Published online Apr 26, 2025. doi: 10.4252/wjsc.v17.i4.100359
Revised: November 29, 2024
Accepted: March 24, 2025
Published online: April 26, 2025
Processing time: 251 Days and 20.8 Hours
Thin endometrium, leading cause of recurrent implantation failure and infertility, has been found to respond to exosomes.
To investigate the efficacy of exosomes in addressing the issue of thin endome
RNA sequencing and reverse transcription-quantitative polymerase chain reaction were employed to identify differentially expressed microRNAs (miRNAs) in human umbilical cord mesenchymal stem cell (hucMSC) treated with exosomes enriched with endometrial cell-derived components. Additionally, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were conducted to highlight significant enrichment in specific biological pathways, molecular functions, and cellular components. Transwell and wound healing assays were performed to assess migratory potential, and western blotting was detected protein level.
A total of 53 differentially expressed miRNAs were identified in hucMSC treated with exosomes enriched with endometrial cell-derived components, comprising 27 upregulated and 26 downregulated miRNAs, which includes miR-137-3p. Enhanced migratory potential was observed in the Transwell and wound healing assays, and western blotting confirmed the epithelial differentiation of hucMSC and the increased p-signal transducer and activator of transcription 3. These effects were attributed to the upregulation of miR-137-3p.
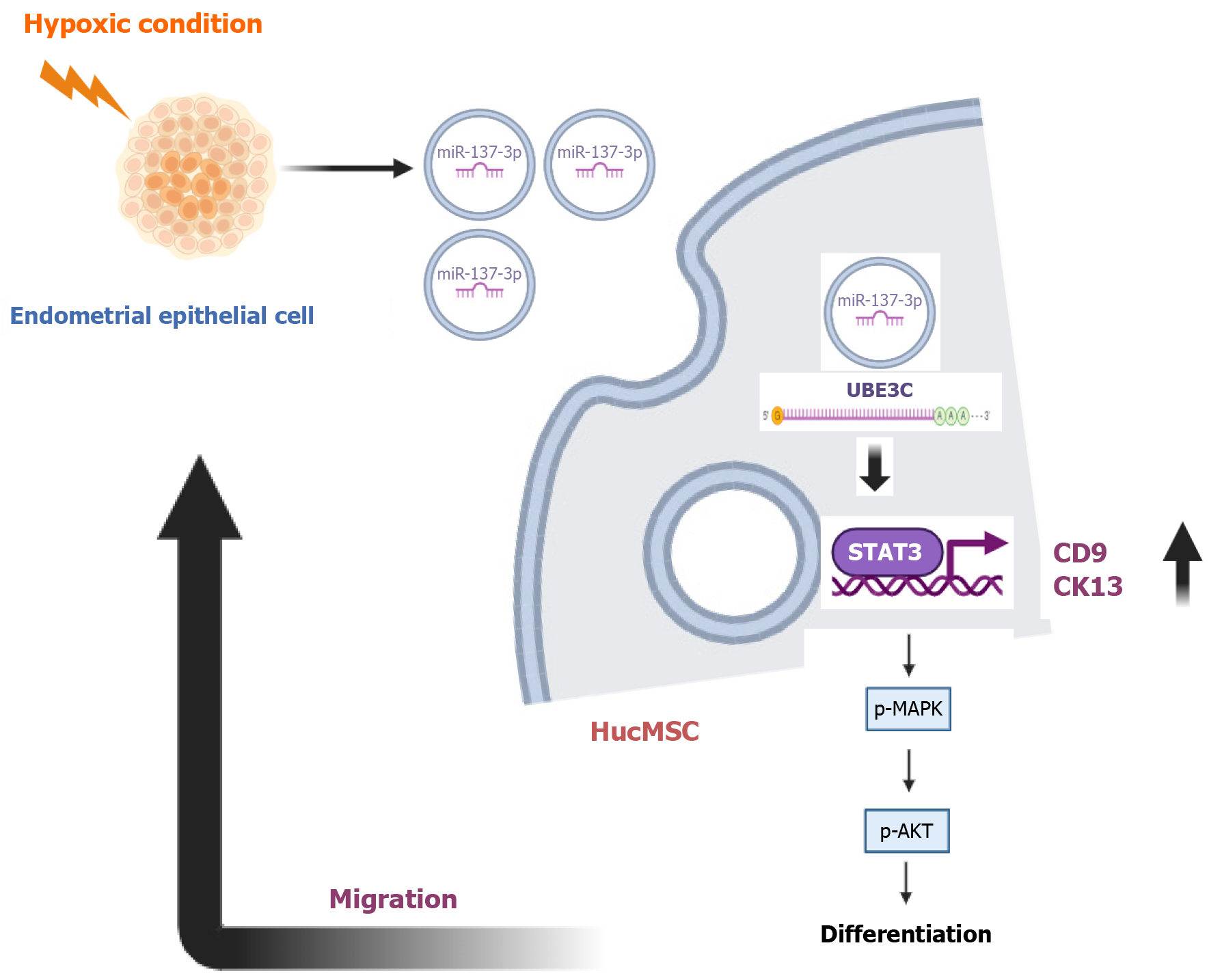
miR-137-3p in exosomes from hypoxia-affected endometrial epithelial cell stimulates the signal transducer and activator of transcription 3 signaling pathway, enhancing the migration and differentiation of hucMSC into endometrial epithelial cell.
Core Tip: This study uncovers that under hypoxic conditions, exosomal miR-137-3p, by targeting ubiquitin protein ligase E3C, activates signal transducer and activator of transcription 3, thereby enhancing the migration and differentiation of human umbilical cord mesenchymal stem cells (hucMSCs) into endometrial epithelial cells. Through microRNA sequencing and reverse transcription-quantitative polymerase chain reaction validation, it was observed that miR-137-3p is significantly upregulated in hucMSCs when co-cultured with endometrial epithelial cells. Additionally, exosome-based therapeutic system was established to evaluate the role of hucMSCs overexpressing miR-137-3p in therapeutic applications. This discovery offers novel insights into cell therapy under hypoxic environments.
- Citation: Zhang WY, Liu SM, Wang HB, Deng CY. Exosomal miR-137-3p targets UBE3C to activate STAT3, promoting migration and differentiation into endometrial epithelial cell of human umbilical cord mesenchymal stem cells under hypoxia. World J Stem Cells 2025; 17(4): 100359
- URL: https://www.wjgnet.com/1948-0210/full/v17/i4/100359.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v17.i4.100359
Human endometrium is characterized by remarkable plasticity, as evidenced by rapid tissue remodeling observed during early pregnancy and swift growth and differentiation that occur during menstrual cycle[1]. Development of thin endo
Exosomes, distinct class of extracellular vesicles secreted by broad array of cells, including endometrial cells[4], facilitate intercellular communication through transport of diverse signaling molecules, such as DNA, mRNAs, microRNAs (miRNAs), proteins, and lipids[5]. They have been implicated in endometrial diseases[6], with endometrial epithelial cell (EEC)-derived exosomes demonstrated to inhibit cell invasion and migration in ovarian endometriosis[7]. Moreover, exosomes from bovine EECs support development of trophoblast cells[8].
Recently, stem cells have emerged as novel therapeutic modality in human regenerative medicine, particularly for regeneration of endometrial disorders, including thin endometrium[9]. Mesenchymal stem cells (MSCs) discern microenvironment at injury site and secrete specific factors that are crucial for repair in multiple physiological processes. Prior research has highlighted efficacy of MSCs in treating thin endometrium, with transplantation of umbilical cord-derived MSCs in rats promoting endometrial recovery[10] and bone marrow-derived MSC transplantation in damaged uterus increasing endometrial thickness and improving fertility[11,12]. Additionally, it has been established that exosomes secreted by human umbilical cord MSCs (hucMSCs) repair hypoxia-induced EEC injury[13], underscoring beneficial effects of MSC-derived exosomes on EEC. Differentiation of human umbilical cord Wharton’s jelly-derived MSCs into EECs has also been documented[14]. Furthermore, our previous findings indicate that exosomes from hypoxia-damaged EECs enhance hucMSC migration and differentiation into EECs, although precise mechanisms governing this process require further elucidation.
MiRNAs, small non-coding RNAs that function as regulators of gene expression[15], have been identified as relevant to endometrial diseases[16] and differentiation of MSCs[17]. A study has conducted integrated miRNA sequencing analyses on adjacent normal endometrium and thin adhesive endometrium samples obtained from patients[18]. Utilising a combination of RNA sequencing and real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR), the present study sought to identify and validate the differential expression of miRNAs in the context of co-culture of hypoxia-injured endometrial epithelial cell (EECD)-derived exosome (EECD-ex) with hucMSC. The analysis revealed a pronounced alteration in the expression of miR-137-3p. Its expression was significantly up-regulated in the EECD-ex + hucMSC group, indicating that it may play a key role in exosome-mediated regulation of hucMSC function. Nonetheless, role of miRNAs in thin endometrium has been examined only sparingly, and mechanisms by which miRNAs modulate recovery of thin endometrium remain to be thoroughly elucidated. This study explores whether exosomes released from hypoxia-damaged EECs influence cellular functions of hucMSCs through delivery of specific signaling molecules, and it also delves into underlying mechanisms.
Human primary EECs, acquired from Cellverse Bioscience Technology (Shanghai, China), were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; 11965092, Gibco, NY, United States) enriched with 100 U/mL streptomycin, 100 ng/mL penicillin, and 10% fetal bovine serum (FBS; SH30406.05, HyClone, Marlborough, MA, United States), within humidified incubator maintained at 37 °C and 5% CO2. HucMSC, similarly sourced from Cellverse Bioscience Tech
Prior to hypoxia exposure, EEC were maintained under conventional conditions. Thereafter, cells were transferred to a hypoxic environment within a modular incubator chamber (94% N2, 5% CO2, 1% O2) at 37 °C for 4 hours. By contrast, control group cells were sustained in a normoxic atmosphere (78% N2, 5% CO2, 21% O2) throughout experimental period.
Culture supernatant from both normal and hypoxia-affected EEC was collected for exosome isolation, referred to as EEC-ex or EECD-ex, respectively, via ultracentrifugation. EEC were initially incubated for 48 hours in exosome-depleted DMEM. Supernatant was then subjected to centrifugation at 2000 r/minute for 20 minutes at 4 °C. To remove cellular debris, a subsequent centrifugation was performed at 10000 r/minute for 30 minutes at 4 °C. Post filtration through a 0.2 μm filter, supernatant underwent two ultracentrifugation rounds at 100000 r/minute for 1 hour at 4 °C. The ultimate precipitates constituted exosomes.
For exosome treatment, hucMSC were first pre-incubated for 24 hours with either EECD-ex or EEC-ex at a concentration of 5 μg/mL, thus forming EECD-ex + hucMSC and EEC-ex + hucMSC groups, respectively. Mimic negative control (NC) and miR-137-3p mimic were sourced from Sangon Biotech (Shanghai, China) and diluted to concentration of 30 nmol/L. Additionally, ubiquitin protein ligase E3C (UBE3C) overexpression plasmid (OE-UBE3C) and Vector-PcDNA3.1(+) were procured. Transfection of hucMSC with OE-NC, OE-UBE3C, mimic NC, or miR-137-3p mimic was performed using jetPRIME® reagent (101000046, Polyplus, Illkrich, France), in strict adherence to manufacturer’s protocol. To suppress function of signal transducer and activator of transcription 3 (STAT3), STAT3 inhibitor Stattic (HY-13818, Med
Upon extraction of total RNA from cells, its quality was assessed using an Agilent 2100 Bioanalyzer. Small RNA libraries were constructed with TruSeq Small RNA Library Prep Kit (RS-200-0036, Illumina, CA, United States). These libraries were then equimolarly combined, denatured, and loaded onto GAIIx flow cell lanes using cBot station and Illumina cluster generation kits. Differentially expressed miRNAs (DEMs) with raw reads ≥ 5 and a P value < 0.05 were identified. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were conducted to evaluate representation of DEM target genes in GO terms and KEGG pathways, with a significance threshold of P value < 0.05.
Total RNA was extracted from cells using TRIzol reagent (R1100, Solarbio, Beijing, China). Reverse transcription was achieved using iScript cDNA Synthesis Kit (KR118, TIANGEN, Beijing, China). SYBR Green (FP205, TIANGEN, Beijing, China) was utilized for RT-qPCR analysis. Relative gene expression was quantified using 2-ΔΔCt method, with U6 (for miRNAs) and GAPDH (for mRNAs) serving as endogenous reference genes. Primer sequences are summarized in Table 1.
| Target gene | Primer (5’-3’) |
| U6-F | ACGATACAGAGAAGATTAGCATGG |
| U6-R | AAATATGGAACGCTTCACGAA |
| hsa-miR-137-3p-F | CTCAACTGGTGTCGTGGAGT |
| hsa-miR-137-3p-R | TCGGCAGGTTATTGCTTAAGAATAC |
| hsa-miR-137-3p-RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCTACGC |
| UBE3C-F | AGCTATGTGGTGTCAGTGATTG |
| UBE3C-R | GAAGGCTGTAAAAACTTGTTGCC |
| STAT3-F | ACCAGCAGTATAGCCGCTTC |
| STAT3-R | GCCACAATCCGGGCAATCT |
| GAPDH-F | ACGGATTTGGTCGTATTGGG |
| GAPDH-R | GGGATCTCGCTCCTGGAAG |
Transwell assay was employed to evaluate migratory potential of hucMSC. Cells in logarithmic growth phase were harvested, washed in phosphate buffered saline, and resuspended in serum-free DMEM/F12. Upper chamber was seeded with 4 × 104 hucMSC, while lower chamber was filled with culture medium rich in 10% FBS. Following a 24-hour incubation, migrated cells were fixed with 2% methanol and stained using a 0.5% crystal violet solution. Cell migration was visualized and documented using a microscope (2103012, AOSVI, Shenzhen, China).
HucMSC were seeded in 6-well plates and incubated in medium devoid of FBS for 24 hours. A linear scratch was created on cell monolayer using a 10 μL pipette tip. Cell migration was observed and imaged at various time points (0 hour, 48 hours) under an inverted microscope. Cell migration rate was quantified using Image J software.
Total protein extraction from hucMSC was achieved through use of RIPA buffer containing protease inhibitor (P0013B, Beyotime, Shanghai, China), with mixture incubated on ice for 30 minutes. Protein samples were subjected to sodium-dodecyl sulfate gel electrophoresis and then transferred to polyvinylidene fluoride membranes (IPVH00010, Millipore, Burlington, MA, United States). After a 2-hour incubation with 5% nonfat milk as a blocking agent, membranes were incubated overnight with primary antibodies targeting CD9 (ab236630, Abcam, United Kingdom), CK19 (ab76539, Abcam, United Kingdom), vimentin (5741, CST, Danvers, MA, United States), CD13 (ab108310, Abcam, United Kingdom), β-Tubulin (CW0098M, CWBIO, Beijing, China), p-STAT3 (9138, CST, Danvers, MA, United States), STAT3 (4904, CST, Danvers, MA, United States), mitogen-activated protein kinase (MAPK; 4695, CST, Danvers, MA, United States), p-MAPK (4370, CST, Danvers, MA, United States), protein kinase B (AKT; 4691, CST, Danvers, MA, United States), p-AKT (13038, CST, Danvers, MA, United States), and UBE3C (ab226173, Abcam, United Kingdom). Membranes were subsequently incubated for 2 hours with horseradish peroxidase-conjugated secondary antibodies. Protein bands were visualized using an ECL reagent (MA0186-2, Meilunbio, Dalian, China).
