Published online Mar 26, 2025. doi: 10.4252/wjsc.v17.i3.99472
Revised: October 24, 2024
Accepted: January 2, 2025
Published online: March 26, 2025
Processing time: 234 Days and 23 Hours
Cervical cancer (CC) stem cell-like cells (CCSLCs), defined by the capacity of differentiation and self-renewal and proliferation, play a significant role in the progression of CC. However, the molecular mechanisms regulating their self-renewal are poorly understood. Therefore, elucidation of the epigenetic mechanisms that drive cancer stem cell self-renewal will enhance our ability to improve the effectiveness of targeted therapies for cancer stem cells.
To explore how DNA methyltransferase 1 (DNMT1)/miR-342-3p/Forkhead box M1 (FoxM1), which have been shown to have abnormal expression in CCSLCs, and their signaling pathways could stimulate self-renewal-related stemness in CCSLCs.
Sphere-forming cells derived from CC cell lines HeLa, SiHa and CaSki served as CCSLCs. Self-renewal-related stemness was identified by determining sphere and colony formation efficiency, CD133 and CD49f protein level, and SRY-box transcription factor 2 and octamer-binding transcription factor 4 mRNA level. The microRNA expression profiles between HeLa cells and HeLa-derived CCSLCs or mRNA expression profiles that HeLa-derived CCSLCs were transfected with or without miR-342-3p mimic were compared using quantitative PCR analysis. The expression levels of DNMT1 mRNA, miR-342-3p, and FoxM1 protein were examined by quantitative real-time PCR and western blotting. In vivo carcinogenicity was assessed using a mouse xenograft model. The functional effects of the DNMT1/miR-342-3p/FoxM1 axis were examined by in vivo and in vitro gain-of-activity and loss-of-activity assessments. Interplay among DNMT1, miR-342-3p, and FoxM1 was tested by methylation-specific PCR and a respective luciferase reporter assay.
CCSLCs derived from the established HeLa cell lines displayed higher self-renewal-related stemness, including enhanced sphere and colony formation efficiency, increased CD133 and CD49f protein level, and heightened transcriptional quantity of stemness-related factors SRY-box transcription factor 2 and octamer-binding transcription factor 4 in vitro as well as a stronger tumorigenic potential in vivo compared to their parental cells. Moreover, quantitative PCR showed that the miR-342-3p level was downregulated in HeLa-derived CCSLCs compared to HeLa cells. Its mimic significantly decreased DNMT1 and FoxM1 mRNA expression levels in CCSLCs. Knockdown of DNMT1 or miR-342-3p mimic transfection suppressed DNMT1 expression, increased miR-342-3p quantity by promoter demethylation, and inhibited CCSLC self-renewal. Inhibition of FoxM1 by shRNA transfection also resulted in the attenuation of CCSLC self-renewal but had little effect on the DNMT1 activity and miR-342-3p expression. Furthermore, the loss of CCSLC self-renewal exerted by miR-342-3p mimic was inverted by the overexpression of DNMT1 or FoxM1. Furthermore, DNMT1 and FoxM1 were recognized as straight targets by miR-342-3p in HeLa-derived CCSLCs.
Our findings suggested that a novel DNMT1/miR-342-3p/FoxM1 signal axis promotes CCSLC self-renewal and presented a potential target for the treatment of CC through suppression of CCSLC self-renewal. However, this pathway has been previously implicated in CC, as evidenced by prior studies showing miR-342-3p-mediated downregulation of FoxM1 in cervical cancer cells. Additionally, research on liver cancer further supports the involvement of miR-342-3p in suppressing FoxM1 expression. While our study contributed to this body of knowledge, we did not present a completely novel axis but reinforced the therapeutic potential of targeting the DNMT1/miR-342-3p/FoxM1 axis to suppress CCSLC self-renewal in CC treatment.
Core Tip: The study revealed a novel DNA methyltransferase 1 (DNMT1)/miR-342-3p/Forkhead box M1 (FoxM1) signaling axis that enhances self-renewal of cervical cancer stem cell-like cells (CCSLCs). Knockdown of DNMT1 or miR-342-3p elevation inhibits CCSLC self-renewal, while FoxM1 suppression also attenuates this process. Overexpression of DNMT1 or FoxM1 reverses the effects of miR-342-3p mimic. This signaling pathway presents a potential therapeutic target for inhibiting CCSLC self-renewal in cervical cancer, offering new insights for targeted treatment strategies.
- Citation: Cao XZ, Zhang YF, Song YW, Yuan L, Tang HL, Li JY, Qiu YB, Lin JZ, Ning YX, Wang XY, Xu Y, Lin SQ. DNA methyltransferase 1/miR-342-3p/Forkhead box M1 signaling axis promotes self-renewal in cervical cancer stem-like cells in vitro and nude mice models. World J Stem Cells 2025; 17(3): 99472
- URL: https://www.wjgnet.com/1948-0210/full/v17/i3/99472.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v17.i3.99472
Cervical cancer (CC) is the fourth most common cancer in the world, representing a severe burden for females in underdeveloped nations[1,2]. Individuals with developed CC are frequently deprived of an operation choice and show poor outcomes after radiation treatment and chemotherapy. In this context, the 5-year survival rate for those with far-off metastasis reaches only 17.1%[3]. In the last decades, much attention has been paid to the cancer stem cell (CSC) hypothesis, which claims that an uncommon type of stem cell-like tumor cell is accountable for tumor progression, resistance, and recurrence[4]. According to this hypothesis, cancer stem-like cells derived from CC (CCSLCs) are a major cause of poor prognosis due to their high potential for inducing tumor formation and progression[5,6]. However, the mechanism that drives the initiation and progression of human CC is not fully understood.
Endogenous noncoding single-stranded RNAs, known as microRNAs (miRNAs), control 30% of the human genes through their union to the 3’-untranslated regions (3’-UTRs) of the miRNAs of interest[7]. In fact, some of the miRNA-based cancer therapeutic strategies have shown promising results even in early-phase human clinical trials[8]. In particular, abnormal miR-342-3p expression has significant associations with progression, proliferation, invasion, and even metastasis in a variety of tumors[9,10]. On the other hand, miR-342-3p reduces the proliferative, migratory, and invasive features of CC cells[11]. In this regard, Tao et al[12] demonstrated that determining the miR-342-3p expression profile might improve disease prognosis and help in decision-making during treatments for colon cancer. However, whether miR-342-3p promotes and maintains CCSLC self-renewal is still unclear.
Epigenetic regulation can promote oncogenes and/or suppress genes that prevent tumor growth, which have an important part in carcinogenesis, progress, invasion, and metastasis[13]. The epigenetic mechanism mainly includes DNA methylation, histone DNA methylation and deacetylation, non-coding RNA regulation, and crosstalk[14]. The main function of DNA methyltransferase 1 (DNMT1) is methylation of DNA[15]. Many reports have indicated that DNMT1 is deeply connected to the incidence and expansion of a variety of tumors. For instance, DNMT1 is crucial for the conservation of breast stem/progenitor cells and their CSCs[16]. The transactivation mechanism of the DNMT1 promoter reprograms the transcription factors octamer-binding transcription factor 4 (OCT4) and SRY-box transcription factor 2 (SOX2) to promote self-renewal of glioblastoma cells and leads to total DNA methylation and DNMT-dependent miRNAs downregulation[17].
Lee et al[18] found that suppressing DNMT1 expression had a relevant role in prostate cancer cells reversing the epithelial-mesenchymal transition and CSC phenotypes. Furthermore, Wang et al[19] showed that DNMT1 was inhibited by miR-342-3p, which signifies stronger proliferation and invasion in colorectal cancer cells. Nonetheless, further work is necessary to understand whether irregular DNMT1 expression leads to the downregulation of miR-342-3p and subsequent induction of CCSLC self-renewal in vitro and in vivo.
