Published online Jul 26, 2012. doi: 10.4252/wjsc.v4.i7.71
Revised: April 16, 2012
Accepted: April 25, 2012
Published online: July 26, 2012
AIM: The generation and characterization of a human embryonic stem cell (hESC) line stably expressing red fluorescent mCherry protein.
METHODS: Lentiviral transduction of a ubiquitously-expressed human EF-1α promoter driven mCherry transgene was performed in MEL2 hESC. Red fluore-scence was assessed by immunofluorescence and flow cytometry. Pluripotency of stably transduced hESC was determined by immunofluorescent pluripotency marker expression, flow cytometry, teratoma assays and embryoid body-based differentiation followed by reverse transcriptase-polymerase chain reaction. Quantification of cell motility and survival was performed with time lapse microscopy.
RESULTS: Constitutively fluorescently-labeled hESCs are useful tools for facile in vitro and in vivo tracking of survival, motility and cell spreading on various surfaces before and after differentiation. Here we describe the generation and characterization of a hESC line (MEL2) stably expressing red fluorescent protein, mCherry. This line was generated by random integration of a fluorescent protein-expressing cassette, driven by the ubiquitously-expressed human EF-1α promoter. Stably transfected MEL2-mCherry hESC were shown to express pluripotency markers in the nucleus (POU5F1/OCT4, NANOG and SOX2) and on the cell surface (SSEA4, TRA1-60 and TG30/CD9) and were shown to maintain a normal karyotype in long-term (for at least 35 passages) culture. MEL2-mCherry hESC further readily differentiated into representative cell types of the three germ layers in embryoid body and teratoma based assays and, importantly, maintained robust mCherry expression throughout differentiation. The cell line was next adapted to single-cell passaging, rendering it compatible with numerous bioengineering applications such as measurement of cell motility and cell spreading on various protein modified surfaces, quantification of cell attachment to nanoparticles and rapid estimation of cell survival.
CONCLUSION: The MEL2-mCherry hESC line conforms to the criteria of bona fide pluripotent stem cells and maintains red fluorescence throughout differentiation, making it a useful tool for bioengineering and in vivo tracking experiments.
- Citation: Ovchinnikov DA, Turner JP, Titmarsh DM, Thakar NY, Sin DC, Cooper-White JJ, Wolvetang EJ. Generation of a human embryonic stem cell line stably expressing high levels of the fluorescent protein mCherry. World J Stem Cells 2012; 4(7): 71-79
- URL: https://www.wjgnet.com/1948-0210/full/v4/i7/71.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v4.i7.71
Human embryonic stem cells (hESC) hold great promise for use in regenerative medicine therapies because of their intrinsic properties of indefinite self-renewal and pluripotency (ability to differentiate into endoderm, ectoderm and mesoderm derivatives). As hESC steadily progress towards clinical applications there is a need to track hESC in vitro and in vivo and to optimise hESC culture expansion and differentiation protocols. To enable this, there is an increasing need for well-characterized, bona fide, genetically stable, fluorescently-tagged hESC lines that both fluoresce as undifferentiated cells and do not silence expression of the fluorescent protein upon differentiation. Unfortunately, one of the properties that distinguishes hESCs from most other cell lines is their tendency to progressively silence exogenous sequences, i.e., transgenes, that are integrated into the cell’s genome[1,2]. Furthermore, because the chromatin in ESCs largely exists in a “poised” state marked by bivalent histone modifications, transgenes are frequently silenced upon differentiation when genetic programs for specific cell lineages are closed as cells progress to a terminally differentiated state. Perhaps for these reasons, at present only a limited number of cell lines with stable robust expression of a constitutively active green fluorescent transgene have been described and are available for use by the hESC research community[3,4]. We therefore set out to produce and characterize a hESC line that robustly expresses high levels of a red fluorescent protein, mCherry, that has an emission spectrum that is spectrally separated from most commonly used fluorophores such as FITC. Here we show that this line, which we have termed MEL2-mCherry, maintains all characteristics of the original MEL2 hESC line, such as expression of pluripotency markers, self renewal over prolonged periods of time, preservation of a normal euploid karyotype and differentiation into representative cell types of the three germ layers, while maintaining mCherry expression. We have further adapted the MEL2-mCherry line to single-cell passaging and demonstrate its utility in cell tracking experiments under various experimental settings. The MEL2-mCherry line, which was purposefully generated in an IP-unencumbered cellular background, will therefore prove a useful tool for the hESC research community.
We chose the MEL2 hESC female (XX) IP-unen-cumbered cell line, generated by Stem Cell Sciences Ltd and previously characterized by the International Stem Cell Initiative[5], as the host for a fluorescent protein-encoding transgene. MEL2 hESC were cultured in KOSR medium on mouse embryonic fibroblasts (MEFs) as described previously. For single cell passage adaptation, MEL2-mCherry hESC were cultured in mTeSR1 medium on Matrigel (1/100 dilution) and passaged weekly using gentle TrypLE (Invitrogen) trypsin substitute digestion (5 min at 25 °C). ROCK inhibitor Y-27632 (Millipore) was included in the medium at 1 μmol/L on each first day after single cell passaging until the culture was single cell-adapted (approximately 3-4 passages).