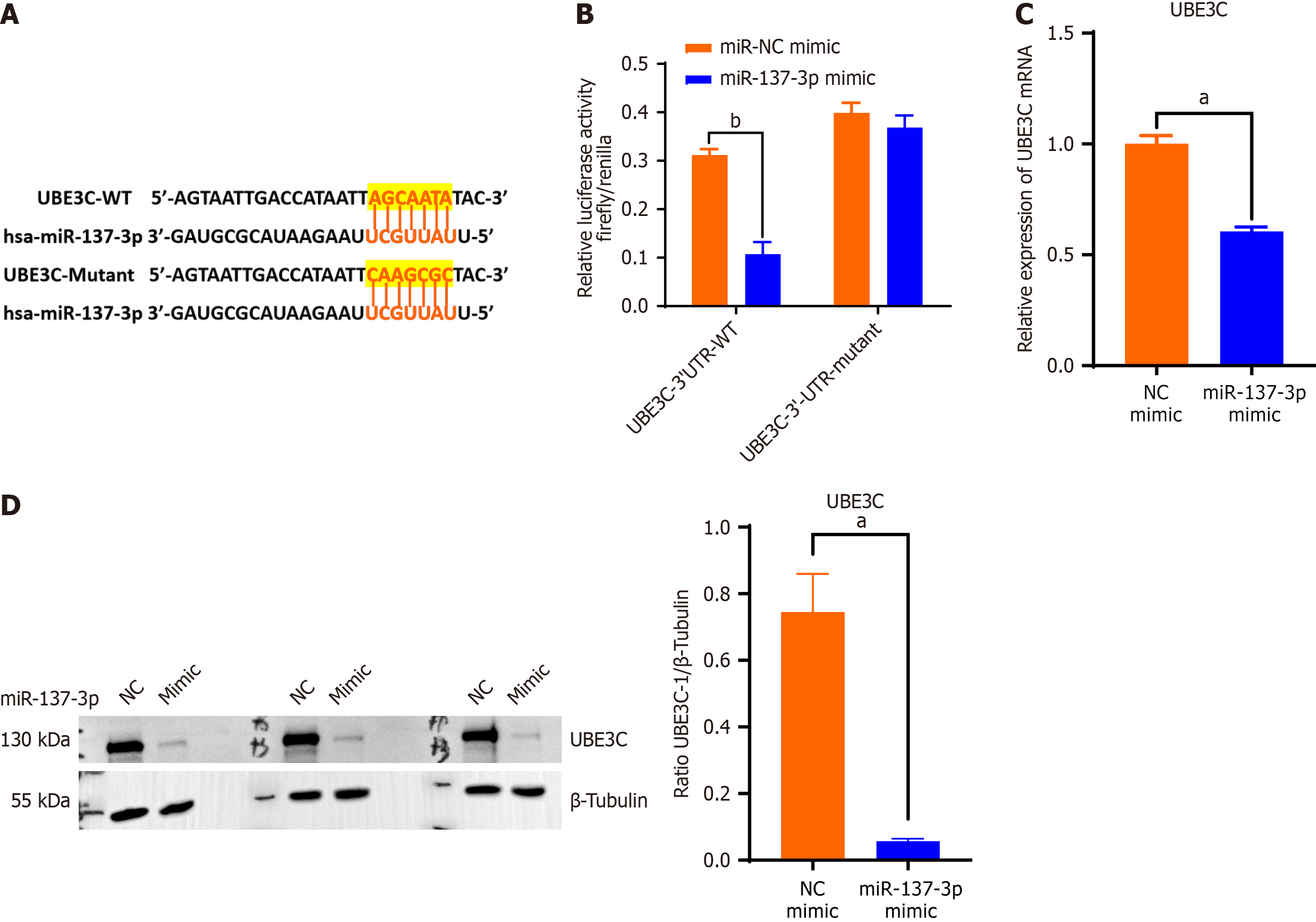
Potential binding of miR-137-3p to UBE3C, as predicted by miRDB, was investigated. A fragment of wild-type (WT) or mutant UBE3C-3’-untranslated region (UBE3C-3’-UTR), encompassing predicted binding site of miR-137-3p, was subcloned into psiCHECK-2 vector (GENERAL BIO, Anhui, China) to generate UBE3C-3’-UTR WT or Mutant constructs. Cells were plated in 24-well plates and co-transfected with miR-137-3p mimic or mimic NC and aforementioned vectors, using jetPRIME® reagent (101000046, Polyplus, Illkrich, France). Forty-eight hours after transfection, luciferase activity was measured using a luciferase reporter assay system (RG028, Beyotime, Shanghai, China).
All experimental data are presented as mean ± SD. One-way ANOVA was employed to compare differences among multiple groups, while Student’s t-test was used for comparisons between two groups. P value < 0.05 was deemed statistically significant. Statistical analyses were conducted using Prism 7.0.
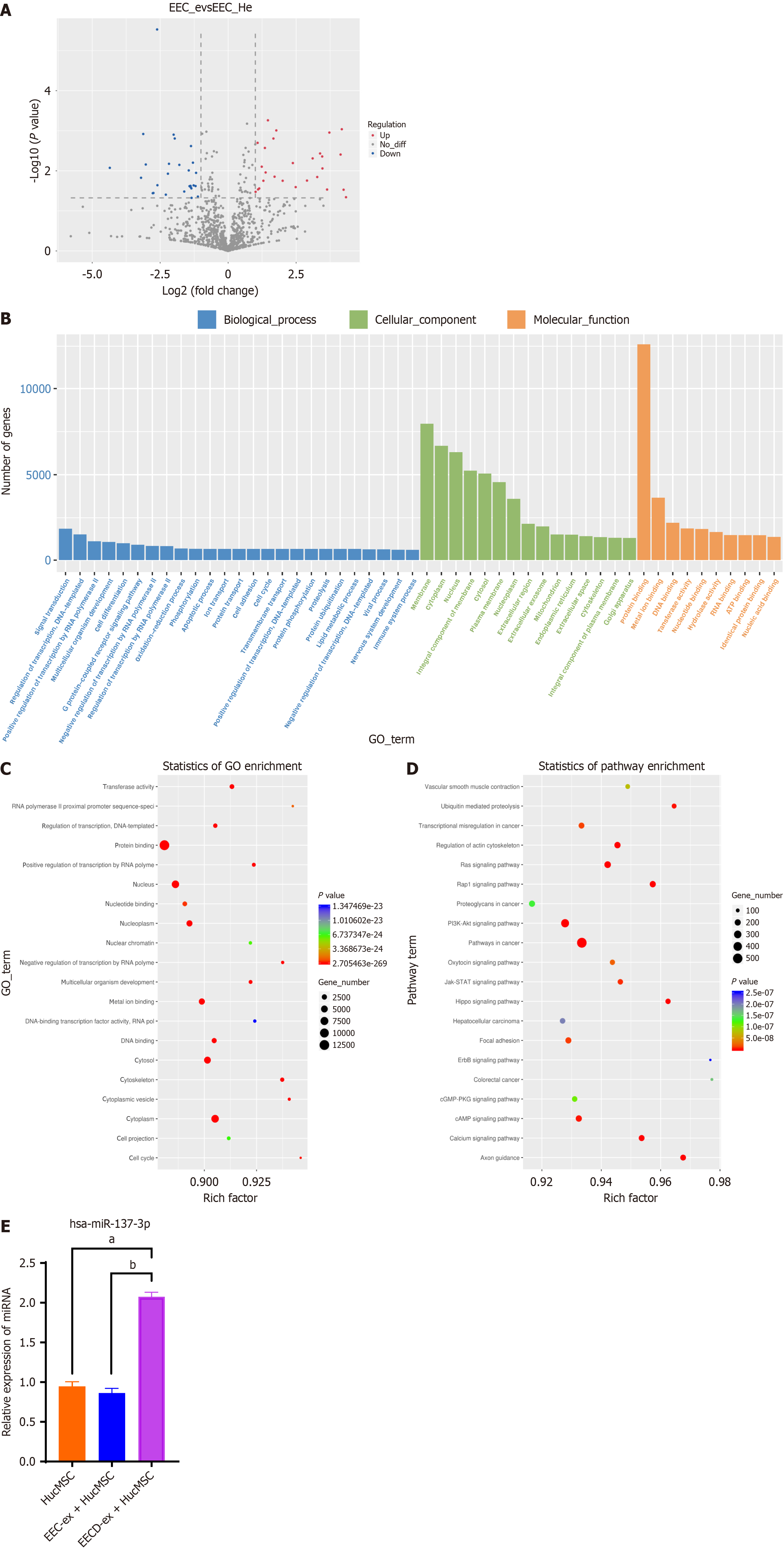
We have previously established that EECD-ex significantly enhances hucMSC migration and differentiation into EECs. Nonetheless, precise underlying mechanisms have yet to be elucidated. In this study, culture media from both normal EECs and hypoxia-induced EEC injury (hypoxia-damaged EEC) were collected for exosome extraction, and these exosomes were subsequently introduced to hucMSCs for coculture. Through miRNA sequencing, we uncovered DEMs in hucMSCs exposed to EEC-ex and EECD-ex. Our analysis revealed identification of 53 DEMs, comprising 26 downregulated and 27 upregulated miRNAs, in EECD-ex + hucMSC group relative to EEC-ex + hucMSC group (Figure 1A). GO and KEGG databases were harnessed to conduct functional analysis of DEM targets, indicating specific biological pathways, molecular functions, and cellular components (Figure 1B and C). Furthermore, KEGG pathway enrichment analysis discerned several pertinent signaling pathways (Figure 1D). RT-qPCR corroborated that, in comparison with hucMSCs and EEC-ex + hucMSC groups, miR-137-3p was upregulated in EECD-ex + hucMSC group, aligning with findings from RNA sequencing (Figure 1E). Consequently, it is proposed that miR-137-3p, released by EECD-ex, may play regulatory role in functional dynamics of hucMSCs.
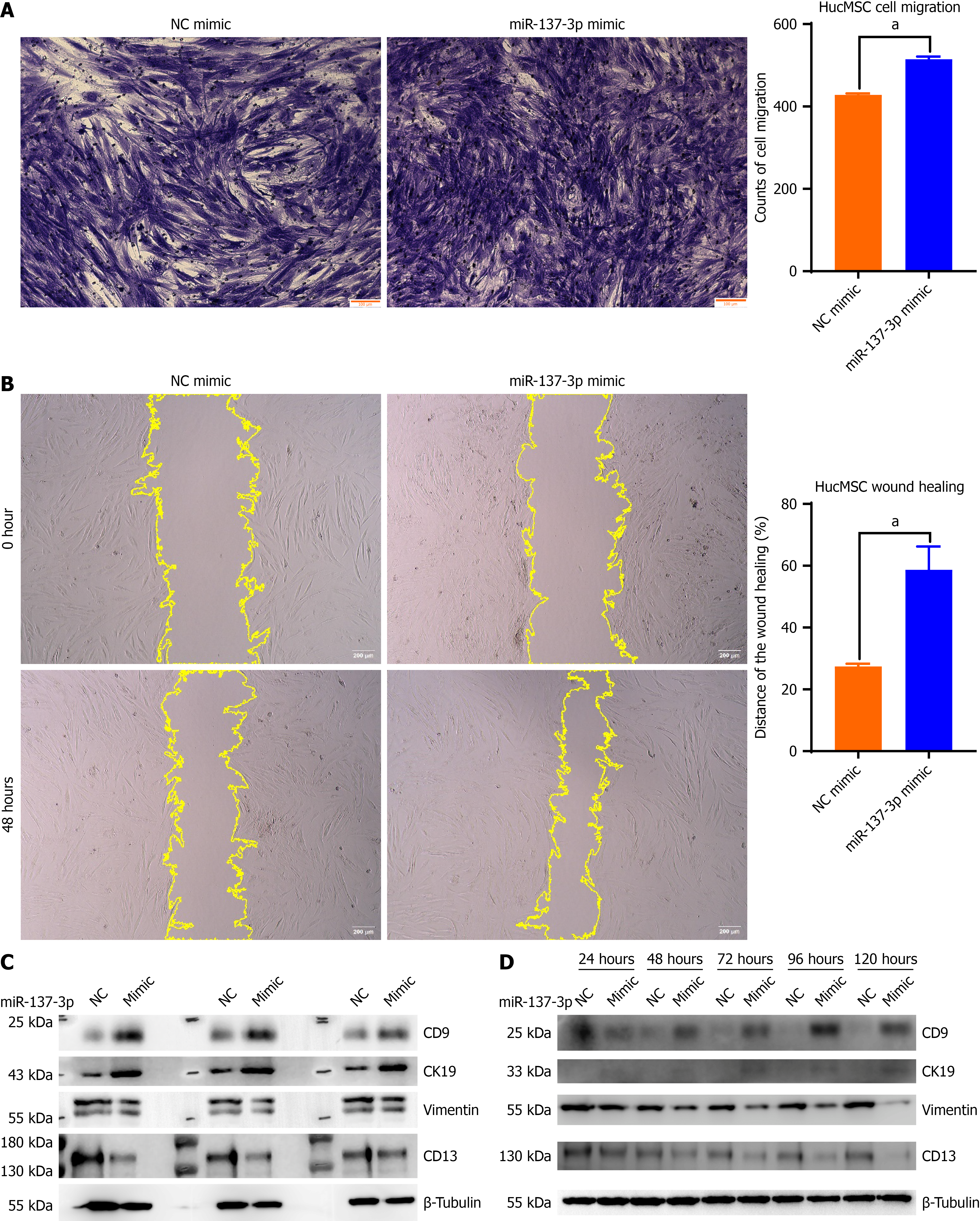
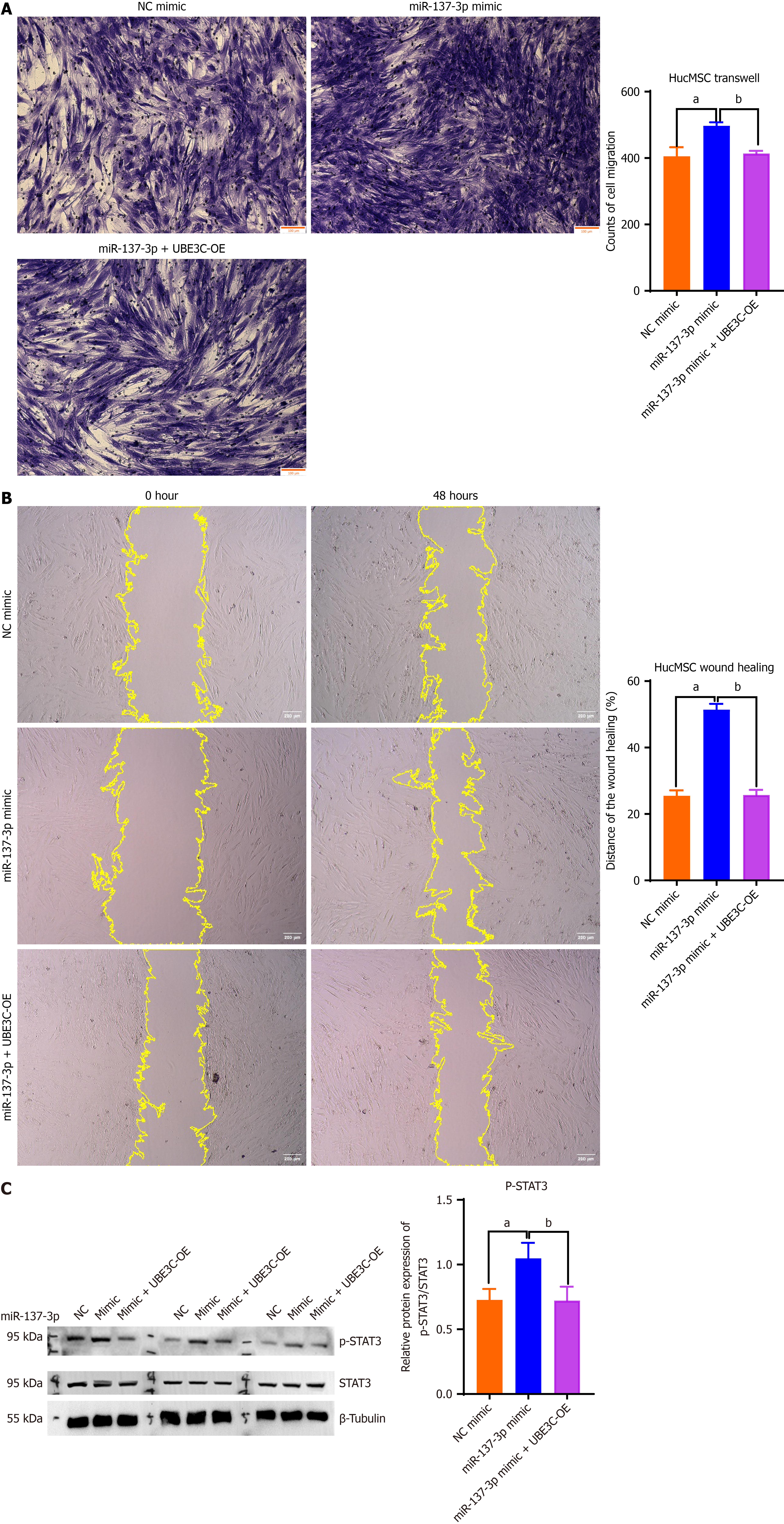
To elucidate role of miR-137-3p in migratory abilities of hucMSCs, we enhanced expression of miR-137-3p via transfection with a miR-137-3p mimic. Transwell assay illustrated that hucMSC migration was markedly amplified in miR-137-3p mimic group when juxtaposed with mimic NC group (Figure 2A). Moreover, wound healing assay indicated that miR-137-3p mimic significantly enhanced migratory distance of hucMSCs (Figure 2B). Aforesaid data suggest facilitative effect of miR-137-3p on migration of hucMSCs. Subsequently, we examined if miR-137-3p modulates differentiation of hucMSCs into EECs. It was observed that, as duration of transfection progressed, miR-137-3p mimic upregulated expression levels of epithelial lineage markers (CK19 and CD9) while simultaneously downregulating levels of stromal lineage markers (CD13 and vimentin) in hucMSCs (Figure 2C). Expression levels of CK19 and CD9 proteins were elevated, whereas those of vimentin and CD13 were reduced in hucMSCs after transfection with miR-137-3p mimic (Figure 2D). These findings further corroborate that miR-137-3p facilitates differentiation of hucMSCs into EECs.
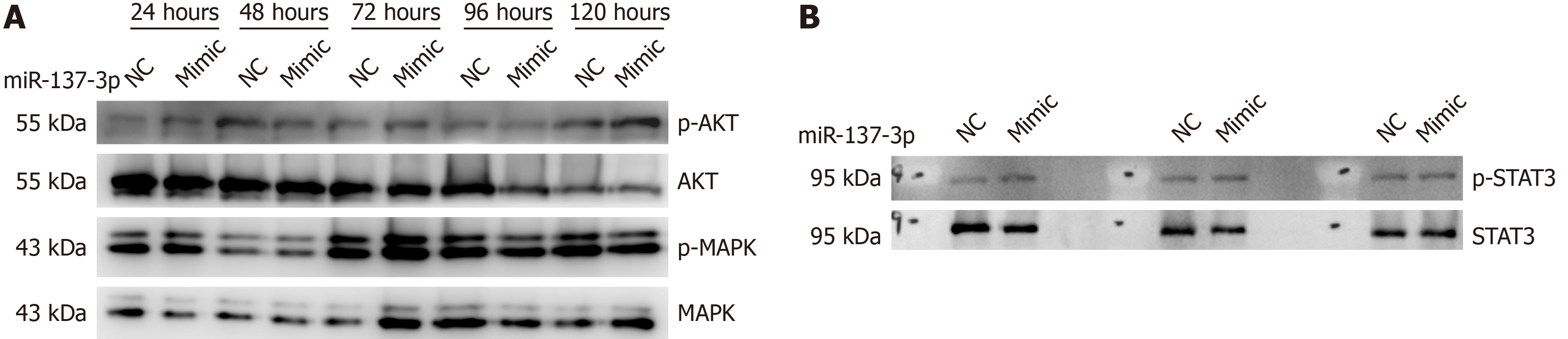