Forkhead box M1 (FoxM1) is necessary not only for cellular cycle advancement but also for apoptosis, angiogenesis, and DNA repair[20]. FoxM1 is significantly upregulated in several cancers, such as CC[21-23]. More importantly, FoxM1 has an important part in maintaining the CSC characteristics. Luo et al[23] found that in nasopharyngeal carcinoma tissues FoxM1 is associated with CSC-related clinicopathological characteristics (late clinical stage, tumor recurrence, and distant metastasis), and the expression of FoxM1 is also closely related to higher levels of the CSC markers Nanog, OCT4, and SOX2. It was also found that FoxM1 stimulates the proliferative, invasive, and stem cell characteristics of nasopharyngeal carcinoma cells[23].
However, more research is needed to evaluate how low miR-342-3p expression and subsequent FoxM1 overexpression in the context of the epigenetic regulation involved in DNMT1 could contribute to enriching CCSLCs and inducing CCSLC self-renewal in CC. In the present study, our objective was to evaluate the possibility that mutual adverse regulation between DNMT1 and miR-342-3p could induce CCSLC self-renewal by upregulating FoxM1 expression in CC cells.
The cell types for the cultures were HeLa, SiHa, and CaSki, together with the control cervical epithelial cell line HcerEpiC from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). For every culture, the medium of choice was Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, CA, United States), which included fetal bovine serum (10%; Gibco), in conditions of 37 °C, humidity, and 5% CO2. To obtain sphere-forming cells (SFCs) of CC, DMEM/F12 including 2% B27, basic fibroblast growth factor (20 ng/mL), epidermal growth factor (20 ng/mL), insulin (4 μg/mL), penicillin G (100 IU/mL), and streptomycin (100 μg/mL) was used as described previously[24]. HeLa cell-derived SFCs were then analyzed as CCSLCs. The SFCs derived from SiHa and CaSki were considered as CCSLCs. All medium reagents were obtained from Invitrogen (United States) except for the insulin (Sigma-Aldrich).
Trizol (Tiangen Biotech, China) was utilized to obtain total RNA from HeLa (1 × 106) or CCSLCs (1 × 106) cells. To extract total miRNA, we worked with the miRcute miRNA isolation Kit (Tiangen Biotech, China). For mRNA quantitation, transcription was performed on total RNA (2 μg) via the SureScript™ First-Strand cDNA Synthesis Kit (GeneCopoeia Inc., MD, United States). cDNA was replicated using the primers shown in Supplementary Table 1. The reaction entailed: Amplification at 95 °C (10 minutes); and 40 cycles at 95 °C (30 seconds), 55 °C (30 seconds), and 70 °C (30 seconds). To quantify miRNA, total miRNA (2 μg) underwent transcription and amplification through the All-in-One™ miRNA quantitative real-time PCR (qPCR) Detection Kit and the TaqMan MicroRNA Assay (Applied Biosystems, CA, United States), as suggested by the provider (GeneCopoeia Inc., MD, United States). U6 was considered the reference. For data analysis, the 2-ΔΔCt method was conducted.
A qPCR array (including 29 miRNAs that changed by flavonoids reported and 3 internal label genes) was used to detect miRNA expression profiles in HeLa-derived CCSLCs and HeLa cells according to the manufacturer’s protocol (Wcgene Biotech, Shanghai, China). Subsequently, the qPCR array (including 92 target genes associated with regulation of stem cell signaling that predicted using miRWalk network) was performed to compare in HeLa-derived CCSLCs transfected with miR-342-3p mimic or with miR-negative control (NC) according to the manufacturer’s protocol (Wcgene Biotech, Shanghai, China). The data underwent analysis utilizing Wcgene Biotech software. MiRNAs or genes displaying fold changes greater than or less than 2.0 were considered to hold biological significance.
DNA from HeLa (1 × 106) or CCSLCs (1 × 106) cells was obtained using the DNA-EZ Reagents V All-DNA-Out (Sangon Biotech, China). The incubation of genomic DNA was conducted with the Methylamp One-Step DNA Modification Kit (Epigentek), as recommended by the manufacturer. For PCR, the HotStar Taq Polymerase (Qiagen, Germany) was applied, together with methylated and unmethylated PCR primers targeting the miR-342-3p promoter (Sangon Biotech; Supplementary Table 2). For imaging of the methylation-specific PCR products, 2.0% agarose gel electrophoresis was used.
HeLa (1 × 106) or SFCs (1 × 106) cells were lysed using cold RIPA buffer (Beyotime Biotechnology, China). Proteins (20 μg) were first quantified with the Bradford assay (Bio-Rad, CA, United States). Then, proteins were separated on a 10% sodium-dodecyl sulfate gel electrophoresis and electrotransferred onto polyvinylidene fluoride membranes (Millipore, MA, United States). The membranes were blocked (2 h, room temperature, 5% BSA in Tris-buffered saline with tween) before overnight incubation with primary antibodies against DNMT1 (1:1000), FOXM1 (1:1000), CD44 (1:1000), and β-actin (1:2000) at 4 °C. Cell Signaling (United States) provided all antibodies. Suitable HRP-conjugated secondary antibodies (Beyotime Biotechnology, China) were included in a further incubation of the membranes (1 h). For detection, the enhanced chemiluminescence kit (Amersham, TN, United States) was implemented.
HeLa, CaSki, and SiHa cells (1 × 103) or their CCSLCs (1 × 103) underwent a sphere generation culture for 6 days. To determine the sphere formation efficiency, we considered the number of spheres generated/cells seeded (1000 cells per well in a 24-well plate) × 100%. Independent triplicates were included.
DMEM (0.8% agarose; Invitrogen, CA, United States) was first placed into 24-well plates (500 μL). Second, HeLa and SiHa cells and their CCSLCs (100 cells/well) were seeded in 24-well plates with CSC-M with 0.4% agarose (top layer) for a 3-week incubation. For colony counting, we employed an IX53 inverted microscope (Olympus, Japan). To know the colony formation efficiency (CFE), we considered the number of colonies generated/cells seeded (100 cells per well in a 24-well plate) × 100%. Independent triplicates were conducted.
HeLa cells (1 × 105) or CCSLCs (1 × 105) were infected for 48 h with lentiviruses bearing DNMT1 shRNA or cDNA or RFP setups in a 6 μg/mL polybrene-complemented medium; afterwards, a second 48 h-culture was carried out. Puromycin (4 μg/mL) was used for the selection assay during the first 7 days, and additional cell supply was conducted with 1 μg/mL puromycin. Transfection of 50 nM micrON™ miR-342 mimic and 100 nM micrOFF™ miR-342 suppressor into HeLa or CCSCs cells was performed using the iboFECT™ CP Reagent, as instructed by the provider (RiboBio, Guangzhou, China). In parallel, the NC of miR-342-3p suppressor (anti-NC) and miR-342-3p mimic (miR-NC) were transfected as well. Cell incubation with small RNA complexes was carried out for 2 h prior to changing the medium. shRNA control (sh-NC) and FOXM1 (shFOXM1) were acquired from Santa Cruz Biotechnology. The respective sequences were shown in Supplementary material. In addition, pcDNA3.1 both control (pcDNA) and FOXM1 (pcDNA-FOXM1) were acquired from Invitrogen. For cell transfection, the opti-MEM Lipofectamine™ 2000 (Invitrogen, CA, United States) was employed as recommended by the provider.
The 3’UTR target sequence of DNMT1 and FOXM1 [wildtype (WT)] that encompassed the ham-miR-342-3p binding spot was introduced into the pGL3 Luciferase vector (Ambion, TX, United States), as recommended by the provider. The control sample was a DNMT1 and FOXM1 3’UTR sequence with a mutation. The transfection of those arrangements (0.2 μg) into HeLa-derived CCSLCs was performed in 24-well microplates together with miR-342-3p or miR-NC using Lipofectamine 2000 (48 h). Finally, the dual luciferase reporter procedure (Promega) was conducted as recommended by the provider.
