Published online Sep 26, 2014. doi: 10.4252/wjsc.v6.i4.485
Revised: August 29, 2014
Accepted: August 30, 2014
Published online: September 26, 2014
Processing time: 61 Days and 20.8 Hours
Neural stem cells (NSCs) contribute to ontogeny by producing neurons at the appropriate time and location. Neurogenesis from NSCs is also involved in various biological functions in adults. Thus, NSCs continue to exert their effects throughout the lifespan of the organism. The mechanism regulating the core functional properties of NSCs is governed by intra- and extracellular signals. Among the transcription factors that serve as molecular switches, Sox2 is considered a key factor in NSCs. Sox2 forms a core network with partner factors, thereby functioning as a molecular switch. This review discusses how the network of Sox2 partner and target genes illustrates the molecular characteristics of the mechanism underlying the self-renewal and multipotency of NSCs.
Core tip: Neural stem cells (NSCs) are cells that are capable of both self-renewal and multipotency. In these two processes, the transcription factor Sox2 serves as a switch for the central molecular mechanism. Sox2 forms complexes with its partner factors to perform its transcription-related functions. This partner switching presumably serves as an important key to the intrinsic functions of NSCs. A detailed understanding of these molecular mechanisms will advance our understanding of basic neuroscience and increase the feasibility of employing cell reprogramming technology in regenerative medicine.
- Citation: Shimozaki K. Sox2 transcription network acts as a molecular switch to regulate properties of neural stem cells. World J Stem Cells 2014; 6(4): 485-490
- URL: https://www.wjgnet.com/1948-0210/full/v6/i4/485.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v6.i4.485
Neural stem cells (NSCs) are cells that are capable of self-renewal and maintaining multipotency[1,2]. NSCs differentiate into neurons, astrocytes, and oligodendrocytes. The cellular origin of mouse NSCs dates back to the initial stage of ontogeny. A blastocyst generates the primitive ectoderm, further differentiating into the neuroectoderm, which serves as a source of primitive NSCs[3,4]. The neuroectoderm then develops and differentiates into the neuroepithelium[3,4]. Primitive NSCs exhibit self-renewal with a rather limited multipotency[5]. On embryonic day 11.5 (E11.5) in the murine fetal period, differentiation into neurons dominates while differentiation into the astrocyte lineage is suppressed by DNA methylation. Then at E14.5, NSCs begin to produce neurons and astrocytes[6-8]. After birth, NSCs manifest their ability to produce oligodendrocytes[8]. NSCs also actively undergo repeated self-renewal in the region of the central nervous system after birth to generate neurons, astrocytes, and oligodendrocytes in a region-dependent manner to build the brain as an organ. It was previously believed that neurons do not regenerate once the brain organogenesis is complete in an adult organism. However, the study[9-13] revised this dogma, and it is now known that neurogenesis takes place even in the adult brain. This process has been best studied in the subventricular zone (SVZ) of the lateral ventricle and the subgranular zone (SGZ) lining the hippocampal region, where NSCs are located and produce new nerve cells through self-renewal[1,14-20]. Understanding the molecular biology underlying the capacity of NSCs to exhibit self-renewal and multipotency is expected to stimulate our exploration of basic neuroscience and lead to practical applications in regenerative medicine, allowing lost neurons to be regenerated as desired. Although some NSCs can be cultured from body tissues as the monolayer[21-23], there are technical challenges as well as issues of productivity and quality related to the practical use of such cultured NSCs in regenerative medicine. However, a new technology was recently developed to reprogram somatic cells through the gene transfer. Using this technology, combinational transfection of the Oct4, Sox2, Klf4, and c-Myc genes into various cells can establish a type of multipotent stem cells, called induced pluripotent stem (iPS) cells[24,25]. By changing the culture conditions under which iPS cells are established, we can artificially induce differentiation into NSCs[26]. This technology has also been utilized to develop induced neuronal (iN) cells, which directly induce differentiation into neurons[27]. iN cells are obtained by transfecting the Ascl1, Brn2, and Myt1l genes into fibroblasts. The gene cluster serving as the switch to precisely regulate cell fate mainly includes transcription factors. One key factor that plays an important role in NSCs is the transcription factor Sox2. Transcription factors bind to response regions in the genome to initiate or terminate the expression of target genes. Concomitantly, they interact with a group of chromatin-regulating factors other than transcription factors to perform various regulatory functions. In this review, I focus on the transcription regulatory network centered around Sox2 to shed light on the molecular regulatory mechanism underlying the biology of NSCs.