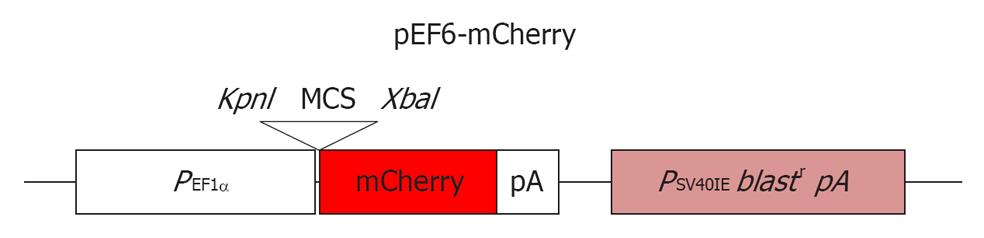
Following standard passaging on MEFs, as described in[6], the MEL2 parental cell line was plated onto a Matrigel (BD Biosciences) substrate (1/100 dilution) and cultured in complete mTeSR1 medium without any additional supplements (Stem Cell Technologies, USA) for 1 d prior to transfection. About 2 μg of pEF6-mCherry plasmid (a gift from Dr. Sweet M, IMB, University of Queensland) (see Figure 1 for plasmid map) was transfected using FuGene6 reagent (1:3 DNA:FuGene6 ratio, Roche Biochemicals) using the manufacturer’s recommendations. Selection with BlasticidinS-HCl (Gibco) at 2 μg/mL was started 3 d after transfection. Resistant colonies uniformly expressing high levels of the mCherry protein were isolated manually and culture expanded. Live FACS sorting of mCherry expressing cells was carried out (using FACSAria, BD Biosciences, USA) and pure sorted cells were reseeded and further expanded for full characterization and cryopreservation.
To confirm appropriate expression of pluripotency-associated genes in the MEL2-mCherry cell line after transgenesis and associated manipulations, we performed immunofluorescence staining as well as flow-cytometric (FACS) analysis, essentially as described previously[7,8]. The primary antibodies used for immunofluorescence were: anti-Oct3/4 (C10, sc5279 SantaCruz, USA) at 1/75 (IF), anti-Sox2 (AB5603, Millipore, USA) at 1/100 (IF), anti-NANOG (9220 Millipore) at 1/150 (IF). Secondary antibodies were used in 1/1000 dilution and were anti-mouse IgG (H + L) AlexaFluor488 (Molecular Probes/Invitrogen). Isotype controls at identical concentrations as the primary antibodies were used to assess non-specific binding. No labeling was detected in isotype control-incubated samples (not shown). Counter-staining of the nuclei was performed using DAPI (4,6-diamidino-2-phenylindole) at 0.1 μg/mL in PBS. For flow cytometry, cells were brought to a single cell suspension using TrypLE (Invitrogen) and stained live with anti-Tra1-60 1/200 (MAB4360, Millipore) or anti-Tra1-81 1/200 (MAB4381 Millipore) or anti-SSEA4 1/400 (MAB4304, Millipore) or anti-TG30 1/400 (MAB4427, Millipore), essentially as described previously (Chung et al[7], 2010). For flow cytometry secondary antibodies, anti-mouse IgG (H + L) AlexaFluor488 or anti-mouse IgM AlexaFluor488 (Molecular Probes/Invitrogen, Carlsbad, USA) were always used in 1/1000 dilution. Isotype control antibodies used at identical concentrations as the primary antibodies were used to set the gates (not shown). Flow cytometry data were collected and analyzed using WEASEL JAVA-based software package (http://www.wehi.edu.au/other_domains/cytometry/).
The differentiation potential of the MEL2-mCherry cell line was assessed using both an in vitro embryoid body formation assay and in vivo teratoma formation. To generate embryoid bodies (EBs), 5 × 104 cells were placed as a single-cell suspension in KOSR medium [20% knockout serum replacement in DMEM/F12 medium (Gibco/Invitrogen, USA)], as described previously[9] in a well of a 6 well ultra low-attachment polystyrene plate (Falcon, USA) and cultured for 2 wk. For the teratoma formation assay, a pellet of 5 × 105 cells was mixed with Matrigel matrix at 1:50 dilution and injected intramuscularly into the thigh muscle of a NOD/SCID mouse. Teratomas were harvested within 4-8 wk; half of it was fixed and processed for paraffin embedding and histological analysis. Haematoxylin/eosin-stained 5 μm sections were permanently mounted, microscopically analysed and imaged on an Olympus IX51 inverted microscope equipped with MicroPublisher 3.3 RTV CCD camera (QImaging, USA). The other half of the teratoma was embedded in OCT compound (Sakura Biotek, USA) via overnight incubations in the 10%-20%-30% gradient of sucrose in PBS and frozen at -80 °C. Sections (6 μm) were cut using a Leica (Leica) cryostat on Superfrost slides (Fisher Scientific). Expression of mCherry was detected using rabbit polyclonal anti-RFP antibody (PM005) from Medical and Biological laboratories (MBL, USA) at 1:500 dilution and secondary anti-rabbit IgG AlexaFluor568 (1:1000 dilution, Molecular Probes/Invitrogen, USA).
To analyse the behavior of the MEL2-mCherry cell line on various substrates, a single-cell suspension of 4 × 104 MEL2-mCherry cells was plated in 100 μL of StemPro (Invitrogen) hES medium in a well of a 96 well plate coated with various protein substrates and on an untreated tissue culture plastic as a control not capable of maintaining efficient hES cell attachment and growth (Substrate 1 in Figure 2). Phase-contrast and fluorescence images were captured using an inverted compound microscope Olympus IX51 (Olympus, Japan) equipped with MicroPublisher 3.3 RTV CCD camera (QImaging. USA).
In order to track and compare hESC colony formation, the MEL2-mCherry cell line was mixed with equal numbers of cells of the parental MEL2 hES line (1 × 104) and seeded into a 6 well plate with either MEFs or Matrigel (BD Biosciences) coating matrix at 1/100 dilution. Images were then captured using Olympus IX81 Corvus-automated microscope equipped with carbon dioxide levels and temperature-controlled chamber (Solent Inc., USA) at 25 min intervals (Figure 2A and B).
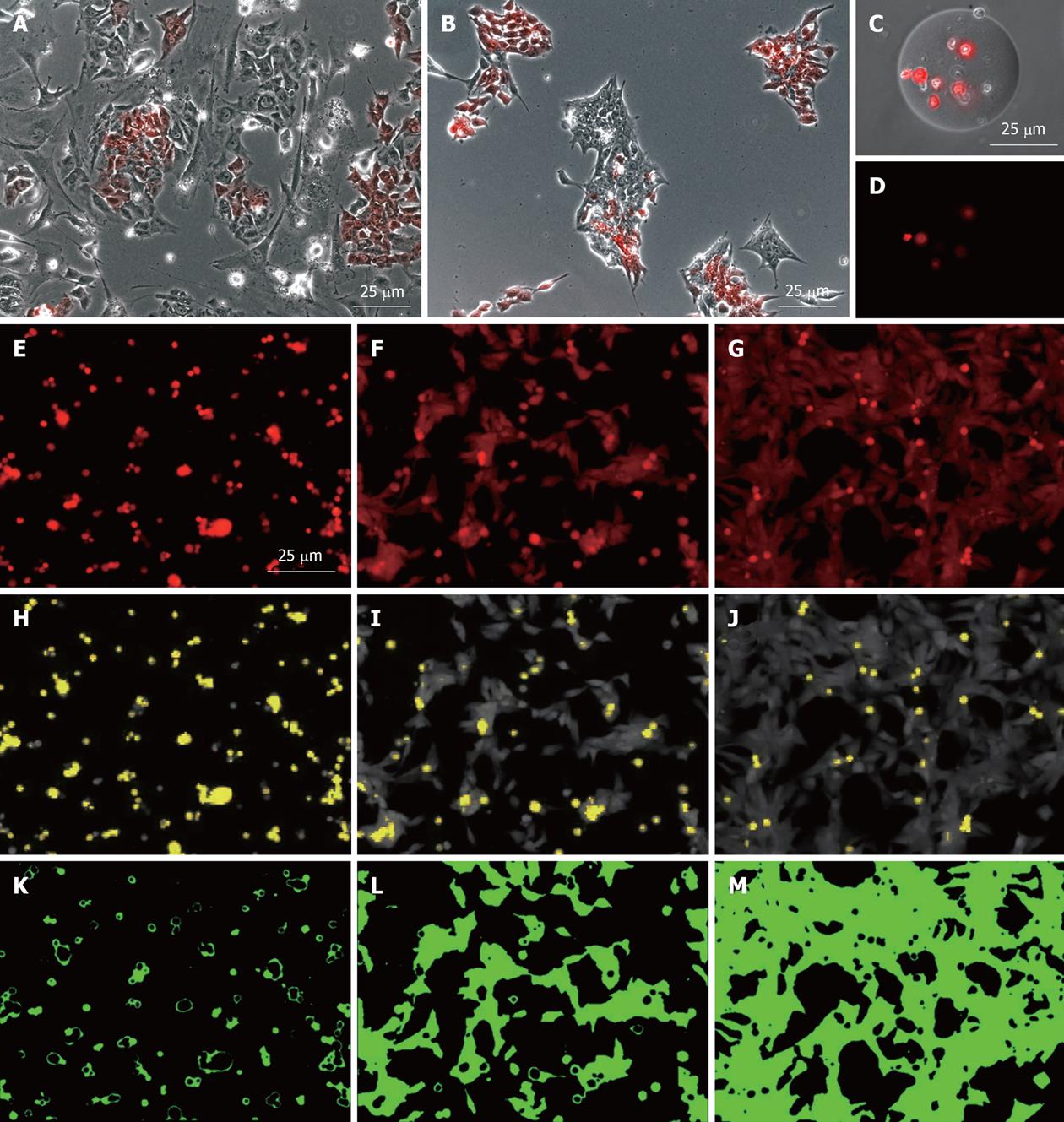
To assess the degree of attachment of cells to various substrates, a simple image analysis algorithm was applied to the analysis of the red channel fluorescent image of the MEL2-mCherry cells 16 h after plating as a single-cell suspension. All analyses were performed using an open-source Java-based freeware ImageJ (v. 1.43 used). Firstly, the area of cell spreading was defined [by utilising the automated background subtraction option (Process > Subtract background), with Rolling Ball diameter set at 40.0 and Smoothing set to “Off”] and manual threshold gating (Figure 2E-M). To define the area occupied by dead cells (retaining high levels of mCherry fluorescence within this time frame), the area occupied by spheroid, and thus perceptually higher mCherry-fluorescent apoptotic or necrotic bodies, was established utilizing the manual threshold gating approach so that normal attached live cells with lower fluorescence density were excluded (Figure 2H-J, shown in yellow).
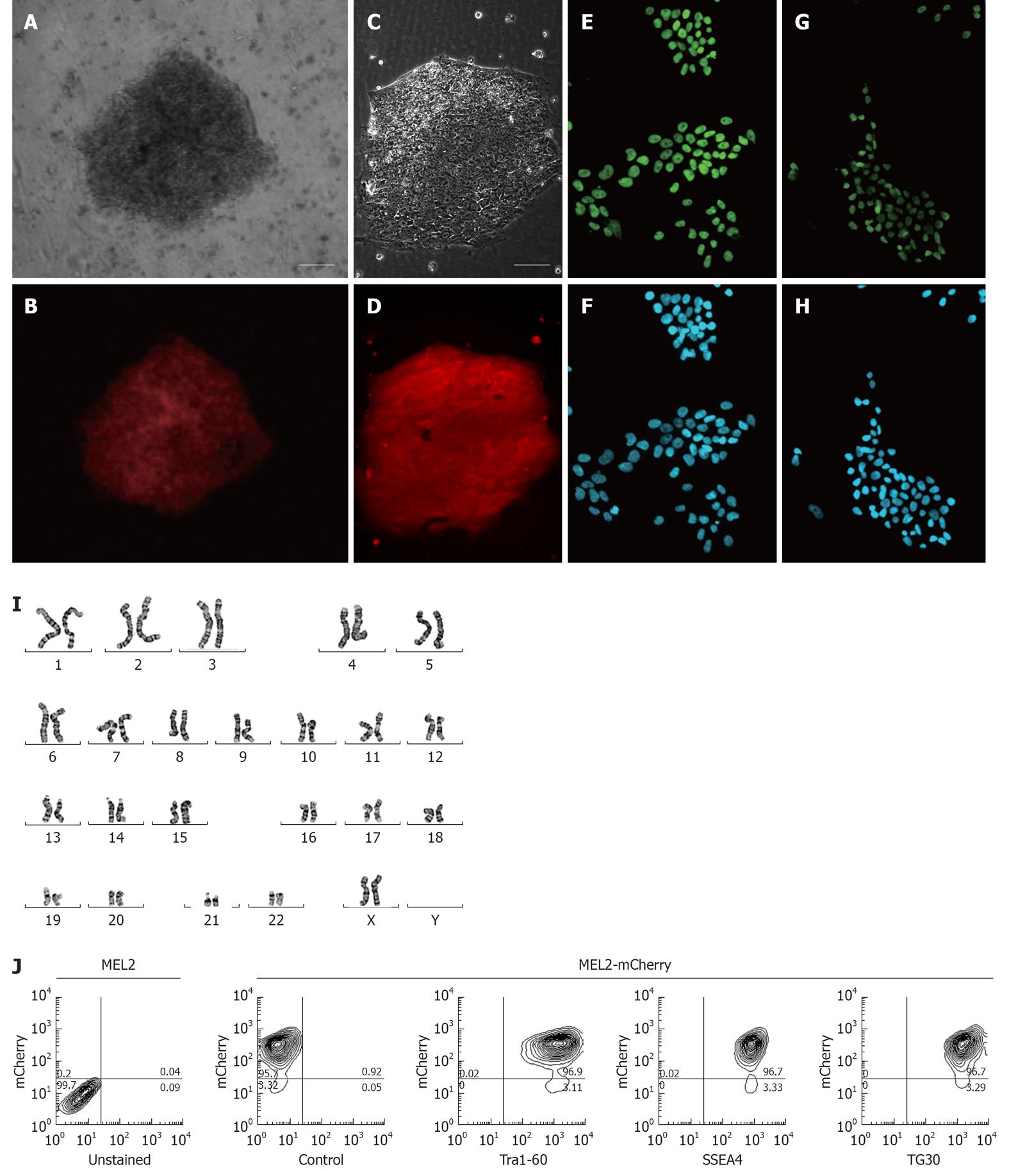
For a fluorescently-tagged hESC line to be useful for cell tracking in vitro and in vivo, it is important to obtain high levels of fluorescent protein expression in both undifferentiated pluripotent and differentiated cells. In order to achieve this, we chose to utilise the promoter of the human translation elongation factor 1 α (EF-1α) gene to drive expression of the red fluorescent mCherry protein. This promoter was previously shown to offer a superior consistency in a wide array of cell types, including hESCs[4], and thus offers the best combination of good expression levels in hESCs and consistent expression across derivatives of the main three basic germ layers[10]. We strategically chose the MEL2 hESC line as a host for transduction with the EF-1α promoter-driven mCherry as this line is IP unencumbered and can be freely distributed. Following transfection of MEL2 hESC with the pEF6-mCherry plasmid, we observed that between 30%-50% of the hESC expressed various levels of mCherry. We next selected highly mCherry expressing clones using Blasticidin resistance and sub-cultured these clones for up to 15 passages. We observed that over these initial passages, individual cells within the colonies inactivated their transgene expression at a low frequency (data not shown), consistent with the previously reported tendency of hESC to inactivate foreign transgenes[1]. However, manual selection of uniformly-red sections of colonies combined with FACS sorting of mCherry-MEL2 cultures allowed us to select subclones that stably and uniformly expressed mCherry fluorescence (Figure 3A-D). In order to make the stable mCherry MEL2 line, which we called MEL2-mCherry, more useful for bioreactor and cell tracking experimentation, we next adapted the line to single cell growth as described in Materials and Methods.
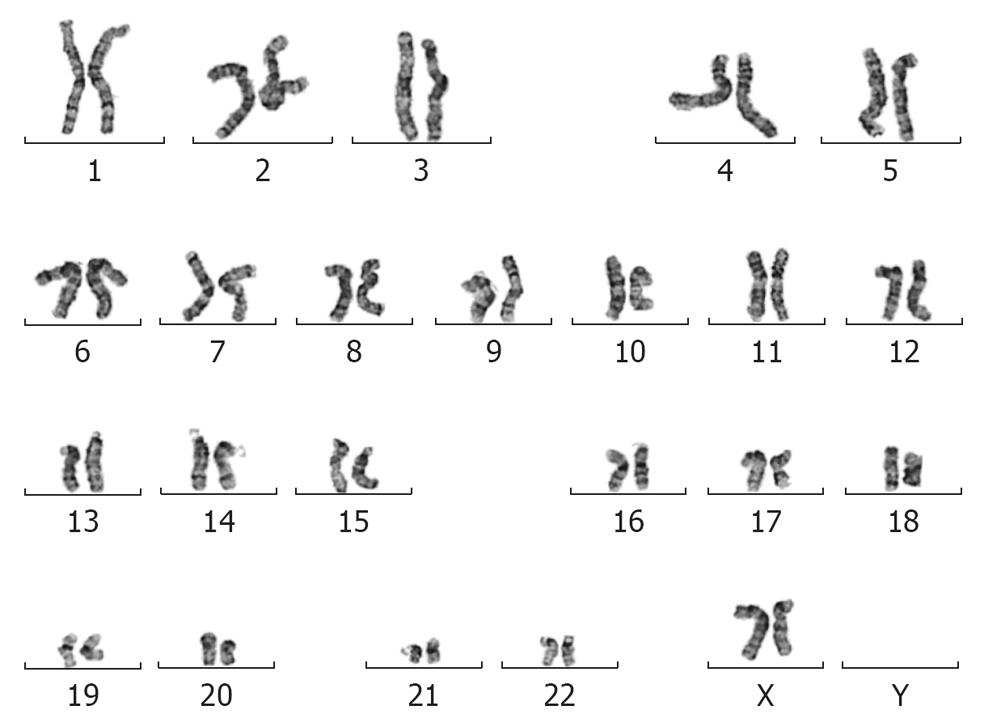
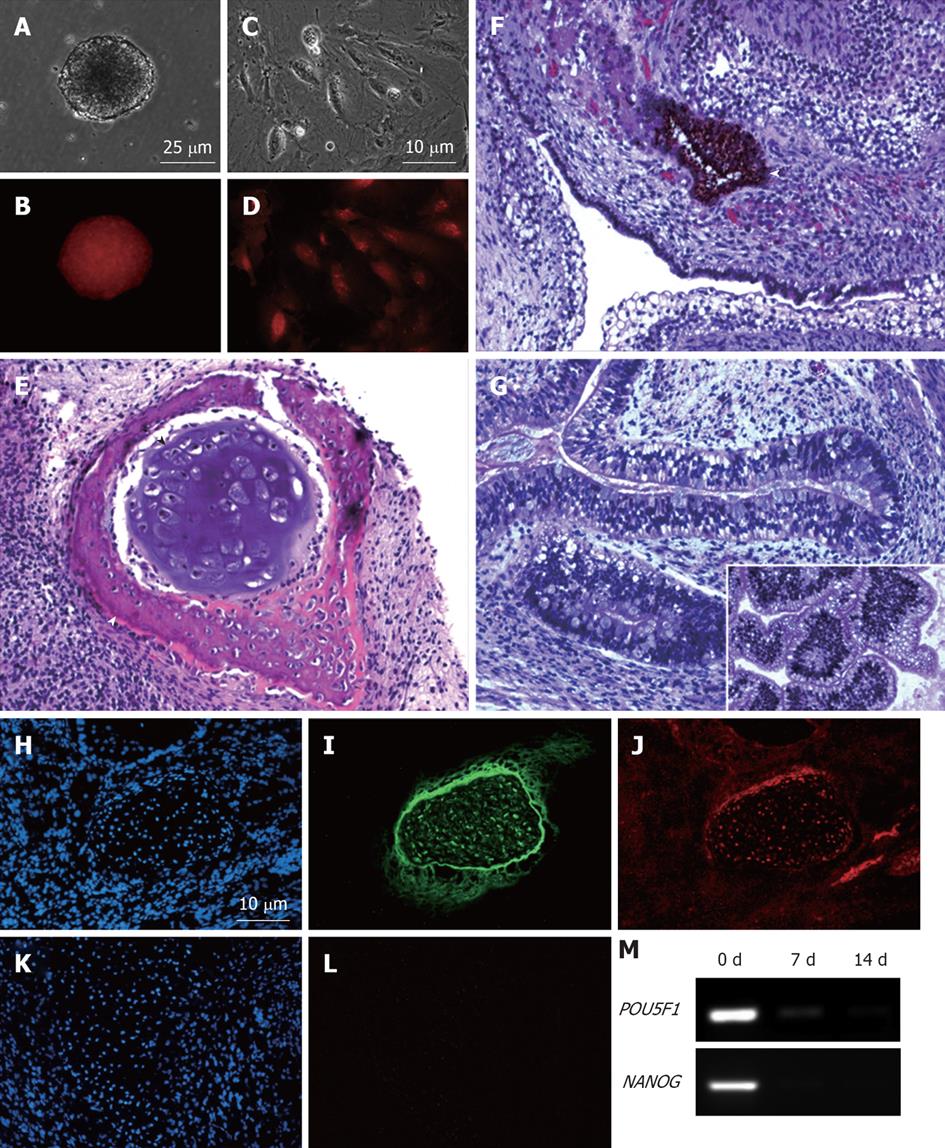
To ascertain whether insertion of the mCherry transgene(s) had interfered with the properties of the parental MEL2 hESC line, we analyzed a number of key pluripotency properties of the newly-generated line. Firstly, we confirmed by immunostaining that the MEL2-mCherry line expresses high levels of nuclear pluripotency-associated transcription factors, such as POU5F1/OCT4 and NANOG (Figure 3E and G). The MEL2-mCherry line also retains high levels of expression of the pluripotency surface markers (Tra1-60, SSEA4, TG30 shown in Figure 3J), as assessed by flow cytometric analysis. MEL2-mCherry hESC display a normal karyotype [analysis of > 15 metaphase spreads at weekly passages 15 (Figure 3I) and 35 (Figure 4)]. Importantly, the MEL2-mCherry cell line continues to express mCherry following extensive differentiation into EBs (Figure 5A and B) and in cells derived from plated EBs (Figure 5C and D). Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of mRNA isolated from EBs differentiated for 0, 1 and 2 wk exemplifies the progressive down-regulation of POU5F1/OCT4 and NANOG mRNA expression and confirms that these genes are no longer detectable in these entirely red fluorescent EBs (Figure 5B and D), suggesting that the line should be useful for in vivo tracking of differentiated hESC derivatives. Furthermore, the MEL2-mCherry line readily formed teratomas in NOD/SCID mice with easily identifiable derivatives of all three primitive germ layers, including ectodermally-derived neural epithelium and melanised retinal epithelium-like structures (Figure 5F), mesoderm-derived cartilage and bone (Figure 5E), and endodermally-derived gut-like epithelium and intestinal crypt-like structures (Figure 5G). Importantly, red fluorescence was maintained throughout the teratoma, indicating that the transgene is not silenced following terminal differentiation into multiple cell types (Figure 5H-L). For example, red fluorescence is clearly detected in chondrocytes (Figure 5I and J), as identified by their morphology and expression of Collagen II (Figure 5I). No significant contributions of undifferentiated cells were observed in these teratomas, as expected for karyotypically-normal hESC. We conclude that MEL2-mCherry hESC retain the key properties of pluripotency, long term self-renewal, differentiation into cell types of the three germ layers and karyotype stability, while maintaining high levels of mCherry expression both before and after differentiation.
Because MEL2-mCherry hESC are highly red fluorescent, this line is extremely well-suited for use in cell-tracking and cell-mixing experiments. It can, for example, be used to explore the mode of colony formation of hESC. At present, it remains to be determined whether hESC form colonies by clonal growth or whether this occurs through cell migration and cell sorting of hESC. This can be elegantly addressed by mixing MEL2-mCherry cells with single cell adapted unlabeled hESC followed by real time assessment of cell behavior over time (Figure 2A and B) (Turner J. Pers comm, manuscript in prep.). The MEL2-mCherry line can also be used to compare and de-convolute cell proliferation, differentiation ability and gene expression of abnormal hESC by simply mixing such cells with the MEL2-mCherry line and then manipulating such mixtures under identical experimental conditions, followed by sorting of the cells on the basis of mCherry expression at the end of each experiment.
MEL2-mCherry hESC are also extremely suitable for analysing hESC behavior and survival after encapsulation within hydrogels or seeding on microcarriers. In such experiments (Figure 2), real time assessment and quantification of cell proliferation, attachment and viability is readily performed (since red fluorescence is rapidly lost following rupture of the plasma membrane) and can be easily quantified by measuring total red fluorescence in culture wells (Sin D. Pers comm, manuscript in prep.). The high level of red fluorescence of the MEL2-mCherry line also greatly aids in the assessment of cellular substrates used in bioengineering applications. As shown in Figure 2, immobilization of MEL2-mCherry cells on engineered surfaces coated with three different extracellular matrix molecules (from left to right) allows for direct, simple and potentially automated assessment of cellular adhesion (Figure 2E-G), cell death (Figure 2H-J) and cell spreading (Figure 2K-M).
HESCs possess great potential for enabling stem cell based therapies and advancing our understanding of very early human development[11]. However, efficient and cost effective methods for expansion and differentiation of these cells compatible with clinical applications are currently lacking, their inherent genetic and epigenetic instability remains a poorly understood problem, immune rejection must be circumvented and their efficacy and long term safety in pre-clinical models still needs to be established[12]. These hurdles are currently being addressed through the development of microcarrier and/or cell encapsulation based culture and differentiation methods[13], engineering of smart surfaces, high throughput screening of small molecules and in depth single cell analysis technologies[14,15]. In order to enable the development of these technologies and allow tracking and interrogation of hESC behavior and gene expression in vitro and in vivo, fluorescently tagged hESC lines are extremely useful as this allows flow cytometric sorting of the cells at will. Here we report on the development of a mCherry expressing MEL hESC line and demonstrate that this line conforms to the criteria expected from bona-fide hESC and maintains red fluorescence both before and after differentiation. As hESC usually efficiently silence exogenous sequences in the undifferentiated state[1,2] and differentiation is known to silence large numbers of genes, e.g.,[16], identifying hESC lines that exhibit stable robust transgene expression in both differentiated and undifferentiated cells, as we have reported here for the MEL2-mCherry line, is a low frequency event. We hypothesise that our protocol has allowed for selection of clones in which transgene(s) insertion(s) into the host DNA is not, or only minimally, detected by the methylation machinery of the hES cell. It will therefore be of interest to map the insertion sites of the E1α-mCherry transgene in the future so as to identify such “safe” harbour sites in the human genome. We next show that the MEL2-mCherry line can be adapted to single cell culture and in this state is extremely useful in examining hESC behavior in both standard culture conditions as well as bioreactor and encapsulation conditions. Indeed, mixing MEL2-mCherry hESC with unlabeled cells allows empirical determination (with potential mathematical deconvolution) of the distribution of compositional variations within hES colonies as a function of the nature of the substrate, plating density, abnormal genotype or other culture conditions. One can also envisage that this line will find utility in toxicity studies, which currently largely rely on staining with viability dyes since red fluorescence is rapidly lost when plasma membrane integrity is lost. By virtue of its persistent red fluorescence following short term in vitro (embryoid body) and long term in vivo differentiation (teratoma), the MEL2-mCherry hESC will further be useful for investigating efficacy, functional integration and safety of hESC based cellular grafts, and constitutes a valuable, freely available IP unencumbered tool for the hESC research community.
We would like to express our gratitude to Rachel Horne and Vani Salvarajan for technical support and Dr Deanne Whitworth for comments on the manuscript. The authors would also like to thank the Australian Stem Cell Centre (ASCC), UQ AIBN Challenge Grant Scheme for financial support and the ASCC’s StemCore pluripotent stem cell core facility for the provision of hES cell lines and logistical support. We would like to thank Matt Sweet for provision of the pEF6-mCherry plasmid.
Human embryonic stem cells (hESCs) are stem cells that can generate every cell type of the human body and can proliferate indefinitely. For these reasons they are attractive stem cells for use in regenerative medicine therapies. To test therapeutic approaches and analyse the behavior of these cells in the dish in real time, one needs to be able to track hESCs in vitro and in vivo. This can be achieved by stably delivering a fluorescent protein to the cells. It is important to show that the expression of such a molecular tracking protein is stable, that it remains expressed after differentiation of the cells and that it does not interfere with the basic properties of hESCs.
hESCs are notoriously difficult to genetically modify and even when an exogenous gene has been inserted the cells have a propensity to silence the delivered transgene. For this reason only very few fluorescently tagged hESC have been reported to date. Here the authors report on the generation of a red fluorescently tagged hESC line that stably expresses high levels of a red fluorescent protein in their undifferentiated and differentiated state.
The immediate advantage of the generation of a red (mCherry)-fluorescently tagged hESC line, as reported in the paper, is that it allows double labeling with commonly used fluorophores such as FITC (green fluorescence). The authors have deliberately chosen to express mCherry in the MEL-2 hESC line that is IP-unencumbered and can therefore be distributed to stem cell researchers worldwide. The authors further demonstrate that this line can be readily adapted to single cell growth, making it extremely useful for analyzing hESC behavior in many applications.
The authors show that the mCherry Mel-2 hESC line can be used for investigating the effect of the cell culture substrate, plating density, genotype or other culture conditions. This line will also find utility in toxicity studies and should prove a be useful tool for investigating efficacy, functional integration and safety of hESC based cellular grafts.
Ovchinnikov et al present the establishment of a hESC line that constitutively expresses mCherry. The advantages of developing such lines are numerous including assessment of proliferation, migration, and in vivo cell tracking. Overall, it is a well organized manuscript with adequate data to support the claims. The hESC line will be a useful tool for the research community.
Peer reviewers: Vladimir Zachar, MD, PhD, Professor, Head, Laboratory for Stem Cell Research, Aalborg University, Fredrik Bajers Vej 3B, 9220 Aalborg Ø, Denmark; Tetsuya Tanaka, PhD, Assistant Professor, Department of animal Sciences, University of Illinois at Urbana-Champaign, 330 ASL, MC-630, 1207 W. Gregory Dr., Urbana, IL 61801, United States; Soo-Hong Lee, PhD, Assistant Professor, College of Life Science, CHA Stem Cell Institute, CHA University, 606-16 Yeoksam 1-dong, Gangnam-gu, Seoul 135-081, South Korea
S- Editor Wang JL L- Editor Roemmele A E- Editor Zheng XM
| 1. | Xia X, Zhang Y, Zieth CR, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Liew CG, Draper JS, Walsh J, Moore H, Andrews PW. Transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1521-1528. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 117] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 3. | Costa M, Dottori M, Ng E, Hawes SM, Sourris K, Jamshidi P, Pera MF, Elefanty AG, Stanley EG. The hESC line Envy expresses high levels of GFP in all differentiated progeny. Nat Methods. 2005;2:259-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 105] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Liu J, Jones KL, Sumer H, Verma PJ. Stable transgene expression in human embryonic stem cells after simple chemical transfection. Mol Reprod Dev. 2009;76:580-586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 793] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 6. | Ellerström C, Strehl R, Noaksson K, Hyllner J, Semb H. Facilitated expansion of human embryonic stem cells by single-cell enzymatic dissociation. Stem Cells. 2007;25:1690-1696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 86] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Chung TL, Turner JP, Thaker NY, Kolle G, Cooper-White JJ, Grimmond SM, Pera MF, Wolvetang EJ. Ascorbate promotes epigenetic activation of CD30 in human embryonic stem cells. Stem Cells. 2010;28:1782-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Whitworth DJ, Ovchinnikov DA, Wolvetang EJ. Generation and Characterization of LIF-dependent Canine Induced Pluripotent Stem Cells from Adult Dermal Fibroblasts. Stem Cells Dev. 2012;21:2288-2297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 9. | Chung TL, Brena RM, Kolle G, Grimmond SM, Berman BP, Laird PW, Pera MF, Wolvetang EJ. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells. 2010;28:1848-1855. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 126] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 10. | Ben-Dor I, Itsykson P, Goldenberg D, Galun E, Reubinoff BE. Lentiviral vectors harboring a dual-gene system allow high and homogeneous transgene expression in selected polyclonal human embryonic stem cells. Mol Ther. 2006;14:255-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713-720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 222] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 12. | Pera MF. Stem cells: The dark side of induced pluripotency. Nature. 2011;471:46-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 199] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 13. | Levenberg S, Huang NF, Lavik E, Rogers AB, Itskovitz-Eldor J, Langer R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Natl Acad Sci USA. 2003;100:12741-12746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 532] [Cited by in RCA: 446] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 14. | Wheeler AR, Throndset WR, Whelan RJ, Leach AM, Zare RN, Liao YH, Farrell K, Manger ID, Daridon A. Microfluidic device for single-cell analysis. Anal Chem. 2003;75:3581-3586. [PubMed] |
| 15. | Di Carlo D, Lee LP. Dynamic single-cell analysis for quantitative biology. Anal Chem. 2006;78:7918-7925. [PubMed] |
| 16. | Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 151] [Article Influence: 7.2] [Reference Citation Analysis (0)] |