Nude mice of 4-5 weeks and 12-14 g (female, nude) were acquired from the Hunan SJA Laboratory Animal Co., Ltd. (certificate No: 43004700050992, China). The mice underwent adaptation for 1 week before assay initiation. Subcutaneous injections with HeLa cells (1 × 103, 1 × 104, 1 × 105) and HeLa-derived CCSLCs cells (1 × 102, 1 × 103, 1 × 104) were administered into the left flank and the right flank of the mice (n = 5/group) for the in vivo tumorigenicity assessment. The animals underwent sacrifice 1 month after the injection, and the weight of the xenografts was determined. Limiting dilutions were calculated using the Extreme Limiting Dilution Analysis software (http://bioinf.wehi.edu.au/software/elda/). The Ethics Committee of Hunan Normal University together with the Committee of Experimental Animal Feeding and Management approved all research assays (Permit No: 2020041).
To assess the in vivo functional impact of DNMT1 and FOXM1 genes on the xenograft expansion of CCSCs, the mice received subcutaneous inoculations (100 μL). HeLa-derived CCSLCs (1 × 105) were transduced with sh-NC, sh-DNMT1, or sh-FOXM1 in DMEM/F12 medium with no serum and matrigel (1:1; BD Biosciences). There were four mice in each group. To evaluate the impact of miR-342-3p on the xenograft expansion of CCSCs in vivo, the mice received sub
Immunohistochemistry was achieved through conventional procedures[24]. Target slides were incubated (4 °C, overnight) using the antibodies anti-DNMT1, FoxM1, and CD133 (all 1:200; Cell Signaling). As a NC, phosphate buffered saline replaced primary antibodies.
The fluorescent in situ hybridization kit (Gene-Pharma, Shanghai, China) was implemented for the fluorescent in situ hybridization experiment as recommended by the provider. The hybridization was executed by means of Cy3-labeled miR-342-3p probes (Gene-Pharma; Supplementary Table 3), and subsequent analysis was performed via an upright fluorescence microscope (ECLIPSE E600, Nikon, Japan).
Measurement data were manifested as mean ± SD, and three or more independent assays were performed. Data analysis employed SPSS 20.0 (SPSS, United States). Additionally, two-tailed Student’s t-tests were carried out for comparisons to control groups. For multiple group comparisons, we applied one-way analysis of variance plus the post-hoc Dunnett’s analysis. Statistical significance was recognized when P < 0.05.
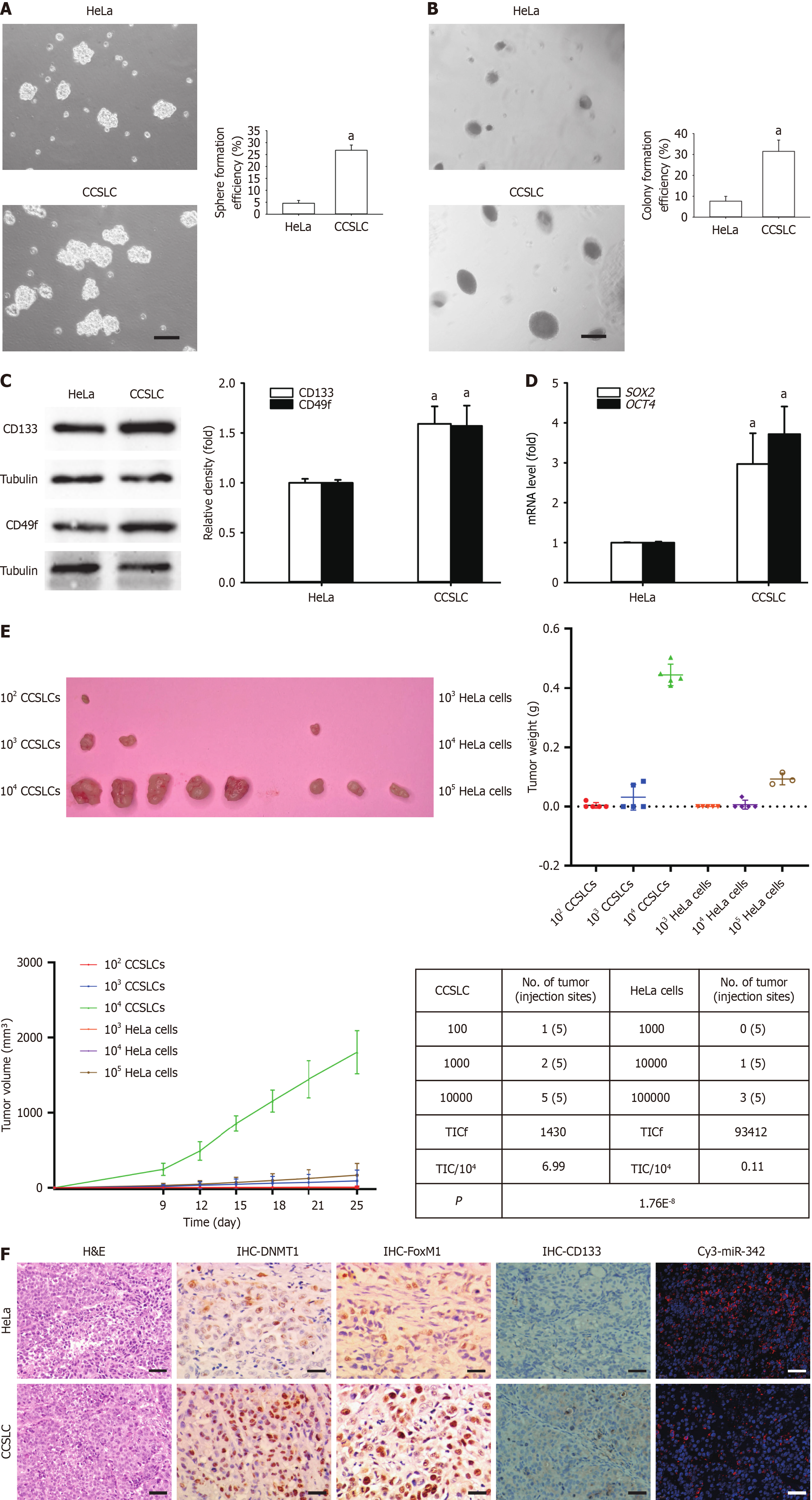
SFCs derived from the HeLa cell line were cultured and characterized for subsequent experiments. These SFCs had stronger self-renewal-related stemness in vitro, involving stronger spheres and CFE (Figure 1A and B), higher CD133 and CD49f protein levels (Figure 1C), and elevated mRNA expression of stemness-related factors SOX2 and OCT4 (Figure 1D). Importantly, SFCs exhibited stronger tumorigenesis in vivo (Figure 1E) and higher protein levels of DNMT1, FoxM1, and CD133 as well as lower miR-342-3p in vivo (Figure 1F). The above indicated that HeLa-derived SFCs had self-renewal-related stemness in vitro and displayed stronger tumorigenesis in vivo.
