brain Res. 2007;176:672-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 78] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 83. | He Y, Sandoval J, Casaccia-Bonnefil P. Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: the role of HDAC and YY1. Neuron Glia Biol. 2007;3:221-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 84. | Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci U S A. 2007;104:14982-14987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 101] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 85. | Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707-719. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2601] [Article Influence: 144.5] [Reference Citation Analysis (0)] |
| 86. | Tsukada Y, Ishitani T, Nakayama KI. KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 2010;24:432-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 87. | Zhang J, Ji F, Liu Y, Lei X, Li H, Ji G, Yuan Z, Jiao J. Ezh2 regulates adult hippocampal neurogenesis and memory. J Neurosci. 2014;34:5184-5199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 132] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 88. | Park DH, Hong SJ, Salinas RD, Liu SJ, Sun SW, Sgualdino J, Testa G, Matzuk MM, Iwamori N, Lim DA. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014;8:1290-1299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 108] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 89. | Foret MR, Sandstrom RS, Rhodes CT, Wang Y, Berger MS, Lin CH. Molecular targets of chromatin repressive mark H3K9me3 in primate progenitor cells within adult neurogenic niches. Front Genet. 2014;5:252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 90. | Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529-533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 298] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 91. | Gonzales-Roybal G, Lim DA. Chromatin-based epigenetics of adult subventricular zone neural stem cells. Front Genet. 2013;4:194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 92. | Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 573] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 93. | Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 307] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 94. | McEwen KR, Ferguson-Smith AC. Distinguishing epigenetic marks of developmental and imprinting regulation. Epigenetics Chromatin. 2010;3:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 95. | Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 96. | Ferrón SR, Charalambous M, Radford E, McEwen K, Wildner H, Hind E, Morante-Redolat JM, Laborda J, Guillemot F, Bauer SR. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature. 2011;475:381-385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 97. | DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849-859. [PubMed] |
| 98. | Giannoukakis N, Deal C, Paquette J, Goodyer CG, Polychronakos C. Parental genomic imprinting of the human IGF2 gene. Nat Genet. 1993;4:98-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 303] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 99. | Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893-905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 536] [Cited by in RCA: 494] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 100. | Krishnakumar R, Blelloch RH. Epigenetics of cellular reprogramming. Curr Opin Genet Dev. 2013;23:548-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 101. | Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1205] [Cited by in RCA: 1112] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 102. | Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111-123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 795] [Cited by in RCA: 730] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 103. | Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324-1343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 241] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 104. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18205] [Article Influence: 958.2] [Reference Citation Analysis (0)] |
| 105. | Hester ME, Song S, Miranda CJ, Eagle A, Schwartz PH, Kaspar BK. Two factor reprogramming of human neural stem cells into pluripotency. PLoS One. 2009;4:e7044. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 106. | Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1254] [Cited by in RCA: 1239] [Article Influence: 72.9] [Reference Citation Analysis (0)] |
| 107. | Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2182] [Cited by in RCA: 2126] [Article Influence: 118.1] [Reference Citation Analysis (0)] |
| 108. | Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 458] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 109. | Aoi T, Yae K, Nakagawa M, Ichisaka T, Okita K, Takahashi K, Chiba T, Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 814] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 110. | Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 662] [Cited by in RCA: 605] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 111. | Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic beta cells into induced pluripotent stem cells. Curr Biol. 2008;18:890-894. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 280] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 112. | Eminli S, Utikal J, Arnold K, Jaenisch R, Hochedlinger K. Reprogramming of neural progenitor cells into induced pluripotent stem cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467-2474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 246] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 113. | Kim JB, Sebastiano V, Wu G, Araúzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 690] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 114. | Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 723] [Cited by in RCA: 697] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 115. | Lin SL, Chang DC, Lin CH, Ying SY, Leu D, Wu DT. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011;39:1054-1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 116. | Lee HJ, Hore TA, Reik W. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell. 2014;14:710-719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 268] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 117. | Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 401] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 118. | Singh RP, Shiue K, Schomberg D, Zhou FC. Cellular epigenetic modifications of neural stem cell differentiation. Cell Transplant. 2009;18:1197-1211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 119. | Shi Y, Desponts C, Do JT, Hahm HS, Schöler HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 666] [Cited by in RCA: 640] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 120. | Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 327] [Cited by in RCA: 303] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 121. | Theunissen TW, Jaenisch R. Molecular control of induced pluripotency. Cell Stem Cell. 2014;14:720-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 122. | Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2048] [Cited by in RCA: 1902] [Article Influence: 105.7] [Reference Citation Analysis (0)] |
| 123. | Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117-1119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 432] [Cited by in RCA: 467] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 124. | Tobin SC, Kim K. Generating pluripotent stem cells: differential epigenetic changes during cellular reprogramming. FEBS Lett. 2012;586:2874-2881. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 125. | Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519-523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2986] [Cited by in RCA: 2655] [Article Influence: 156.2] [Reference Citation Analysis (0)] |
| 126. | Ficz G, Hore TA, Santos F, Lee HJ, Dean W, Arand J, Krueger F, Oxley D, Paul YL, Walter J. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem Cell. 2013;13:351-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 320] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 127. | Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 359] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 128. | Leitch HG, Tang WW, Surani MA. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr Top Dev Biol. 2013;104:149-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |