INTRODUCTION: KIDNEY DISEASES AND THE NOTION OF THERAPEUTIC USES OF INDUCED PLURIPOTENT CELLS FOR RENAL REPLACEMENT THERAPY
Kidney organs perform essential physiological roles in excretion and homeostasis[1]. Kidney diseases can arise during development, juvenile, or adult life. Types of renal disease include acute kidney injury (AKI), which is the abrupt loss of renal function that can often become permanent, and chronic kidney disease (CKD), the progressive loss of renal function that culminates in organ failure known as end stage renal disease (ESRD)[1,2]. The need for new treatments for kidney disease, the 8th leading cause of death in the United States[3], is a growing concern in the medical field. For example, there are approximately 31 million people in the United States diagnosed with CKD[4]. Unfortunately, kidney diseases are a global health problem as well, and have continued to increase in incidence in correlation with the rise in aged populations and escalation in conditions like diabetes that often negatively impact renal health[5-8]. At present, kidney disease treatments deal with symptom management through the renal replacement therapies of dialysis or organ transplant. However, formulating therapies that repair kidney structure and restore compromised functionality is of the upmost importance considering the limited numbers of viable kidneys available for transplant, as well as the complications that can arise in organ recipients[9-13]. To identify innovative ways to combat kidney diseases, numerous research groups have focused their energies on the identification of adult renal stem cells[14-17]. However, this has remained a controversial topic despite the multitude of studies performed to date[14-17]. In addition to the search for endogenous renal stem cells, the study of renal lineage specification during kidney organogenesis has been pursued-knowledge which can be applied toward the development of cell-based therapies for the purpose of kidney regeneration that would not necessitate the employment of adult stem cells.
One such cell-based alternative is the use of induced pluripotent stem cells (iPS) derived from the patient’s own tissue. iPS cells can be used to study development and cell differentiation without the need for embryonic stem (ES) cell lines, whose cell source carries with it a surplus of ethical concerns, and can provide a resource to help researchers with disease modeling and drug development[9]. Using iPS cells from the patient’s renal tissue can serve to circumvent the need for a kidney transplant and avoid the use of lifelong immunosuppressant drug treatments. Thus, the notion of iPS-based regenerative medicine has many appealing benefits if the paramount challenges associated with realization of such cell-based therapies can be overcome. Utilizing integrating viral vectors containing the “Yamanaka factors” to reprogram cells has shown substantial success in generating iPS cells (approximately 0.1%), but the fact that these viral vectors integrate into the genome (sometimes in large copy number) has been a serious cause for debate as to their toxicity and their relative capability to be used in a clinical setting. Researchers have also investigated other avenues such as the use of non-integrating vectors so as to make the iPS cells safer to use in cell therapies, but with limited success, as evidenced by the very low induction rates and relative efficiency of the reprogramming method (approximately 0.001%). Making safer and more controllable iPS cells is an integral part of developing cell-based therapies for the treatment of diseases and injuries. For example, Abad et al[18] shows evidence of how uncontrolled reprogramming can affect the body in the form of teratomas developing in multiple organs of transgenic mice transiently expressing the four “Yamanaka factors”. Other alternatives to the use of reprogramming factors are also being investigated, such as the use of microRNAs (miRNAs) to generate iPS cells. This method shows much promise, even though the cells’ behavior in vivo still has to be controlled (approximately 10% efficiency reported in previous studies)[19].
For the purposes of treating kidney disease, researchers have been assessing different ways of obtaining renal progenitor cells, and one such way involves partial reprogramming of differentiated renal cells into a renal progenitor state. Experimental evidence has supported the notion that the more closely related the start and end cells types are, the more efficient the reprogramming process will be. Although the method proved to be better than most at producing reprogrammed cells (approximately 0.875%)[20], the overall amount of progenitors produced is still not cost-effective enough to be of applicable merit for therapeutic purposes. Another drawback to this partial reprogramming method is the thorough screening process that has to be applied in order to find the adequate combination of genes that will successfully reprogram the kidney cells into a progenitor-like state, which would be both time-consuming and costly. A method of obtaining renal progenitors that has received significant attention is the directed differentiation of iPS cells. Typically done with growth factors (which are rather expensive), exciting recent reports have now suggested that certain low-cost chemical compounds can be used to achieve the same goal of directing iPS cells towards a specific renal cell lineage with an approximate 90% conversion rate in one week. Although still dependent on the production of iPS cells, directed differentiation into renal progenitors is still a promising method that can be applied in tandem with a more optimized, efficient, and safer reprogramming protocols. In the following sections we further discuss these and other recent advances, as well as their general impact in the medical field.
REPROGRAMMING METHODS: REVERSE ENGINEERING TO OBTAIN STEM AND OTHER PROGENITOR CELLS FROM DIFFERENTIATED CELLS
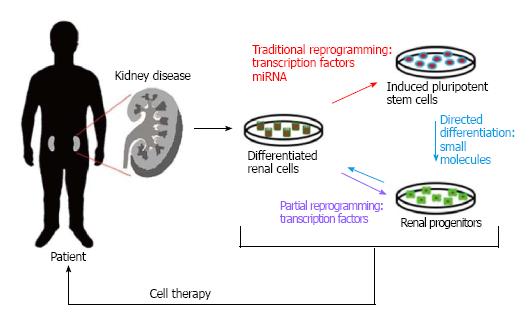
Current therapies directed towards the treatment of kidney disease focus on symptom management instead of treating and hopefully curing the overall condition, and because of this researchers are working on alternatives that may now aid in the restoration of normal kidney function. As aforementioned, one alternative to current methods is the use of reprogrammed cell-based therapies in order to restore damaged or diseased kidneys. Two of the most prominent reprogramming strategies currently being used involve either the conversion of different sources of stem cells into renal progenitors, or the reprogramming of differentiated renal cell populations into a more pluripotent state (Figure 1).
Figure 1 Renal cell reprogramming methods.
(Red) Traditional reprogramming involving the use of transcription factors or miRNAs to generate pluripotent stem cells; (Purple) partial reprogramming with transcription factors to obtain multipotent progenitors; (Blue) directed differentiation into cells with a specific phenotype by treating induced pluripotent stem cells with small molecules. These newly reprogrammed/differentiated cells can then be used therapeutically to replace lost cell types within the injured kidney.
Traditional cell reprogramming involves the overexpression of developmental genes in differentiated adult cells in order to induce an earlier developmental and pluripotent phenotype. The typical factors that are overexpressed for cell reprogramming, discovered by Takahashi et al[22] and Yamanaka et al[22] back in 2006, are OCT4, SOX2, c-MYC, and KLF4 (now deemed “Yamanaka factors”), these factors are typically transfected into cells through the use of lentiviral vectors, which insert these exogenous genes into the host genome. At first, a cocktail of four viral vectors, each one containing one of the previously mentioned “Yamanaka factors” was introduced into the cell in order to promote a change in cell phenotype. However, these techniques lacked efficiency due to many non-specific genomic integrations, as well as the heterogeneous population that resulted from the process (some cells were only partially reprogrammed because not all of the vectors integrated)[21,22]. In terms of kidney disease, producing iPS cells from cells of renal origin would contribute greatly to the development of cell therapies and treatments as they would be predicted to integrate more readily into the diseased kidney due to their conserved epigenetic memory[23].
Interestingly, Zhou et al[24,25] were able to generate iPS cells from human exfoliated renal epithelial cells found in urine, something that can be collected without the need for surgical intervention, which would help in the development of therapies for kidney disease due to their epigenetic memory of renal origin. Using a cocktail of four different retroviruses containing the four “Yamanaka factors” they were able to create iPS cells (in about 16-25 d after transduction) from the previously mentioned cell source (cultured for about a week from 13 different test subjects, which means that the complete protocol would last about a month to produce iPS cells) with varying degrees of reprogramming efficiency (0.01%-4.0%) and ability to differentiate, something that is to be expected when you use multiple integration vectors, mainly because the researcher cannot assess if the cells have incorporated all of the four reprogramming factors, or if the integrated vector copy number is low enough for the reprogrammed cells to be viable for therapeutic purposes (something that was not investigated in this study). Although a reprogramming efficiency of 4.0% is relatively high compared to other studies, the fact that there is great variation between the iPS cells produced (evidenced by the varying degrees of reprogramming efficiencies between the different donors, and that not a lot of the iPS cells produced could differentiate into other cells types) greatly diminishes their clinical applications, and provides further evidence that utilizing multiple integration vectors to reprogram cells is not an effective method for producing iPS cells for therapeutic purposes.
Researchers have tried to address this issue by trying create a better method of reprogramming that could decrease the number of genomic integrations, and assure that all of the factors necessary for reprogramming are being expressed within the cell. Sommer et al[26] managed to do just this by creating a single lentiviral vector containing all of the “Yamanaka factors” which is able to convert mouse postnatal fibroblasts into iPS cells. Not only were they able to ensure that all four transcription factors were integrated into the cell’s genome, but they were able to reduce the amount of genomic integrations to a mean of about 1.5-2.8 proviral copies[26]. Compared to the multiple vector approach, this single vector method has an increased efficiency of 0.5%, which is about 50 times more efficient (relative efficiency of the multiple vector method is about 0.01%-0.05%), but this number might vary between cell types due to the many unique properties found in the various tissues throughout the body. Fortunately iPS cells have already been created from cell sources from cells of distinct embryonic origins (endoderm, mesoderm, ectoderm). This will benefit the development of regeneration cell therapies as iPS cells derived from the affected or injured organ will work more effectively in cell therapies that intend to regenerate the particular organ of interest, because of the genome-wide epigenetic memory of the differentiated adult cell that is to be reprogrammed[27].
Working under the previously stated premise (specifically in the case of renal disease), Wang et al[23] were able to generate iPS cells from mouse renal tubular epithelial cells (RTECs) using a single lentiviral vector containing the previously mentioned “Yamanaka factors” in about 21 d. The cells produced in this study were relatively indistinguishable from mouse ES cells, as confirmed by morphological, immunocytochemical, genetic expression, and karyotype analysis[23]. Not only did these cells adopt an ES-like morphology and were able to express undifferentiated ES cell markers such as fibroblast growth factor 4 (FGF-4) and NANOG, but they were also able to differentiate into cell types of all three germ layers, as evidenced by the presence of AFP, desmin, and nestin (endoderm, mesoderm, and ectoderm markers respectively) in embryoid bodies formed from said cells[23]. The cells also exhibited a normal 40XY karyotype and once reprogrammed, the viral transgenes were largely silenced which is necessary if there is any chance of applying this method in clinical applications, mainly to avoid problems during differentiation that might result in tumor development; the relative efficiency of this method, however, leaves something to be desired (0.1%)[23].
Another reprogramming strategy that researchers have pursued is partial reprogramming of cells into a more multipotent phenotype that can produce cell lineages of a specific organ structure, which in the case of Hendry et al[20] would be embryonic nephron progenitors (NPs). The efficiency of the reprogramming process is correlated to the lineage relationship between the start and end cell types, in other words, the more closely related the start and end cells types are, the more efficient the reprogramming process[20]. Hendry and colleagues investigated this premise by trying to generate NPs from HK2 cells line (human kidney cell line; adult proximal tubule cells)[20]. Through combinatorial screening of 15 different transcription factors associated with the specification of the nephron progenitor phenotype they were able to identify 6 (SIX1, SIX2, OSR1, EYA1, HOXA11, SNAI2) genes that would recapitulate the network of genes associated with the cap mesenchyme (CM)[20]. Each factor was packed into individual lentiviral constructs accompanied by green fluorescent protein (GFP) to identify successfully infected cells, and successful reprogramming events were defined by significant morphological changes as well as robust expression of SIX2 and Cbp/P300-interacting transactivator 1 (CITED1) protein, CM-specific markers[20]. Reprogrammed cells showed upregulation of Matrix metalloproteinase 9 and 2 (MMP9 and MMP2), epithelial-to-mesenchymal transition (EMT) markers, as well as repressed expression of epithelial cadherin (E-CADHERIN), which suggests the occurrence of an EMT event within these cells[20].
Further evidence of the cells’ conversion into nephron progenitors can be seen in a recombination assay that was developed to test the induced NPs’ potential in ex vivo organoid cultures[28,29]. They found that the induced progenitors were able to integrate with the endogenous NP field, and failed to integrate into the uretic bud compartments (a cellular population that the CM does not make). The overall efficiency of this partial reprogramming process is about 0.875%, which is substantially better than many of the techniques discussed so far (most likely due to the close relation between adult proximal tubule cells and NPs), however, the use of multiple lentiviral constructs makes the use of these cells quite toxic; therefore integrating all of these factors into a single construct might increase the efficiency of the reprogramming quite drastically, as well as their potential for use in therapies.
TANGENTIAL AND NON-INTEGRATION METHODS OF REPROGRAMMING
The use of integrating viral vectors has become quite widespread in the field of cell reprogramming, but because of various concerns that have arisen during their use (interruption of the cell’s genome and/or the risk spontaneous reactivation of the viral genome that might lead to tumor formation) researchers are actively looking for different alternatives so as to decrease the risk of using reprogrammed cells in the treatments of diseases such as end-stage renal disease (ESRD). Nightingale et al[30] (2006) were able to produce a non-integrating lentiviral vector that was able to transiently express GFP in about 90% of cultured human T lymphoid cells for approximately 20 d, which speaks to the potential of non-integrating vectors[30]. In 2008, Stadtfeld et al[31] were able to generate mouse iPS cells from fibroblasts and liver cells by using non-integrating adenoviruses that transiently expressed the four “Yamanaka factors”. The cells were showed distinct characteristics of pluripotency such as the expression of endogenous pluripotency genes, demethylation of Oct4 and Nanog promoters, and the ability to produce teratomas in vivo and contribute to all three germ layers[31]. Even though the infection efficiency of the adenoviral vectors was relatively high (50%-60% for quadruple infected cells), the overall reprogramming efficiency this non-integrating method was between 0.0001% to 0.001% (significantly lower than integrating viral methods; 0.1% on average)[31], something that is probably due to the rapid dilution of the adenoviruses during cell division which results in the cells not being exposed to the reprogramming factors for an adequate amount of time so as to induce a successful change in phenotype.
Another example of iPS cells created by non-integrating vectors can be seen in Guarino et al[32]. Yu and colleagues were able to create human iPS cells by utilizing three modified episomal vectors containing different combinations of six reprogramming factors (OCT4, SOX2, NANOG, LIN28, c-MYC, and KLF4) and the SV40 large T gene (SV40LT) to counteract the toxic effects of c-MYC expression, a cis-acting oriP element and an Epstein-Barr nuclear antigen 1 (EBNA1) gene[32]. The latter of these elements provided the vector with the ability to be transfected without the need for viral packaging and to be stably replicated outside of the chromosome[32]. The factors packaged inside the vectors were linked by the internal ribosome entry site 2 (IRES2), and this was done in order to increase reprogramming efficiency by coexpressing them[32]. Utilizing these vectors researchers were able to make iPS cells that exhibited typical ES cell colony morphology and gene expression profile, and they were able to produce teratomas in vivo that contained differentiated derivatives of all three germ layers[32]. Subclones of the reprogrammed cells showed no signs of the vector or transgene sequences other than the change in phenotype, which is an incredible accomplishment, the reprogramming efficiency of the method however, is rather low (about three to six colonies per 106 input cells)[32].
Although non-integrating vectors are a good alternative in order to produce safer iPS cells for use in treatments, they are not very cost-efficient considering that these methods and vectors produce very low amounts of reprogrammed cells. Another alternate method that has seen a lot of attention in recent years is the use of miRNAs instead of exogenous transcription factors as a means of reprogramming[19]. Wang et al[19] used a lentiviral vector containing miR302/367, a unique cluster of miRNAs that is highly expressed in EM cells, in order to produce iPS cells from human embryonic kidney (HEK) 293T cells and found that these reprogrammed cells generated ES-like colonies, showed increased expression of ES cell markers (SOX2, KLF4, c-MYC, OCT4, LIN28 and NANOG), could form embryoid bodies, and could differentiate into germ-like cells in vitro and in vivo. So as to improve the differentiation potential of the miRNA-induced iPS cells researchers cultured the HEK293T cells in serum-free media, as well as in the presence of two small molecules: vitamin C and fibroblast growth factor (bFGF) so as to better shape the morphology of the reprogrammed cells[19]. Although the overall efficiency of the reprogramming method described is yet to be determined, previous reprogramming studies with miRNAs have demonstrated this type of approach to be more efficient than the standard reprogramming factor methods (10% vs 0.1%, respectively)[33], making this type of method a promising candidate for further studies.
FORWARD THINKING: OBTAINING RENAL PROGENITORS USING LOW-COST AND EFFICACIOUS SMALL MOLECULES
In the endeavor to create renal progenitors, controlled differentiation of iPS cells has become a good alternative to partial reprogramming of differentiated cells. One particular technique that stands out is the use of small molecules in order to induce a more renal-specific pluripotent state. Lam et al[34] created an intermediate mesoderm (IM)-specific differentiation platform around the small molecule CHIR99021, a glycogen synthase kinase-3β inhibitor (CHIR). This inhibitor manages to recapitulate mesendoderm formation during development in human pluripotent stem cells (hPSCs), as evidenced by the compounds ability to produce cell lineages that transiently expressed various primitive streak genes such as BRACHY, MIXL1, FOXA2, EOMES, and GSC[34]. The transient expression pattern of these genes in the CHIR-treated cells during a 72 h period is also consistent with that found in cells during the course of gastrulation, which means that CHIR99021 imitates normal developmental mimetics[34].
Utilizing this compound researchers were able to screen various exogenous factors in order to determine the minimum requirements needed promote differentiation of these CHIR-induced mesendoderm-like cells toward IM[34]. They reported that fibroblast growth factor-2 (FGF2) in combination with retinoic acid (RA) was able to induce IM differentiation in the mesendoderm-like cells. This conclusion was drawn from the fact that the treated cells were both PAX2 and LHX1 positive, two markers for which coexpression in the same domain has only been described in the developing kidney and dorsal spinal cord. Further evidence that these PAX2+LHX1+ cells were directed to an IM state and that they could produce IM-derived cell populations and tissues came in the form of tubule structures (with primary cilia) expressing proximal tubular markers once the exogenous FGF2 and RA were removed from the culture media. One of the many differentiated kidney markers whose expression was evaluated in the PAX2+LHX1+ cells was SIX2, a multipotent nephron progenitor cell marker. This nephron progenitor population composes what is known as the CM and these give rise to nearly all epithelial cell types in the nephron tubule, with the exception of those from the collecting duct. Lam et al[34] were able to use the double positive IM-like cells in order to screen different growth factors so as to identify the conditions that promote and sustain a SIX2+ cell population, and they were able to determine that the addition of FGF9 and Activin A could do just this, as well as induce the expression of other CM markers such as SALL1 and WT1. Although researchers were able to effectively produce IM-like cells that are able to differentiate into subsequent renal cell populations, the need for exogenous growth factors is still an issue due to that fact that these very same growth factors are incredibly expensive, and therefore not very cost-effective to use in clinical applications.
Araoka et al[35] on the other hand, utilized a combination consisting of only small molecules, as opposed to small molecules and growth factors, in order reach the same goal. In this particular strategy the mesendoderm stage is skipped altogether and the hPSCs are differentiated directly into an IM state. Using high throughput chemical screening they were able to identify two compounds that increased induction of Odd-skipped related 1 (Osr1), a transcriptional regulator that is expressed in the embryonic day 7.5 IM until kidney organogenesis and therefore a good marker to utilize in order to identify IM cells. The two compounds identified were AM580 and TTNPB, RA receptor antagonists (RAR) that induce differentiation of hPSCs into OSR1+ IM cells with relatively high efficiency (> 60% and > 50% respectively) when compared to positive controls. To further optimize OSR1 induction researchers combined each RAR with CHIR, which resulted in an increased induction rate of around 80% in only 5 d utilizing only two chemicals in a serum –free environment.
As mentioned before, one of the main differences between the methods used Araoka et al[35] and Lam et al[34] is that the former can skip the mesendoderm stage altogether. This was demonstrated when researchers analyzed mesendoderm markers (BRACHYURY, GOOSECOID, and MIXL1) in the small molecule-treated hPSCs, and found that the induction rate for BRACHYURY+ cells was around 6%, and that expression levels for said markers were very low in cells produced from the small molecule method when compared to cells produced with CHIR and growth factor activin A. The ability of these IM-like cells to produce the various IM-derivative cell types was also evaluated, and after additional days of differentiation researchers found that these induced IM cells did in fact produce cells expressing marker genes for various IM-derivative cell types such as FOXD1, SALL4, GATA4, among others. These cells also had the ability to give rise to the derivative cell types in vivo, as well as renal tubule-like structures positive for renal tubule markers such as Lotus tetragonolobus lectin (LTL), E-CADHERIN, and laminin in vitro.
Both of these studies[34,35] provide evidence that utilizing small molecules in order to produce renal progenitors for cell therapies is a viable option in the field of regenerative medicine, and the various benefits that this type of method provides makes it a good alternative to explore. Utilizing these chemical compounds is not only less costly, but more efficient in terms of number of cells converted to the desired phenotype (even though the reprogramming efficiencies for the template iPS cells were not stated in either one of the studies). Unfortunately there is still some variability between studies that needs to be addressed before any progress can be made on any viable therapeutic solution.
Both of the methods described above are highly efficient for IM differentiation of hPSCs in terms of the time the procedure takes, the markers analyzed, and the compounds used[34,35]. Lam et al[34] utilizes a method that has both chemical compounds and growth factors, but only takes 3 d to produce IM cells. Araoka et al[35] on the other hand only use chemical compounds, but take about two more days in order to reach the same goal. In terms of markers utilized the former uses a combination of LHX1 and PAX2 (a pair of markers that, as stated previously, are only found in the developing kidney and dorsal spinal cord), while the latter uses an engineered OSR1-GFP human iPS cell line to verify if the cells have reached an IM state, a gene that is also expressed in the lateral plate mesoderm and can therefore provide some heterogeneity to the sample that might alter the results of future studies.
CONCLUSION
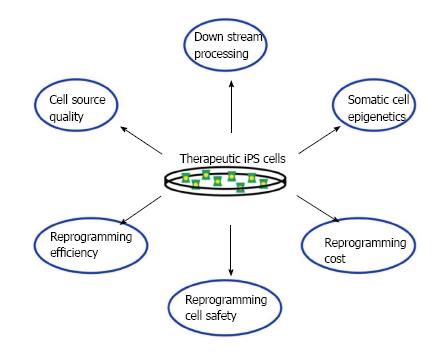
Recent progress in knowledge about cellular reprogramming has rapidly advanced prospects for the development of regenerative therapies for the medical treatment of many conditions, among them being kidney diseases, making this a very exciting time in the field of nephrology. Here, we discussed a number of research studies in the field of stem cell reprogramming. We explored how such methods have been utilized to reprogram renal lineages, and thus might be used to develop therapies to treat kidney disease. Additionally, iPS cells can be used for disease modeling to identify targeted therapeutics for heritable conditions[36]. Moving forward, there are a number of complex issues to further resolve about the therapeutic application of iPS cells for disease treatment, and most assuredly other issues yet to be identified, which apply both to the kidney and other organs within the body (Figure 2).
Figure 2 Challenges faced when developing induced pluripotent stem cell-based therapies.
Many issues still have to be overcome before any effective cell therapies can be used.
Issues involved in the therapeutic application of reprogrammed cells include the number and type of cells needed, along with the identification of an appropriate delivery system for the condition to be treated. Currently, there are various ongoing clinical trials in the United States that are using stem cells to treat a wide range of conditions such as age-related macular degeneration to polycystic kidney disease[37]. The amount of cells utilized by these studies can fluctuate between the stem cells type and the way they are used (50000-200000 human embryonic stem cells in retinal cell transplants and 2 × 106 mesenchymal stem cells (MSCs)/kg of patient’s weight in kidney disease treatments[37,38]), but even so the amount of pluripotent cells produced by the methods mentioned in this review are still relatively low when compared to the amount used in the before-mentioned trial therapies.
Other issues that still need to be addressed are cell quality (can you isolate healthy renal cells to reprogram as opposed to the diseased ones?) and downstream processing, a problem because, due to ethical reasons, many of the pluripotency tests that are usually performed on reprogrammed cells can’t be done with human iPS cells, which might create some heterogeneity within the human iPS cell lines. Also, we have barely scratched the surface of how epigenetic memory affects iPS cell differentiation patterns. All of these concerns still need to be investigated before adequate therapies can be developed (Figure 2).
Although there remains a sizable amount of work to be done in order to optimize the efficiency of these methods, they still represent a promising alternative to current therapies, mainly because they have the potential to provide the affected patient with the means to regain kidney function without the need for a kidney transplant or dialysis. It would be interesting to see how these methods would be affected if they were done with other animal models, such as in the zebrafish, an organism that has the capacity to regenerate renal tissue[39-41], and what type of information can be learned from animal models about how reprogramming methods can be optimized or the nature of renal progenitors[42]. As more insights continue to be gathered about the genetic mechanisms of renal lineage development and regeneration in various vertebrate models, as represented for example by recent reports in the zebrafish[43-45], frog[46], and mouse[47], crucial information may be elucidated about potent methods to regulate renal reprogramming or even to promote pathways of endogenous cell regeneration in the damaged kidney.
Moving forward, there may be significant challenges for cell-based therapies posed by the microenvironment in the damaged kidney-termed by some as the “seed and soil” dilemma. Namely, the importance of an appropriate microenvironment, or niche, the so-called “soil”, is essential for the prosperity and normal growth of the stem cells, or “seeds”, to be administered in a putative treatment[48]. The complexity of renal anatomy and composition alone may pose significant hurdles to cell-based therapies, and can be further complicated if the environment due to the disease state is refractory to the success of the regenerative therapy. In sum, altering the microenvironment to facilitate success of the cellular therapy is likely to be vital.
One promising avenue is the utilization of other stem cells, e.g., MSCs, which have been shown in a number of contexts to stimulate a local, if not organismal, humoral environment that facilitates regeneration[37,38]. The kidney is in fact among such organs whose status can be improved by MSCs in some disease settings[49]. In animal models of AKI, administration of MSCs has provided renoprotective effects[50-53]. Notably, a limitation that has been recognized is the inability of MSCs to mediate improvements in chronic renal disease states[54]. These observations indicate that much remains to be learned about how to facilitate cell-based therapies with approaches that address the complex variables associated with any given disease state. Thus, it is imperative that future research is performed to better understand the relationships and physiological impacts of disease states within organisms. Nevertheless, the progress in stem cell biology to date continues to fuel enthusiasm that methods like reprogramming can be harnessed to improve quality of life and relieve suffering in the decades to come.
ACKNOWLEDGMENTS
We thank the staffs of the Notre Dame Department of Biological Sciences and Office of Research for their support, and thank the Center for Zebrafish Research at Notre Dame for their outstanding dedication in the care and welfare of our zebrafish colony. Finally, we thank our research lab for their comments, discussions, and insights about this work, and our families for their constant love and support.