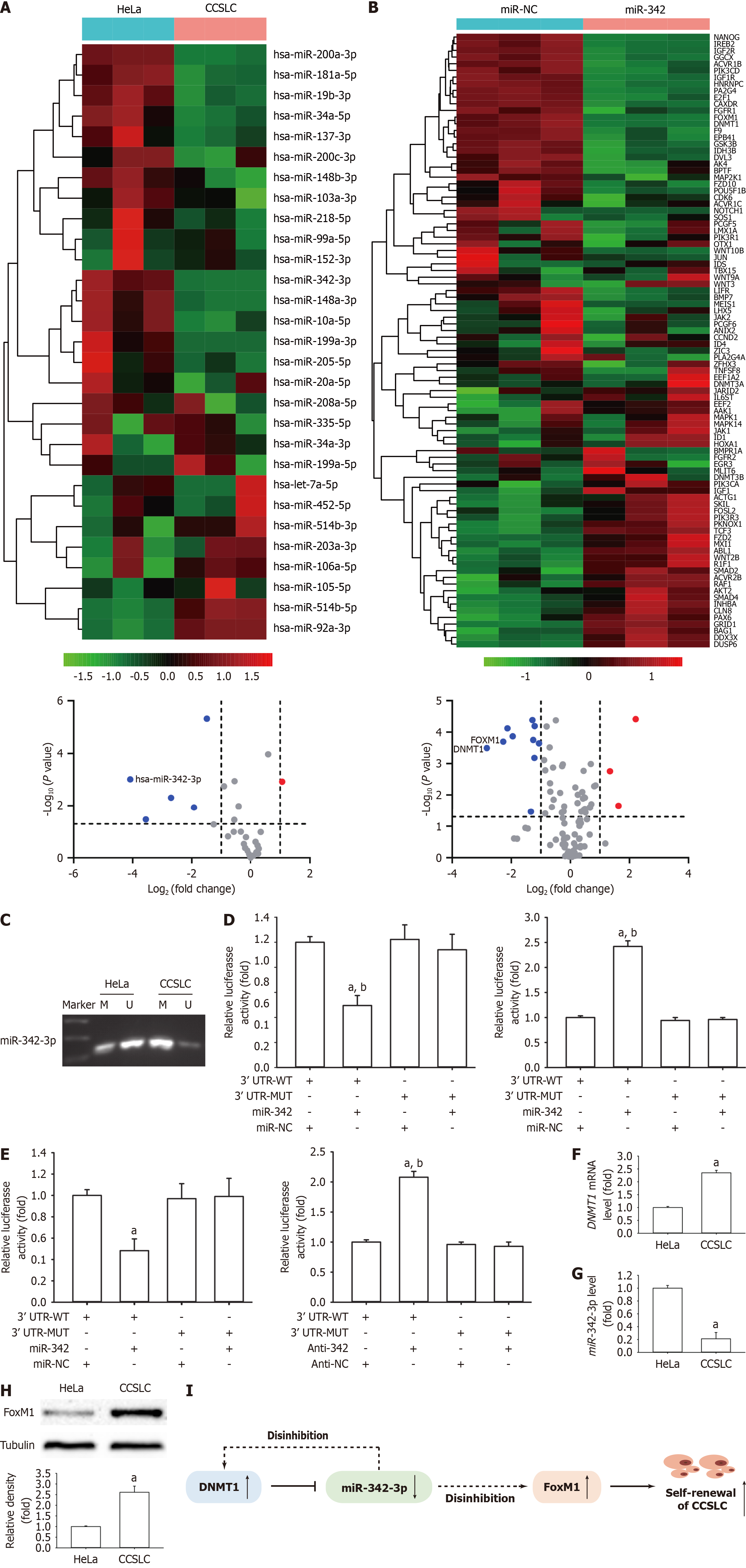
MiRNAs have a critical role in regulating CSCs including their stemness characteristics. To prioritize miRNAs with the greatest difference in expression between CCSLCs and HeLa cells (control), the mRNA level was compared using a qPCR array. The results demonstrated that hsa-miR-342-3p, hsa-miR-199a-3p, hsa-miR-148a-3p, hsa-miR-34a-5p, and hsa-miR-200a-3p were significantly lower in CCSLCs compared with HeLa cells (Figure 2A; Supplementary Table 4).
To further explore the molecular mechanism by which miR-342-3p inhibited self-renewal-related stemness in CCSLCs, we used a qPCR array to compare the differential expression profiles of gene mRNAs in CCSLCs transfected with miR-342-3p mimic (miR-342) and miR-342-3p mimic-NC oligonucleotide (miR-NC). Transfection with miR-342-3p mimics was found to downregulate the mRNA expression of seven target genes, including DNMT1, FOXM1, and insulin-like growth factor-1 receptor etc. (Figure 2B; Supplementary Table 5).
Notably, the methylation level of the miR-342-3p promoter was higher in the HeLa-derived CCSLCs vs the HeLa cells (Figure 2C). The luciferase reporter assay showed that the luciferase activity was repressed or stimulated after cotransfection with the miR-342-3p mimic or inhibitor and DNMT1-3’-UTR-WT (Figure 2D). Nevertheless, this outcome was not replicated after cotransfection with miR-342-3p mimic or suppressor and DNMT1-3’-UTR-Mutation (Figure 2D). Our findings indicated that DNMT1 was a plain target of miR-342-3p in HeLa-derived CCSLCs.
High FoxM1 levels and downregulation of miR-342-3p altogether hold a relevant role in CC advancement[11]. Moreover, previous reports, including ours, have indicated that FoxM1 is a crucial modulator of CSLC characteristics in a variety of cancer cells[11,21-23]. We found the luciferase activity was repressed or raised after cotransfection with miR-342-3p mimic or suppressor and FOXM1-3’-UTR-WT (Figure 2E). Nevertheless, this outcome was not present after cotransfection with miR-342-3p mimic or suppressor and FOXM1-3’-UTR-Mutation (Figure 2E).
We next determined whether SFCs from HeLa cells were considered HeLa-derived CCSLCs with the phenotypes of DNMT1 and FoxM1 upregulation as well as low miR-342-3p levels. As expected, CCSLCs from HeLa cells shared the expression pattern of the above three for DNMT1, FoxM1, and miR-342-3p (Figure 2F-H). Accordingly, the aforementioned results suggested that constitutive activation of DNMT1 silences miR-342-3p expression by its promoter methylation. Low expression of miR-342-3p activates DNMT1 by ceasing inhibition. This reciprocal negative feedback regulation leads to higher expression of DNMT1 and lower expression of miR-342-3p. Lower expression of miR-342-3p caused FoxM1 upregulation by ceasing inhibition and subsequently promoted and maintained the CCSLC self-renewal in CC (Figure 2I).
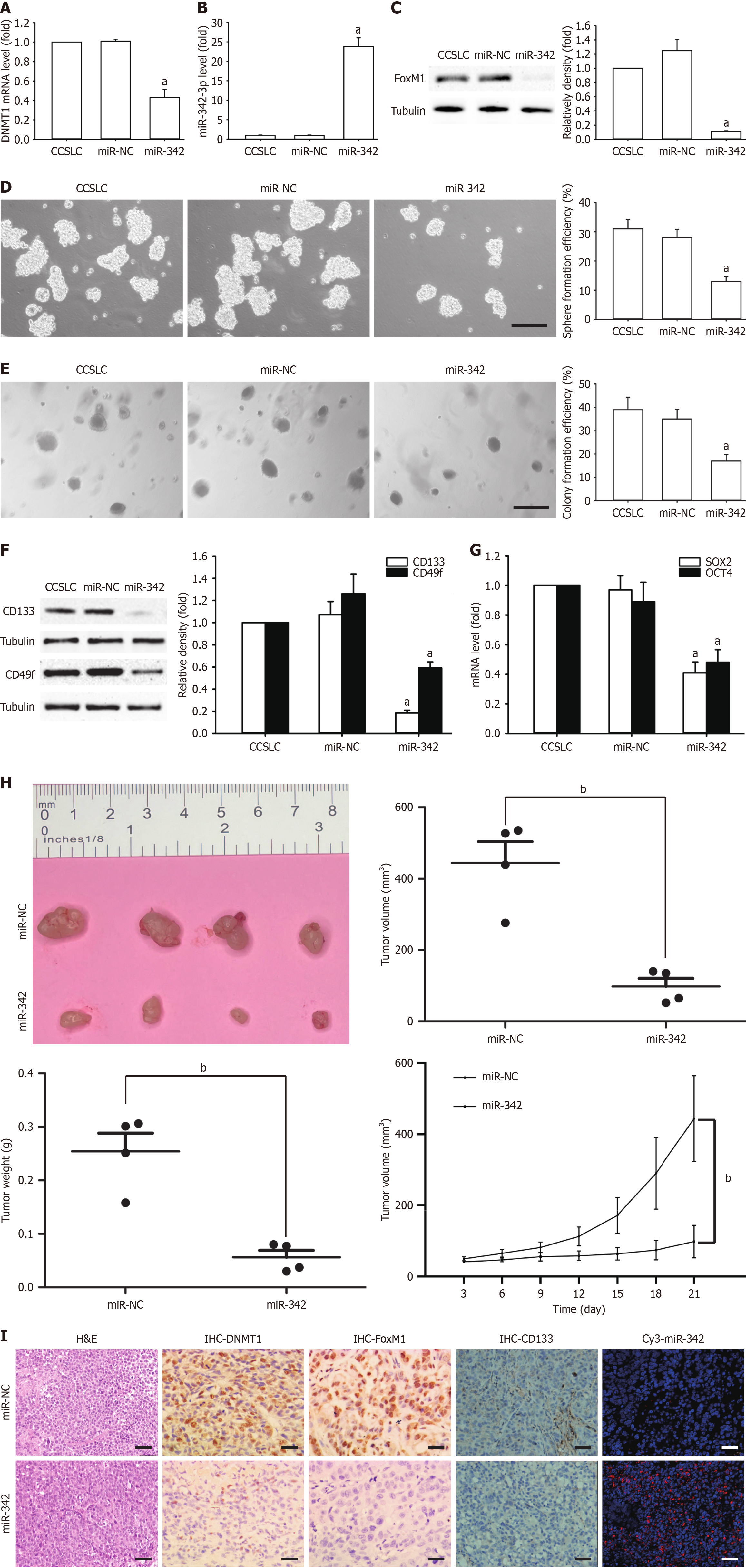
To study the in vitro impact of miR-342-3p on self-renewal-related stemness, HeLa-derived CCSLCs were transfected using miR-342-3p mimic (miR-342) or miR-NC. Compared to the miR-NC transfection group, DNMT1 mRNA level in CCSLCs was markedly decreased in CCSLCs transfection with miR-342-3p mimic (Figure 3A), and miR-342-3p was higher in CCSLCs after transfecting with miR-342-3p mimic (Figure 3B). In addition, FoxM1 protein amounts in CCSLCs were remarkably decreased (Figure 3C). Furthermore, the miR-342-3p mimic transfection attenuated sphere and CFE in CCSLCs vs the miR-NC transfection block or the non-transfected cells (Figure 3D and E). Moreover, the levels of CD133 and CD49f protein (Figure 3F) and SOX2 and OCT4 mRNA (Figure 3G) diminished after transfection of CCSLCs with miR-342-3p mimic.
Next, we tested the effects of agomir-342 (a miR-342-3p mimic) in xenografts of nude mice. The result showed that agomir-342 efficiently decreased the volume and weight of the tumor (Figure 3H). Most importantly, agomir-342 effectively inhibited tumor growth and appeared to downregulate DNMT1, FoxM1, and CD133 protein expression and elevated miR-342-3p levels in individuals bearing HeLa-derived CCSLCs (Figure 3I). Our results imply that miR-342-3p could reduce DNMT1 and FoxM1 expression to attenuate self-renewal-related stemness, suggesting that mutual negative regulation between DNMT1 and miR-342-3p leading to the regulation of FoxM1 might be involved in the maintenance and promotion of self-renewal-related stemness.
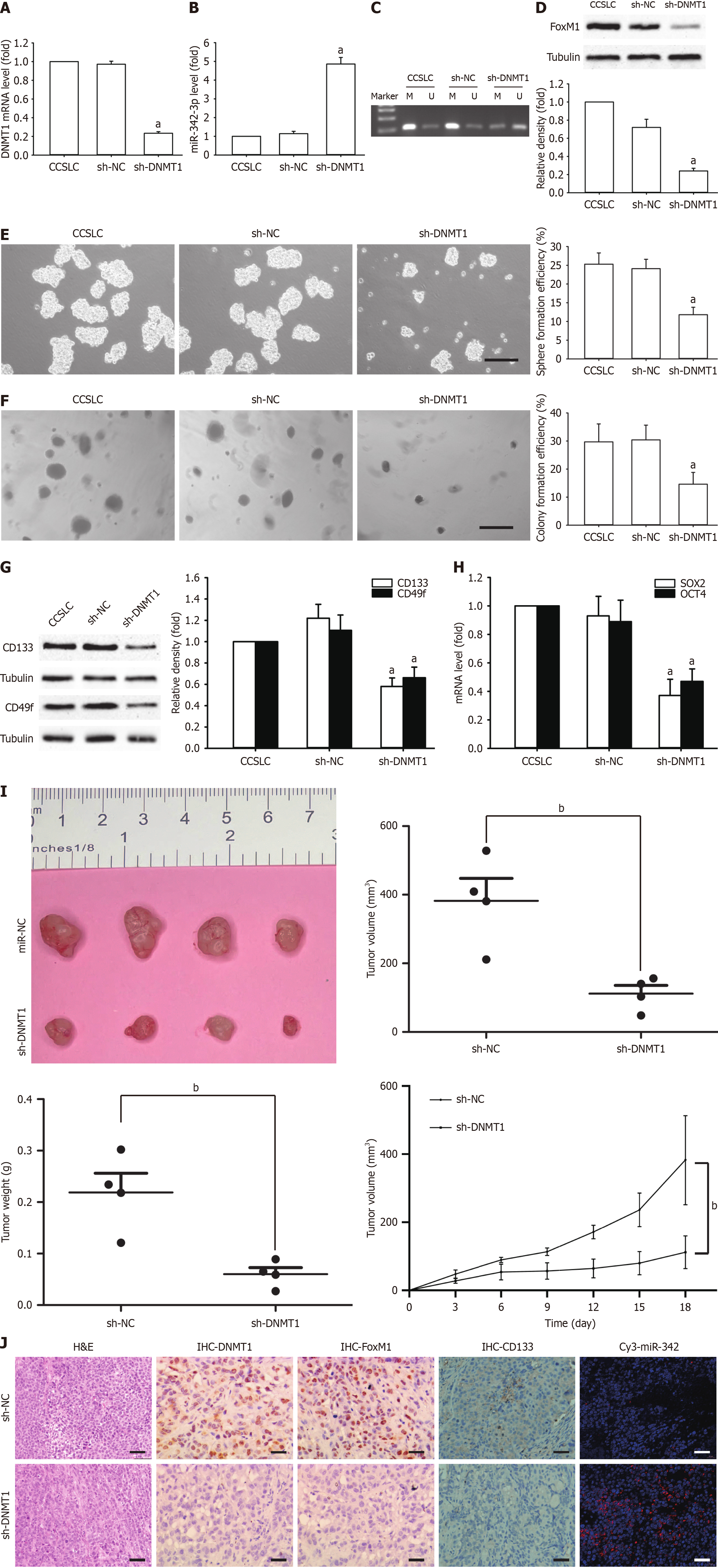
To evaluate the increased function of DNMT1 in preserving CSLC characteristics, DNMT1 shRNA was implemented for the knockdown of DNMT1 in HeLa-derived CCSLCs (sh-DNMT1). DNMT1 mRNA level were remarkably reduced in the knocked-down DNMT1 CCSLCs (Figure 4A). Meanwhile, miR-342-3p was higher and had lower miR-342-3p promoter methylation vs the sh-NC control cells (Figure 4B and C). FoxM1 protein levels in CCSLCs were remarkably reduced (Figure 4D). Among CCSLCs, DNMT1 knockdown decreased spheres and CFE (Figure 4E and F). Additionally, the CD133 and CD49f protein quantities decreased in CCSLCs bearing DNMT1 shRNA (Figure 4G). Moreover, SOX2 and OCT4 gene expression in CCSLCs bearing DNMT1 shRNA were lower vs the HeLa-derived CCSCs transfected with sh-NC (Figure 4H). The knockdown of DNMT1 blocked tumor growth (Figure 4I) and appeared to reduce DNMT1, FoxM1, and CD133 protein expression as well as elevated miR-342-3p levels in our xenograft samples (Figure 4J). These findings suggest that the knockdown of DNMT1 repressed self-renewal-related stemness in HeLa-derived CCSLCs, probably by upregulation of miR-342-3p after its promoter hypomethylation.
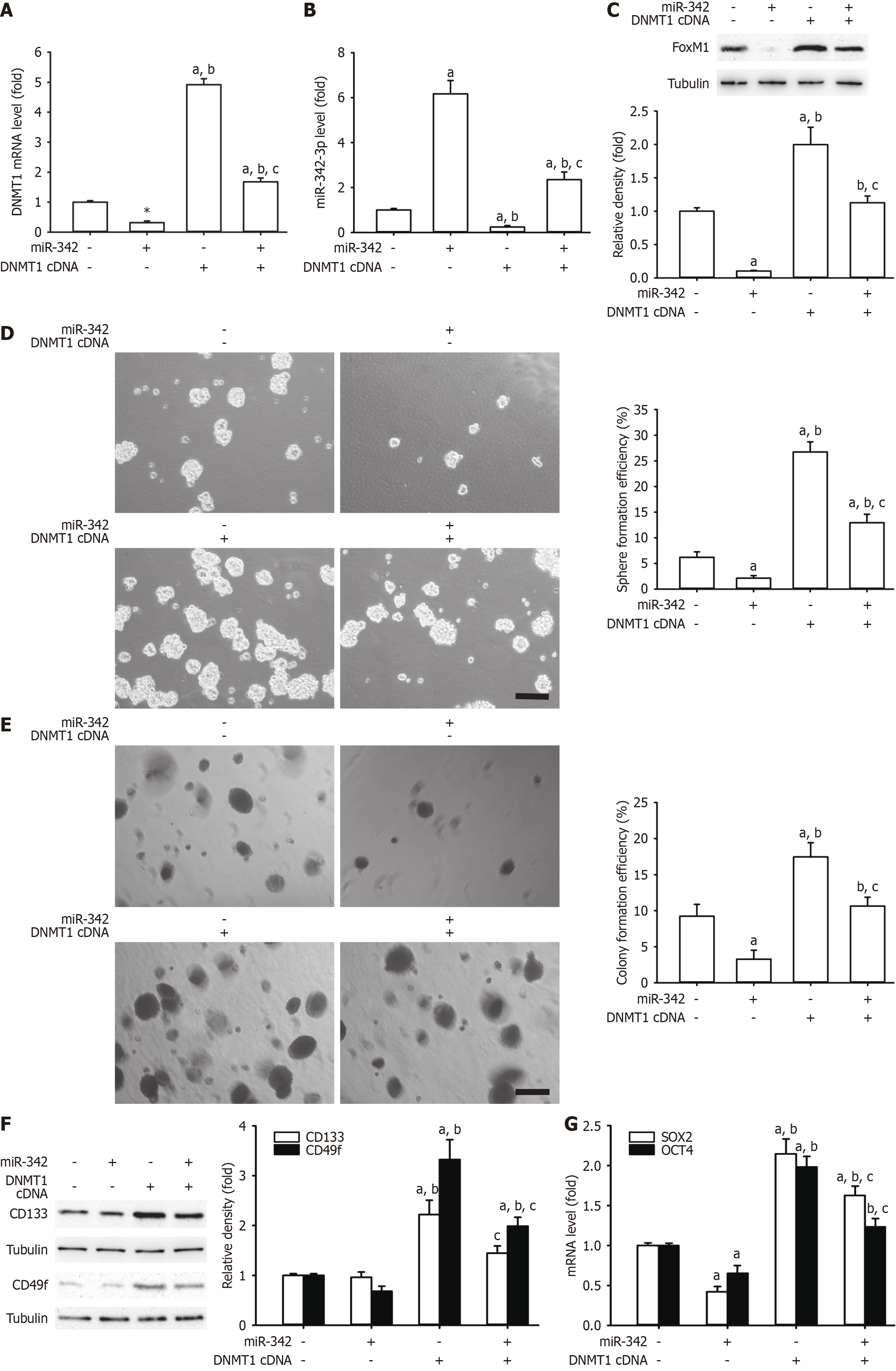
DNMT1 is responsible for self-renewal-related stemness in a variety of cancers and may be subject to miRNA regulation[15,25,26]. To investigate the interplay between miR-342-3p and DNMT1, we constructed DNMT1 expression plasmids to perform a rescue experiment. Among our results, overexpression of DNMT1 reestablished the DNMT1 mRNA level that had been diminished by miR-342-3p and that had been increased by miR-342-3p mimics (Figure 5A and B). Additionally, restoration of DNMT1 abrogated effects of miR-342-3p mimic on the FoxM1 protein expression, the spheres, CFE, and the CD133 and CD49f protein levels (Figure 5C-F). The SOX2 and OCT4 mRNA amounts were lower in miR-342-3p mimics (Figure 5G). The above suggested that DNMT1 might be a straight functional target gene of miR-342-3p. Moreover, miR-342-3p operated as a tumor suppressor via the mutual adverse modulation between miR-342-3p and DNMT1.
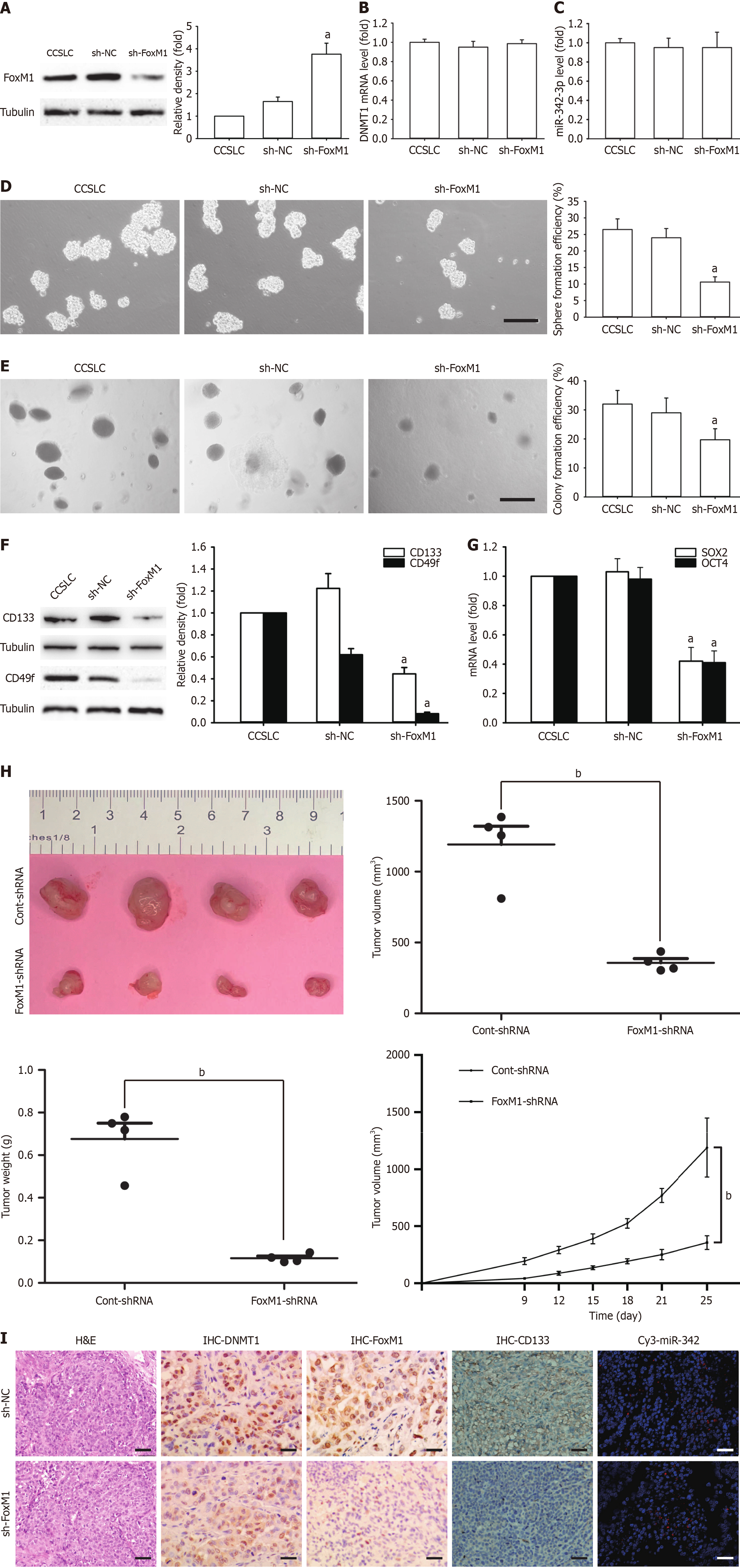
To understand how FoxM1 maintained self-renewal-related stemness in CCSLCs, FOXM1 was downregulated using shRNA in HeLa-derived CCSLCs (shFOXM1). The resulting FoxM1 protein amounts were markedly decreased (Figure 6A). However, DNMT1 mRNA and miR-342-3p levels were not significantly altered in comparison to the non-transduced cells and the sh-NC control block (Figure 6B and C). In CCSLCs, FOXM1 knockdown diminished spheres and CFE (Figure 6D and E). In CCSLCs bearing FOXM1 shRNA, the CD133 and CD49f protein levels were decreased (Figure 6F). On the other hand, in CCSCs bearing FOXM1 shRNA, SOX2 and OCT4 gene expression levels were lower to those transfected with sh-NC (Figure 6G). Moreover, knockdown of FOXM1 inhibited tumor expansion (Figure 6H) and appeared to downregulate FoxM1 and CD133 protein expressions but did not affect DNMT1 and miR-342-3p expression in our xenograft model (Figure 6I). These findings suggest that knockdown of FOXM1 inhibited self-renewal-related stemness, mimicked the impact of miR-342-3p upregulation, and indicated that the reciprocal regulation of DNMT1 and miR-342-3p was upstream of FoxM1.