Sox2 belongs to the Sry gene family and contains a DNA-binding domain referred to as a high-mobility group (HMG) domain, which is highly conserved across the family. To date, more than 20 genes have been identified in the Sox gene family[28,29]. Sox2 is a maternal factor that is specifically expressed in the inner cell mass (ICM) and primitive ectoderm[30]. Sox2 expression is widely observed among the cells within the neural tube at early stages of neurodevelopment[31]. Its expression is subsequently localized to the ventricular layer in the neuronal cortex, where NSCs and their precursor cells are present after the mid-fetal period. During this period, Sox2 is not expressed in layers where terminally differentiated neurons are present[32]. In the adult brain, NSCs are localized to the SVZ of the lateral ventricle and the SGZ lining the hippocampal region, where they undergo self-renewal and perform neurogenesis[1,14]. All of such self-renewing cells express Sox2. Sox2 plays an important role in maintaining the functions of NSCs[32-35]. It has been reported that SoxB1 family members, Sox1 and Sox3, which show high sequence homology to Sox2, exhibit similar functions[36]. Sox2 functions as a maternal factor in pre-implantation embryos[30]. Zygotic knock-down of Sox2 using a specific siRNA resulted in an incomplete trophectoderm (TE) in fertilized embryos, which failed to progress beyond the morula stage[30]. Sox2 expression is detected in both the ICM and TE, and its expression becomes restricted to the ICM[29]. During embryogenesis, the ICM becomes the embryo, and the TE forms the placenta. A high level of Sox2 gene expression has been confirmed in the neuroectoderm that gives rise to NSCs[31]. During embryogenesis, Sox2 promotes neuroectoderm cell fate by suppressing the mesodermal cell fate[37]. Moreover, Sox2 plays important roles in the differentiation of the central nervous system and peripheral nervous system during embryogenesis by controlling the proliferation and differentiation of neural stem/progenitor cells[32]. Sox2 deficiency is embryonically lethal in mice because the fetus fails to form embryonic stem (ES) cells from the ICM or produce trophoblast stem cells[30,38]. Sox2 conditional knock out (KO) mice have been reported to undergo neurodegeneration leading to dysfunctional neuronal differentiation in the adult brain[35,39]. Various research approaches have been employed to demonstrate that Sox2 expression is localized to NSCs and that its function is essential for these cells.
Sox2 collaborates with other transcription factors[40,41]. In ES cells and NSCs, Sox2 regulates the self-renewal mechanism and suppresses differentiation in a dosage-sensitive manner[42,43]. Sox2 and a POU factor known as Oct4 form a specific partnership to coordinately regulate the mechanism that maintains undifferentiated ES cells[44,45]. The target genes of this partnership include Nanog, Utf1, and Fgf4[4]. Sox family members form partner complexes with POU factors, but the partnership assumes various forms depending on the cell type[41]. In NSCs, Sox2 interacts with POU factors such as Pax6, Brn1, and Brn2, where Pax6 forms complexes with Sox2 to regulate the differentiation of cells of the optic nerve and lens[46-49]. Pax6 is coexpressed in Sox2-positive cells and reportedly regulates the self-renewal and neurogenesis of NSCs in the hippocampus in the adult brain[50]. The expression of Nestin, a marker for NSCs, is coordinately regulated by Sox2 and POU factors[47,51]. Sox2 and the partner code of Brn1 and Brn2 bind to the regulatory region of the Nestin and Sox2 genes to perform an important function in the regulation of gene expression[47,51-53]. Furthermore, Sox2 can bind to Prx1 (MHox1/Prrx1) and function as its partner[54]. Because Prx1 and Sox2 are coexpressed in certain cells in the NSC region, they are expected to coordinately activate the target genes, and they have been suggested to be involved in the regulatory mechanism that maintains the undifferentiated state of NSCs.
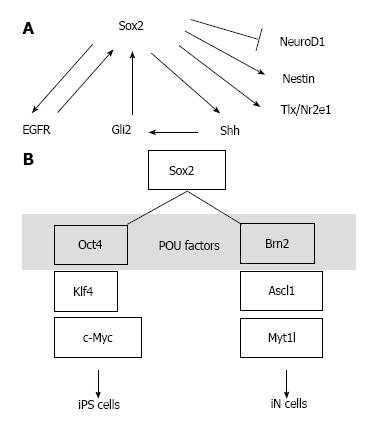
Sox2 is a transcription factor, and many reports have been published describing analyses of its target genes. Sox2 regulates the expression of its target gene called Sonic hedgehog (Shh) to regulate NSCs in the hippocampus[39,55,56]. Shh is a humoral factor that transmits outside signals from outside into the cell via its receptor Patched, and induces Smo/Gli signal activation[57-59]. Another transcription factor, Gli2, is a downstream target of Shh and regulates Sox2 gene expression[60]. Therefore, these factors may constitute a positive feedback loop. Additionally, Notch and the epidermal growth factor receptor (EGFR) pathway regulate the number of NSCs and their self-renewal[61]. EGF stimulation can turn neural progenitors into multipotent NSCs through the receptor, EGFR[62]. Whereas EGFR signaling increases Sox2 expression, Sox2 enhances Egfr expression, which suggests a positive feedback mechanism[63] (Figure 1A). The nuclear receptor, Tlx (Nr2e1), is an essential factor in the mechanism that maintains undifferentiated NSCs[64-66]. A possible negative feedback model of Tlx gene expression has been reported, in which Sox2 binds to Tlx to regulate its transcription[67]. Based on these findings, it is conceivable that the Sox2-centered feedback loop mechanism involving Sox2 target genes serves as an important system for the self-renewal mechanisms of NSCs.
It was recently reported that the crosstalk between Sox2 and Wnt signaling regulates the switching during the differentiation of NSCs to neurons[68]. Sox2 and Tcf act as molecular switches thus interacting with the overlap sequence, and this process, in turn, regulates NeuroD1 expression[68]. Although the mechanisms underlying the molecular switching of numerous genes are being increasingly revealed, it remains unknown how such mechanisms activate differentiation switches at the appropriate times and locations in response to intra- and extracellular changes, while suppressing the expression of genes other than those involved in neuronal differentiation.
Combined transfection of the Oct4, Sox2, Klf4, and c-Myc genes transforms somatic cells into pluripotent stem cells[24,25]. In this process, the transcriptional network is switched on to generate multipotent stem cells. It is likely that the partnership between Sox2 and Oct4 functions as the core switch[4]. The addition of Klf4 to the partner complexes presumably allows for multidimensional regulation of various modes of switching. In the multipotency induction process, the use of serum-free culture medium with EGF actively induces the formation of NSCs[26]. Conversely, induction of the iN cell phenotype is conducted using a cell engineering technology that directly transdifferentiates somatic cells into neurons[27]. Forced expression of the Ascl1, Brn2, and Myt1l genes can induce neuronal differentiation. However, this method is not intended for the maintenance of NSCs. Brn2 is a partner factor of Sox2[51,53]. When Sox2 is added to the group of iN-factors and cells are cultured in EGF- or bFGF-containing medium, combinations other than Oct4, Klf4, and c-Myc may be able to produce artificial NSCs. Moreover, based on the concept of the Sox2 partner code[41], the establishment of neuronal subtype-specific NSCs also seems possible, using combinations of Pax6 and Prx1 or other POU factors (Figure 1B).
I have reviewed the link between the molecular mechanisms at work in NSCs and properties of stem cells, with a focus on the network involving the Sox2-centered partner code and its target genes. The localized expression of Sox2 in NSCs/neural progenitors enhances its molecular specificity. By forming complexes with its partner factors, Sox2 exerts its transcriptional-regulation function. The partner factors involved vary depending on the molecular context of the stem cell lineage. Sox2 target genes include molecular switches controlling the NeuroD1 gene (which is capable of inducing neuronal differentiation) as well as the feedback loop with the factors involved in self-renewal such as members of the EGFR signaling pathway. By manipulating Sox2 and its partner factors, researchers can now artificially induce differentiation into pluripotent, or multipotent stem cells and into neurons. Nevertheless, many questions remain unanswered regarding the Sox2-based self-renewal mechanism and the regulatory mechanism underlying multipotency. Further research using conditional KO mice is needed to explore functions of Sox2, its partner factors, and chromatin-regulating factors that interact with Sox2 and its partner factors as well as to identify the entire panel of Sox2 target genes.
I thank the members of the Center for Frontier Life Science for helpful discussions. Crimson Interactive Pvt. Ltd. (Ulatus) and NPG Language Editing are acknowledged for their assistance in translating and editing the manuscript.