Expressions of p-AKT and p-MAPK proteins in hucMSCs were, respectively, elevated and diminished following transfection with miR-137-3p mimic (Figure 3A). This suggests that miR-137-3p may exert its influence through AKT pathway rather than MAPK pathway. Additionally, miR-137-3p mimic augmented expression of p-STAT3 protein in hucMSCs (Figure 3B), indicating possible involvement of STAT3 signaling in role of miR-137-3p.
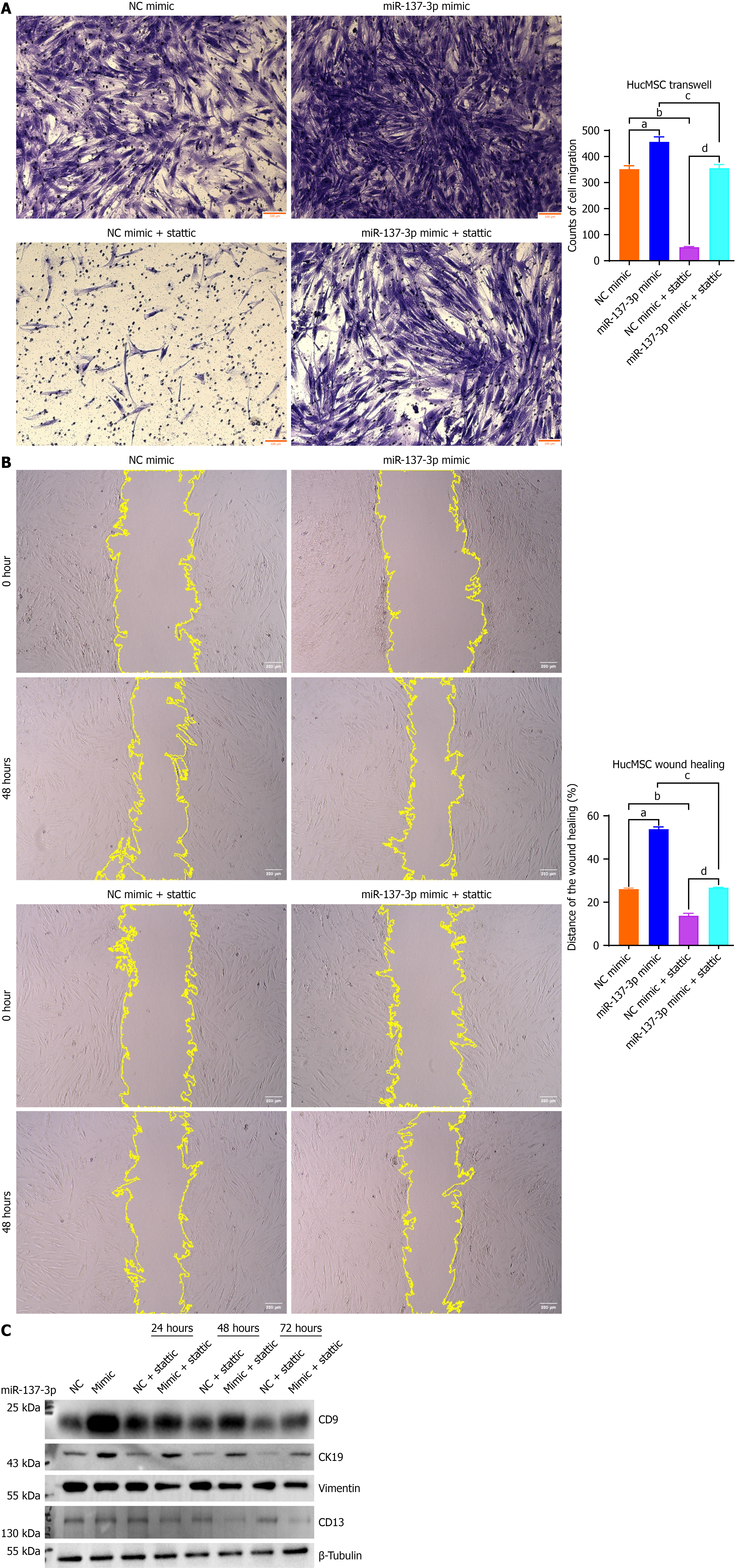
To ascertain whether miR-137-3p mediated functional modifications of hucMSCs by modulating STAT3, hucMSCs were transfected with miR-137-3p mimic, followed by administration of STAT3 inhibitor (Stattic). Transwell assay demonstrated that miR-137-3p mimic potentiated hucMSC migration, response that was attenuated upon introduction of STAT3 inhibitor (Figure 4A). In accordance with these findings, wound healing assay revealed that while miR-137-3p mimic augmented migratory distance of hucMSCs, this effect was negated by STAT3 inhibitor (Figure 4B). Western blot analysis of epithelial and stromal lineage markers further supported notion that miR-137-3p mimic enhanced hucMSC differentiation into EECs, an effect that was inhibited by STAT3 inhibitor (Figure 4C). Collectively, these data suggest that STAT3 inhibition can counteract stimulatory effects of miR-137-3p on hucMSC function.
We posited that miR-137-3p might target UBE3C, hypothesis supported by predictions from miRDB (Figure 5A). Subsequent dual-luciferase reporter assays confirmed that miR-137-3p specifically binds to 3’-UTR of UBE3C; however, this interaction was markedly weakened when UBE3C-3’-UTR was mutated (Figure 5B). Transfection of HucMSCs with miR-137-3p mimic led to reduction in UBE3C mRNA levels (Figure 5C). Notably, protein levels of UBE3C were observed to decrease, whereas levels of STAT3 were seen to increase in hucMSCs following miR-137-3p mimic transfection (Figure 5D). This evidence implies that miR-137-3p downregulates UBE3C to promote STAT3 activation.
In a subsequent experiment, to explore if UBE3C is implicated in miR-137-3p-regulated functional variations of hucMSCs, hucMSCs were transfected with combination of miR-137-3p mimic and overexpression vector of UBE3C. Transwell assay revealed that migratory capacity of hucMSCs, when overexpressing UBE3C, was significantly impaired in miR-137-3p mimic + OE-UBE3C group compared to miR-137-3p mimic alone group (Figure 6A). Wound healing assay provided corroborative results, showing that hucMSC migration was effectively restrained in miR-137-3p mimic + OE-UBE3C group (Figure 6B). Moreover, in miR-137-3p mimic + OE-UBE3C group, expression of p-STAT3 protein was observed to decrease (Figure 6C). These outcomes suggest that overexpression of UBE3C reverses facilitative effects of miR-137-3p on hucMSC function.
Thin endometrium is prevalent cause of chronic infertility and is associated with adverse outcomes[20]. It has been substantiated that exosomes derived from hucMSCs ameliorate hypoxia-induced injury in EECs[13]. Notably, our prior research has elucidated that EECD-ex augment hucMSC migration and differentiation into EECs. In this contribution, we have further unveiled that miR-137-3p is upregulated in hucMSCs co-cultured with EECD-ex. miR-137-3p facilitates hucMSC migration and differentiation into EECs. Furthermore, miR-137-3p targets UBE3C to promote STAT3 activation, thereby enhancing hucMSC migration and differentiation into EECs.
The genesis of exosomes commences with biogenesis of multi-vesicular bodies within cellular interior. Organelles within EECs damaged by hypoxia can encapsulate molecular cargoes within intra-luminal vesicles of exosomes, which are subsequently liberated into extracellular milieu upon multi-vesicular body fusion with plasma membrane[21]. Exosomes, once disseminated into extracellular environment, can be conveyed to distant tissues or cells via circulatory system, lymphatic system, or other bodily fluids. Exosomal surface is adorned with specific proteins and lipids, which facilitate their recognition and binding to target cells, thereby influencing tropism of exosomes. Target cells can internalize exosomes through variety of mechanisms, including phagocytosis, endocytosis, and receptor-mediated endocytosis. Once internalized, miRNAs harbored by exosomes are released into cytoplasm and modulate gene expression within target cell. MiRNAs exert regulatory control over biological functions, such as cell proliferation, differentiation, and apoptosis, by modulating stability or translational efficiency of specific mRNAs through binding[22].
Within scope of this investigation, exosomes characterized by high expression of miR-137-3p catalyze repair of endometrium through multitude of processes. These exosomes facilitate differentiation of stem cells into endometrial cells, critical step in regeneration of endometrium. Exosomes, enriched with elevated levels of miR-137-3p, convey molecular signals that induce differentiation to their target sites, guiding transformation of stem cells into necessary cell types and expediting repair of endometrium. By transferring miRNAs to umbilical cord MSCs, exosomes modulate gene expression profiles of these target cells, either promoting or inhibiting expression of specific genes. This capability is of paramount importance in exosomes’ role in regeneration of endometrium. By modulating expression of genes associated with repair and regeneration, exosomes contribute to enhancing functional status of endometrium. The bioactive molecules borne by exosomes with high miR-137-3p expression stimulate proliferation and migration of endometrial cells, thereby accelerating repair process. This is exceedingly beneficial for restoring integrity and thickness of endometrium, particularly in scenarios of endometrial damage, such as those caused by Asherman syndrome or endometriosis. It is conceivable that endometrial repair may also occur through alternative pathways. Exosomes contain growth factors and miRNAs that foster angiogenesis, facilitating formation of new blood vessels within endometrium to provide essential nutrients and oxygen, thus promoting its regeneration. Specific constituents within exosomes modulate inflammatory responses, mitigating harmful inflammation while fostering beneficial inflammation that participates in repair process. miR-137-3p derived from exosomes can enter umbilical cord blood MSCs (UCB-MSCs), enhancing their migratory capacity and promoting their differentiation into epithelial cells, while simultaneously inhibiting differentiation of mesenchymal cells. In UCB-MSCs with elevated miR-137-3p expression, overexpression of UBE3C further inhibits enhancement of migratory ability and differentiation potential toward epithelial cells. Post administration of a STAT3 inhibitor, migratory capacity and differentiation potential of UCB-MSCs overexpressing miR-137-3p into epithelial cells are also attenuated. It is plausible that exosomal miR-137-3p regulates biological functions of UCB-MSCs through downstream molecules, such as UBE3C and STAT3.
In total, 72 DEMs have been identified in thin, adhered endometrium of patients, as compared to adjacent normal endometrium, through miRNA sequencing analysis[18]. MiRNA microarray chip analysis reveals that in damaged endometrium of rats, 39 miRNAs are upregulated, and 45 miRNAs are downregulated, effect which is overturned by transplantation of hucMSCs[10]. In current study, miRNA sequencing was conducted to screen for DEMs in hucMSCs incubated with EEC-ex or EECD-ex. We observed that 53 DEMs (27 upregulated and 26 downregulated) were identified in hucMSCs pre-treated with EECD-ex. GO analysis demonstrated enrichment of DEM targets in specific cellular components, molecular functions, and biological pathways. Moreover, several pertinent signaling pathways were elucidated through KEGG analysis. DEMs were further validated using RT-qPCR, and our findings revealed pronounced upregulation of miR-137-3p in hucMSCs co-cultured with EECD-ex.
Compared to other delivery methods, such as transfection, exosome-mediated miRNA delivery exhibits several advantages: (1) Superior biocompatibility and relatively diminished immunogenicity. Exosomes, derived from living cells, inherit numerous intrinsic attributes of their parent cells, including specific protein markers on cell surface. This property enables exosomes to be tailored for targeted delivery, thereby enhancing selectivity for target cells. They demonstrate excellent biocompatibility in vivo and present a reduced risk of eliciting immune responses compared to synthetic carriers, such as liposomes or polymer nanoparticles[23]; (2) Enhanced permeability through biological barriers and markedly low RNA degradation rate. Exosomes possess capability to penetrate stringent biological barriers, such as blood-brain barrier, rendering them highly promising vehicle for treatment of central nervous system disorders. Internal microenvironment of exosomes shields carried RNA from degradation by cellular RNA enzymes, ensuring RNA integrity upon arrival at target cells[24]; and (3) Efficient uptake by target cells and a minimal side effect profile. Exosomes can be internalized by target cells via various mechanisms, including phagocytosis, endocytosis, and receptor-mediated endocytosis. In contrast to certain chemical carriers, exosomes exhibit lower toxicity and side effects, which enhances their safety as therapeutic vehicles[25].
It has been reported that in HTR-8/SVneo trophoblast cells stimulated by high glucose, miR-137-3p plays a role in regulating cell viability, migration, and invasion[26], osteogenic differentiation of bone marrow-derived MSCs[27], and myogenic differentiation of endometrial mesenchymal stem/stromal cells[28]. Here, we have revealed that overexpression of miR-137-3p not only facilitated migration of hucMSCs but also promoted their differentiation into EECs. To best of our knowledge, this study is first to validate biological function of miR-137-3p in modulating differentiation of hucMSCs into EECs.
STAT proteins are pivotal in a myriad of biological functions, including migration, proliferation, and differentiation[29]. Augmenting phosphorylation of STAT3 in bone marrow-derived MSCs can bolster osteogenic differentiation[30]. Icaritin enhances both proliferation and osteogenic differentiation of MSCs via activation of STAT3 pathway[31]. Hypoxia-induced migration and osteogenic differentiation of bone marrow-derived MSCs occur through STAT3 phosphorylation[32]. Myofibroblast differentiation of endometrial MSCs, driven by activin A, is also STAT3-dependent[33]. In this study, we identified that overexpression of miR-137-3p increased expression of p-STAT3 in hucMSCs, indicative of STAT3 signaling pathway activation. Notably, miR-137-3p accentuated migration and differentiation of hucMSCs into EECs, effect that was mitigated by STAT3 inhibitor, Stattic. Our findings thus conclude that miR-137-3p activates STAT3 signaling pathway to enhance migration and differentiation of hucMSCs into EECs. In this paper, although the mRNA level of STAT3 was not significantly up-regulated, the protein expression was remarkably elevated. It was found that miR-137-3p targeted the expression of ubiquitination-related proteins. We questioned whether miR-137-3p could inhibit the ubiquitination of STAT3 by suppressing the expression of ubiquitinated proteins. Moreover, we wondered if ubiquitination could occur, leading to a phenomenon where the antagonistic sites of STAT3 phosphorylation show higher-intensity phosphorylation. Subsequently, whether STAT3 with such modified phosphorylation status could enter the nucleus and influence downstream transcription factors was also under consideration. This line of speculation was repeated as we further explored if ubiquitination might take place, resulting in the appearance of the antagonistic site phenomenon of STAT3 phosphorylation with higher-intensity phosphorylation, and then enabling STAT3 to enter the nucleus and exert an effect on downstream transcription factors.
UBE3C is a member of HECT ubiquitin protein ligase family, which modulates crucial cellular signaling pathways[34]. UBE3C is essential in maintaining stem cell properties in lung cancer[35]. Particularly, UBE3C is key mediator in miRNA-regulated cell migration and proliferation. Prior studies have established that UBE3C is targeted by miR-542-3p and miR-30a-5p, which influences gene expression in hepatocellular carcinoma and breast cancer[36]. In present investigation, miRDB predicted that miR-137-3p might bind to UBE3C, and this direct interaction was subsequently confirmed using luciferase reporter assay. Our research unveiled that miR-137-3p targets UBE3C to potentiate STAT3 activation. More
Although in vitro experiments can reveal the mechanisms of miR-137-3p at the cellular level, their results may not fully reflect the complex in vivo environment. Therefore, in vivo experiments are crucial for validating the actual effects of miR-137-3p in animal models, particularly in tissue repair, regeneration, or disease models. By injecting miR-137-3p mimics or inhibitors into animal models, one can observe their impact on tissue repair, inflammatory responses[37], or tumor growth[38], while also assessing their stability, biodistribution, and potential toxicity or side effects in living organisms. Additionally, in the in vivo environment, miR-137-3p may interact with other miRNAs, signaling pathways, or cytokines, forming a complex regulatory network, which further highlights the importance of in vivo experiments. Clinical trials are the critical step in evaluating the therapeutic potential of miR-137-3p, such as in wound healing, tissue regeneration, or the treatment of certain diseases, where miR-137-3p may serve as a potential biomarker or therapeutic target. Through clinical trials, the safety, efficacy, and optimal delivery methods of miR-137-3p in patients can be assessed. To successfully apply miR-137-3p in clinical settings, it is also necessary to develop effective delivery systems[39] to ensure its targeted delivery to specific tissues or cells and its functional efficacy.
In summary, our study has elucidated that miR-137-3p, sourced from EECD-exosomes, specifically targets UBE3C, thereby activating STAT3 signaling pathway. This activation notably facilitated migration and differentiation of hucMSC into EEC. These findings may serve as a foundational cornerstone for innovative therapeutic strategies aimed at add
| 1. | Sternberg AK, Buck VU, Classen-Linke I, Leube RE. How Mechanical Forces Change the Human Endometrium during the Menstrual Cycle in Preparation for Embryo Implantation. Cells. 2021;10:2008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 2. | Tabeeva G, Silachev D, Vishnyakova P, Asaturova A, Fatkhudinov T, Smetnik A, Dumanovskaya M. The Therapeutic Potential of Multipotent Mesenchymal Stromal Cell-Derived Extracellular Vesicles in Endometrial Regeneration. Int J Mol Sci. 2023;24:9431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 3. | Gharibeh N, Aghebati-Maleki L, Madani J, Pourakbari R, Yousefi M, Ahmadian Heris J. Cell-based therapy in thin endometrium and Asherman syndrome. Stem Cell Res Ther. 2022;13:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 4. | Homer H, Rice GE, Salomon C. Review: Embryo- and endometrium-derived exosomes and their potential role in assisted reproductive treatments-liquid biopsies for endometrial receptivity. Placenta. 2017;54:89-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 5. | Vyas N, Dhawan J. Exosomes: mobile platforms for targeted and synergistic signaling across cell boundaries. Cell Mol Life Sci. 2017;74:1567-1576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 6. | Mishra A, Ashary N, Sharma R, Modi D. Extracellular vesicles in embryo implantation and disorders of the endometrium. Am J Reprod Immunol. 2021;85:e13360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 7. | Zhang M, Wang X, Xia X, Fang X, Zhang T, Huang F. Endometrial epithelial cells-derived exosomes deliver microRNA-30c to block the BCL9/Wnt/CD44 signaling and inhibit cell invasion and migration in ovarian endometriosis. Cell Death Discov. 2022;8:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 8. | Wang X, Li Q, Xie T, Yuan M, Sheng X, Qi X, Xing K, Liu F, Guo Y, Xiao L, Ni H. Exosomes from bovine endometrial epithelial cells ensure trophoblast cell development by miR-218 targeting secreted frizzled related protein 2. J Cell Physiol. 2021;236:4565-4579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 9. | Azizi R, Aghebati-Maleki L, Nouri M, Marofi F, Negargar S, Yousefi M. Stem cell therapy in Asherman syndrome and thin endometrium: Stem cell- based therapy. Biomed Pharmacother. 2018;102:333-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 128] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 10. | Zhang L, Li Y, Dong YC, Guan CY, Tian S, Lv XD, Li JH, Su X, Xia HF, Ma X. Transplantation of umbilical cord-derived mesenchymal stem cells promotes the recovery of thin endometrium in rats. Sci Rep. 2022;12:412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 11. | Yi KW, Mamillapalli R, Sahin C, Song J, Tal R, Taylor HS. Bone marrow-derived cells or C-X-C motif chemokine 12 (CXCL12) treatment improve thin endometrium in a mouse model. Biol Reprod. 2019;100:61-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Xia L, Meng Q, Xi J, Han Q, Cheng J, Shen J, Xia Y, Shi L. The synergistic effect of electroacupuncture and bone mesenchymal stem cell transplantation on repairing thin endometrial injury in rats. Stem Cell Res Ther. 2019;10:244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 13. | Wang H, Liu S, Zhang W, Liu M, Deng C. Human Umbilical Cord Mesenchymal Stem Cell-Derived Exosome Repairs Endometrial Epithelial Cells Injury Induced by Hypoxia via Regulating miR-663a/CDKN2A Axis. Oxid Med Cell Longev. 2022;2022:3082969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Shi Q, Gao J, Jiang Y, Sun B, Lu W, Su M, Xu Y, Yang X, Zhang Y. Differentiation of human umbilical cord Wharton's jelly-derived mesenchymal stem cells into endometrial cells. Stem Cell Res Ther. 2017;8:246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Bjorkman S, Taylor HS. MicroRNAs in endometriosis: biological function and emerging biomarker candidates†. Biol Reprod. 2019;100:1135-1146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 16. | Kolanska K, Bendifallah S, Canlorbe G, Mekinian A, Touboul C, Aractingi S, Chabbert-Buffet N, Daraï E. Role of miRNAs in Normal Endometrium and in Endometrial Disorders: Comprehensive Review. J Clin Med. 2021;10:3457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 17. | Yang C, Luo M, Chen Y, You M, Chen Q. MicroRNAs as Important Regulators Mediate the Multiple Differentiation of Mesenchymal Stromal Cells. Front Cell Dev Biol. 2021;9:619842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Zong L, Zheng S, Meng Y, Tang W, Li D, Wang Z, Tong X, Xu B. Integrated Transcriptomic Analysis of the miRNA-mRNA Interaction Network in Thin Endometrium. Front Genet. 2021;12:589408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Zhang JW. Impact of miR-214-5p and miR-21-5p from hypoxic endometrial exosomes on human umbilical cord mesenchymal stem cell function. World J Stem Cells. 17:102404. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 20. | Lin J, Wang Z, Huang J, Tang S, Saiding Q, Zhu Q, Cui W. Microenvironment-Protected Exosome-Hydrogel for Facilitating Endometrial Regeneration, Fertility Restoration, and Live Birth of Offspring. Small. 2021;17:e2007235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 21. | Arya SB, Collie SP, Parent CA. The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. 2024;34:90-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 154] [Article Influence: 154.0] [Reference Citation Analysis (0)] |
| 22. | Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1443] [Cited by in RCA: 1556] [Article Influence: 155.6] [Reference Citation Analysis (0)] |
| 23. | Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, Hu BY, Qin W, Zou TT, Fu Y, Shen XT, Zhu WW, Geng Y, Lu L, Jia HL, Qin LX, Dong QZ. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 324] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 24. | Chu X, Yang Y, Tian X. Crosstalk between Pancreatic Cancer Cells and Cancer-Associated Fibroblasts in the Tumor Microenvironment Mediated by Exosomal MicroRNAs. Int J Mol Sci. 2022;23:9512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 25. | Fröhlich E. Cellular elimination of nanoparticles. Environ Toxicol Pharmacol. 2016;46:90-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Li Y, Liu L. LncRNA OIP5-AS1 Signatures as a Biomarker of Gestational Diabetes Mellitus and a Regulator on Trophoblast Cells. Gynecol Obstet Invest. 2021;86:509-517. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Kong L, Zuo R, Wang M, Wang W, Xu J, Chai Y, Guan J, Kang Q. Silencing MicroRNA-137-3p, which Targets RUNX2 and CXCL12 Prevents Steroid-induced Osteonecrosis of the Femoral Head by Facilitating Osteogenesis and Angiogenesis. Int J Biol Sci. 2020;16:655-670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 28. | Chen HS, Hsu CY, Chang YC, Chuang HY, Long CY, Hsieh TH, Tsai EM. Benzyl butyl phthalate decreases myogenic differentiation of endometrial mesenchymal stem/stromal cells through miR-137-mediated regulation of PITX2. Sci Rep. 2017;7:186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 29. | Rao D, Macias E, Carbajal S, Kiguchi K, DiGiovanni J. Constitutive Stat3 activation alters behavior of hair follicle stem and progenitor cell populations. Mol Carcinog. 2015;54:121-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | Chen L, Zhang RY, Xie J, Yang JY, Fang KH, Hong CX, Yang RB, Bsoul N, Yang L. STAT3 activation by catalpol promotes osteogenesis-angiogenesis coupling, thus accelerating osteoporotic bone repair. Stem Cell Res Ther. 2021;12:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 31. | Lim RZL, Li L, Yong EL, Chew N. STAT-3 regulation of CXCR4 is necessary for the prenylflavonoid Icaritin to enhance mesenchymal stem cell proliferation, migration and osteogenic differentiation. Biochim Biophys Acta Gen Subj. 2018;1862:1680-1692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 32. | Yu X, Wan Q, Cheng G, Cheng X, Zhang J, Pathak JL, Li Z. CoCl(2) , a mimic of hypoxia, enhances bone marrow mesenchymal stem cells migration and osteogenic differentiation via STAT3 signaling pathway. Cell Biol Int. 2018;42:1321-1329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Zhang Z, Wang J, Chen Y, Suo L, Chen H, Zhu L, Wan G, Han X. Activin a promotes myofibroblast differentiation of endometrial mesenchymal stem cells via STAT3-dependent Smad/CTGF pathway. Cell Commun Signal. 2019;17:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 34. | Singh S, Ng J, Sivaraman J. Exploring the "Other" subfamily of HECT E3-ligases for therapeutic intervention. Pharmacol Ther. 2021;224:107809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 35. | Gu J, Mao W, Ren W, Xu F, Zhu Q, Lu C, Lin Z, Zhang Z, Chu Y, Liu R, Ge D. Ubiquitin-protein ligase E3C maintains non-small-cell lung cancer stemness by targeting AHNAK-p53 complex. Cancer Lett. 2019;443:125-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 36. | Tao J, Liu Z, Wang Y, Wang L, Yao B, Li Q, Wang C, Tu K, Liu Q. MiR-542-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting UBE3C. Biomed Pharmacother. 2017;93:420-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 37. | Xiong J, Wei B, Ye Q, Liu W. MiR-30a-5p/UBE3C axis regulates breast cancer cell proliferation and migration. Biochem Biophys Res Commun. 2019;516:1013-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 38. | Hu S, Ma J, Su C, Chen Y, Shu Y, Qi Z, Zhang B, Shi G, Zhang Y, Zhang Y, Huang A, Kuang Y, Cheng P. Engineered exosome-like nanovesicles suppress tumor growth by reprogramming tumor microenvironment and promoting tumor ferroptosis. Acta Biomater. 2021;135:567-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 39. | Kooijmans SA, Vader P, van Dommelen SM, van Solinge WW, Schiffelers RM. Exosome mimetics: a novel class of drug delivery systems. Int J Nanomedicine. 2012;7:1525-1541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 236] [Article Influence: 18.2] [Reference Citation Analysis (0)] |