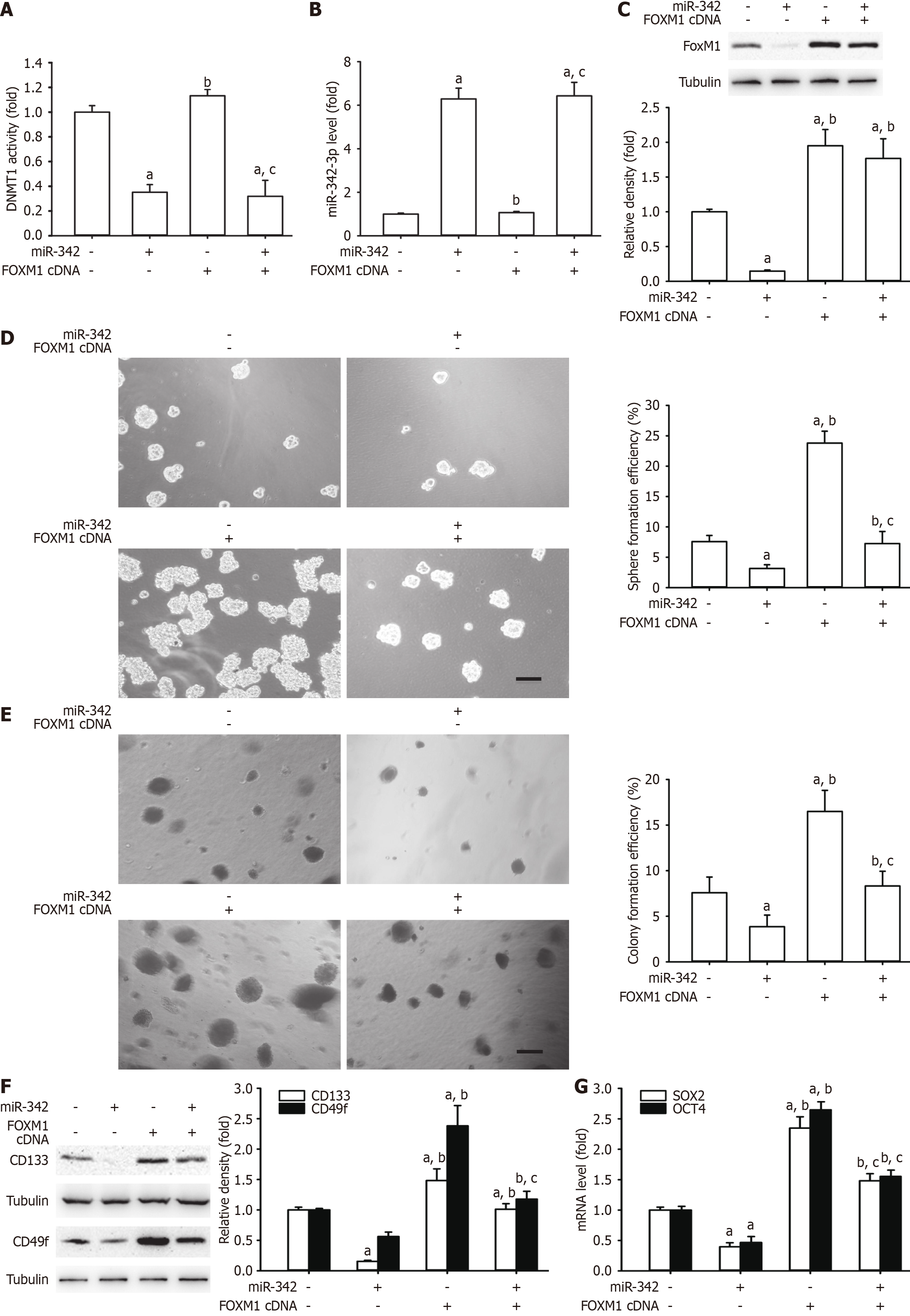
FoxM1 has a pivotal function in the generation of self-renewal-related stemness, and it may be subject to miRNA regulation[22,27,28]. To explore whether miR-342-3p plays an inhibitory role in self-renewal-related stemness in CCSLCs via FoxM1, we created FOXM1 plasmids to perform a rescue experiment. As a result, overexpression of FOXM1 did not change DNMT1 mRNA and miR-342-3p by miR-342-3p mimics (Figure 7A and B). Furthermore, restoration of FOXM1 abrogated the inhibitory effects of miR-342-3p mimics on FoxM1 protein levels, spheres, CFE, and CD133 and CD49f protein (Figure 7C-F). The SOX2 and OCT4 mRNA quantities decreased by miR-342-3p mimics, and these effects were rescued by restoration of FOXM1 (Figure 7G). The above suggested that FoxM1 might be a straight functional target gene of miR-342-3p. Moreover, miR-342-3p operated as a tumor suppressor via FoxM1.