P- Reviewer: Abdelalim EM, Fukuda T, Lu F S- Editor: Song XX L- Editor: A E- Editor: Lu YJ
| 1. | Gage FH. Mammalian neural stem cells. Science. 2000;287:1433-1438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3484] [Cited by in RCA: 3412] [Article Influence: 136.5] [Reference Citation Analysis (0)] |
| 2. | Yao J, Mu Y, Gage FH. Neural stem cells: mechanisms and modeling. Protein Cell. 2012;3:251-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Schoenwolf GC, Smith JL. Mechanisms of neurulation: traditional viewpoint and recent advances. Development. 1990;109:243-270. [PubMed] |
| 4. | Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 207] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 5. | Hitoshi S, Ishino Y, Kumar A, Jasmine S, Tanaka KF, Kondo T, Kato S, Hosoya T, Hotta Y, Ikenaka K. Mammalian Gcm genes induce Hes5 expression by active DNA demethylation and induce neural stem cells. Nat Neurosci. 2011;14:957-964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 6. | Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 481] [Cited by in RCA: 456] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 7. | Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 258] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 8. | Kondo T, Raff M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004;18:2963-2972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 9. | Altman J. Autoradiographic investigation of cell proliferation in the brains of rats and cats. Anat Rec. 1963;145:573-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 361] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 10. | Altman J, Das GD. Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J Comp Neurol. 1965;124:319-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2456] [Cited by in RCA: 2481] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 11. | Paton JA, Nottebohm FN. Neurons generated in the adult brain are recruited into functional circuits. Science. 1984;225:1046-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 306] [Cited by in RCA: 292] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 12. | Eriksson PS, Perfilieva E, Björk-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313-1317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4410] [Cited by in RCA: 4295] [Article Influence: 159.1] [Reference Citation Analysis (0)] |
| 13. | Gerber GB, Aldridge WG, Gerber G, Altman KI, Hempelmann LH. The catabolism of nucleic acids. V. Autoradiographic studies on the replacement of DNA in normal and x-irradiated rats. Int J Radiat Biol. 1963;6:23-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 14. | Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 992] [Cited by in RCA: 1018] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 15. | Suh H, Deng W, Gage FH. Signaling in adult neurogenesis. Annu Rev Cell Dev Biol. 2009;25:253-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 260] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 16. | Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2152] [Cited by in RCA: 2330] [Article Influence: 137.1] [Reference Citation Analysis (0)] |
| 17. | Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 140] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 18. | Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687-702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2119] [Cited by in RCA: 1971] [Article Influence: 140.8] [Reference Citation Analysis (0)] |
| 19. | Duan X, Kang E, Liu CY, Ming GL, Song H. Development of neural stem cell in the adult brain. Curr Opin Neurobiol. 2008;18:108-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 233] [Cited by in RCA: 217] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 20. | Kempermann G, Jessberger S, Steiner B, Kronenberg G. Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 2004;27:447-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Pollard SM, Conti L, Sun Y, Goffredo D, Smith A. Adherent neural stem (NS) cells from fetal and adult forebrain. Cereb Cortex. 2006;16 Suppl 1:i112-i120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 203] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 22. | Palmer TD, Ray J, Gage FH. FGF-2-responsive neuronal progenitors reside in proliferative and quiescent regions of the adult rodent brain. Mol Cell Neurosci. 1995;6:474-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 494] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 23. | Ray J, Gage FH. Differential properties of adult rat and mouse brain-derived neural stem/progenitor cells. Mol Cell Neurosci. 2006;31:560-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 145] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 24. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14316] [Article Influence: 842.1] [Reference Citation Analysis (0)] |
| 25. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18196] [Article Influence: 957.7] [Reference Citation Analysis (0)] |
| 26. | Matsui T, Takano M, Yoshida K, Ono S, Fujisaki C, Matsuzaki Y, Toyama Y, Nakamura M, Okano H, Akamatsu W. Neural stem cells directly differentiated from partially reprogrammed fibroblasts rapidly acquire gliogenic competency. Stem Cells. 2012;30:1109-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035-1041. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2235] [Cited by in RCA: 2274] [Article Influence: 151.6] [Reference Citation Analysis (0)] |
| 28. | Miyagi S, Kato H, Okuda A. Role of SoxB1 transcription factors in development. Cell Mol Life Sci. 2009;66:3675-3684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 29. | Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 594] [Cited by in RCA: 725] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 30. | Keramari M, Razavi J, Ingman KA, Patsch C, Edenhofer F, Ward CM, Kimber SJ. Sox2 is essential for formation of trophectoderm in the preimplantation embryo. PLoS One. 2010;5:e13952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 163] [Cited by in RCA: 162] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 31. | Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1695] [Cited by in RCA: 1696] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 32. | Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. Int J Biochem Cell Biol. 2010;42:421-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 264] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 33. | Suh H, Consiglio A, Ray J, Sawai T, D’Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515-528. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 656] [Cited by in RCA: 651] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 34. | Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 980] [Cited by in RCA: 1030] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 35. | Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805-3819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 36. | Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 37. | Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 386] [Cited by in RCA: 417] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 38. | Miyagi S, Masui S, Niwa H, Saito T, Shimazaki T, Okano H, Nishimoto M, Muramatsu M, Iwama A, Okuda A. Consequence of the loss of Sox2 in the developing brain of the mouse. FEBS Lett. 2008;582:2811-2815. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 71] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 39. | Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nat Neurosci. 2009;12:1248-1256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 396] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 40. | Kiefer JC. Back to basics: Sox genes. Dev Dyn. 2007;236:2356-2366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 160] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 41. | Kondoh H, Kamachi Y. SOX-partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. Int J Biochem Cell Biol. 2010;42:391-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 153] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 42. | Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells. 2008;26:903-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 249] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 43. | Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 420] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 44. | Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635-2645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 572] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 45. | Tomioka M, Nishimoto M, Miyagi S, Katayanagi T, Fukui N, Niwa H, Muramatsu M, Okuda A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002;30:3202-3213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 236] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 46. | Inoue M, Kamachi Y, Matsunami H, Imada K, Uchikawa M, Kondoh H. PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development. Genes Cells. 2007;12:1049-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 47. | Josephson R, Müller T, Pickel J, Okabe S, Reynolds K, Turner PA, Zimmer A, McKay RD. POU transcription factors control expression of CNS stem cell-specific genes. Development. 1998;125:3087-3100. [PubMed] |
| 48. | Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510-3519. [PubMed] |
| 49. | Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 285] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 50. | Maekawa M, Takashima N, Arai Y, Nomura T, Inokuchi K, Yuasa S, Osumi N. Pax6 is required for production and maintenance of progenitor cells in postnatal hippocampal neurogenesis. Genes Cells. 2005;10:1001-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 131] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 51. | Tanaka S, Kamachi Y, Tanouchi A, Hamada H, Jing N, Kondoh H. Interplay of SOX and POU factors in regulation of the Nestin gene in neural primordial cells. Mol Cell Biol. 2004;24:8834-8846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 52. | Zappone MV, Galli R, Catena R, Meani N, De Biasi S, Mattei E, Tiveron C, Vescovi AL, Lovell-Badge R, Ottolenghi S. Sox2 regulatory sequences direct expression of a (beta)-geo transgene to telencephalic neural stem cells and precursors of the mouse embryo, revealing regionalization of gene expression in CNS stem cells. Development. 2000;127:2367-2382. [PubMed] |
| 53. | Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, Reinbold R. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846-41857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 54. | Shimozaki K, Clemenson GD, Gage FH. Paired related homeobox protein 1 is a regulator of stemness in adult neural stem/progenitor cells. J Neurosci. 2013;33:4066-4075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 55. | Palma V, Lim DA, Dahmane N, Sánchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Alvarez-Buylla A, Ruiz i Altaba A. Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development. 2005;132:335-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 448] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 56. | Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, Schneider-Maunoury S, Alvarez-Buylla A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat Neurosci. 2008;11:277-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 388] [Cited by in RCA: 407] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 57. | Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743-2748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1185] [Cited by in RCA: 1170] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 58. | Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418:892-897. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 59. | Rahnama F, Shimokawa T, Lauth M, Finta C, Kogerman P, Teglund S, Toftgård R, Zaphiropoulos PG. Inhibition of GLI1 gene activation by Patched1. Biochem J. 2006;394:19-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 60. | Takanaga H, Tsuchida-Straeten N, Nishide K, Watanabe A, Aburatani H, Kondo T. Gli2 is a novel regulator of sox2 expression in telencephalic neuroepithelial cells. Stem Cells. 2009;27:165-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 61. | Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 385] [Cited by in RCA: 361] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 62. | Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36:1021-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 806] [Cited by in RCA: 843] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 63. | Hu Q, Zhang L, Wen J, Wang S, Li M, Feng R, Yang X, Li L. The EGF receptor-sox2-EGF receptor feedback loop positively regulates the self-renewal of neural precursor cells. Stem Cells. 2010;28:279-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 64. | Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 417] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 65. | Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 66. | Yu RT, McKeown M, Evans RM, Umesono K. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature. 1994;370:375-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 161] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 67. | Shimozaki K, Zhang CL, Suh H, Denli AM, Evans RM, Gage FH. SRY-box-containing gene 2 regulation of nuclear receptor tailless (Tlx) transcription in adult neural stem cells. J Biol Chem. 2012;287:5969-5978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 68. | Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, Moore L, Nakashima K, Asashima M, Gage FH. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12:1097-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 504] [Article Influence: 31.5] [Reference Citation Analysis (0)] |