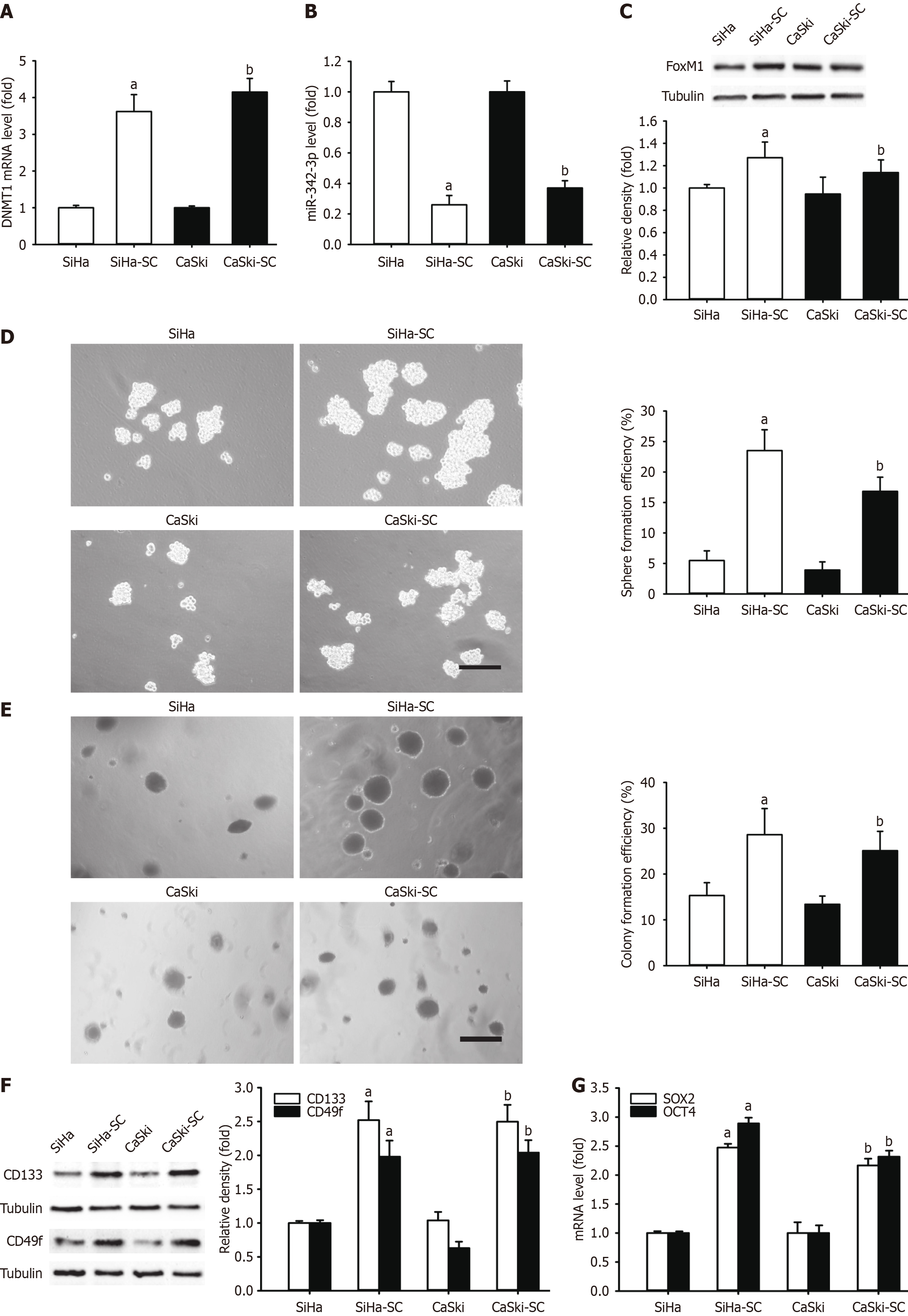
Two additional CC cell lines, CasKi and SiHa cells, were chosen to contrast the level of DNMT1 mRNA, miR-342-3p, and FoxM1, and self-renewal-related stemness. The comparison was performed between the cellular monolayer and the respective SFCs. As a result, DNMT1 mRNA level increased (Figure 8A), while miR-342-3p levels were significantly decreased (Figure 8B). In addition, FoxM1 protein expression (Figure 8C), spheres, and CFE (Figure 8D and E) were considerably amplified in the respective CCSLCs. We also assessed CD133 and CD49f levels in CasKi and SiHa cells in relation to the respective CCSLCs, with increasing results (Figure 8F). The SOX2 and OCT4 mRNA quantities were enhanced as well (Figure 8G). The present outcome indicated that the DNMT1/miR-342-3p/FoxM1 axis might promote self-renewal-related stemness in CasKi and SiHa cell lines. Finally, the DNMT1/miR-342-3p/FoxM1 axis could be a new target to mimic self-renewal-related stemness in CCSLCs for CC treatment.
We aimed to study the functional linkage between DNMT1, miR-342-3p, and FoxM1, which regulates CCSLC self-renewal in CC. As far as we know, this is the first time that the CCSLC self-renewal in CC in vitro and in vivo was enhanced by the mutual negative regulation between DNMT1 and miR-342-3p, with the subsequent increase in FoxM1 levels by reactivation via miR-342-3p silencing. These results seem important not only because they describe the links among DNMT1, miR-342-3p, and FoxM1 in a cell subpopulation with CCSLC self-renewal but also because they might provide ideas for the development of novel therapies against CCSLCs, which are causal cells in CC.
In recent years, much attention has been paid to silencing tumor suppressors or miRNAs inhibitors in precancerous cells via epigenetic regulation[29]. DNMT1 is unusually triggered in tumors and CSC, which catalyze DNA methylation[30-32]. Moreover, DNMT1 is essential to keep CSLC characteristics[16]. In breast cancer, epigenetic altering factors, including 5-azacytidine, can significantly shrink the CSC subgroup[33]. Based on the GEO analyses, DNMT1 and FOXM1 transcript levels were increased in CC tissues compared with paracarcinoma tissues. With respect to the positive correlation between the expression levels of DNMT1 and FOXM1 in CC progression, we hypothesized that DNMT1 (an epigenetic modification enzyme) activates FoxM1 in an epigenetic regulatory event, which simultaneously affects DNMT1 and FoxM1. The above seems to play a relevant role in CC progression, particularly by promoting the CCSLC self-renewal in CC.
Increasing evidence shows that miRNAs are implicated in abnormal machinery of DNA hypermethylation via DNMT regulation. In particular, miR-342-3p works as a tumor inhibitor and has low levels in various cancers, such as human acute myeloid leukemia and CC[10,11,19,34]. Interestingly, it has been suggested that miR-342-3p is modulated by DNMT1 via promoter methylation in cancerous cells or CSLCs[19,35]. Our study provided evidence that the mutual negative interplay between DNMT1 and miR-342-3p stimulated the emergence of CCSLC self-renewal and carcinogenicity in CCSLCs. Knockdown of DNMT1 elevated miR-342-3p levels and decreased CCSLC self-renewal and carcinogenicity. MiR-342-3p mimic transfection repressed both the transcriptional and posttranscriptional expression of DNMT1. More notably, the overexpression of DNMT1 reversed the blocking impact of overexpressing miR-342-3p on CCSLC self-renewal and carcinogenicity in HeLa cell lines. Furthermore, the luciferase assay showed DNMT1 as a target gene of miR-342-3p, which would signify that miR-342-3p is particularly implicated in the modulation of DNMT1. These findings suggest that the mutual negative modulation between DNMT1 and miR-342-3p could become a prospective therapeutic approach for treatment through CCSLCs targeting. The above could assist in the search for a demethylating agent in the field of epigenetic cancer therapy.
FoxM1 is a well-known cancerogenic transcription element that plays a role in numerous biological routes, including the generation of CSLC characteristics[21-23]. However, in CCSLCs, the FoxM1-linked upstream modulatory processes remain unclear. This study is consistent with this hypothesis since we observed that knockdown of FOXM1 can replicate the inhibitory impact of miR-342-3p on CCSLC self-renewal. Importantly, increased expression of FOXM1 reversed the suppressive effects of overexpressing miR-342-3p on CCSLC self-renewal and carcinogenicity but did not affect DNMT1 and miR-342-3p expression in HeLa cells. However, one limitation should be noted. Differential self-renewal capability between the CC cell lines HeLa, SiHa, and CasKi used in our study were observed, and the reason was unclear. This may be attributed to the different sensitivities of the cell lines and their differentially expressed genes[36,37].
Our results in this work provided a rationale for FoxM1 promoting CCSLC self-renewal by miR-342-3p downregulation, which functioned through the mutual negative modulation between DNMT1 and miR-342-3p in CC. Interestingly, we also demonstrated that overexpression of FOXM1 is able to reverse the suppressive impact of miR-342-3p over CCSLC self-renewal without influencing the expression of miR-342-3p and DNMT1. As far as we know, this is the first report to demonstrate miR-342-3p downregulation via upregulation of FOXM1 by DNMT1 activation in promoting CCSLC self-renewal of CC.
The authors would like to acknowledge Prof. Jian-Guo Cao (Hunan Normal University, Changsha, Hunan) for his enthusiastic support and strong guidance and Prof. Ying-Jun Chen (Guangdong Pharmaceutical University, Guangzhou, Guangdong) for her professional review of the statistical methods of this study.
| 1. | Liu Y, Shi W, Mubarik S, Wang F. Assessment of secular trends of three major gynecologic cancers burden and attributable risk factors from 1990 to 2019: an age period cohort analysis. BMC Public Health. 2024;24:1349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 2. | Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2279] [Cited by in RCA: 3618] [Article Influence: 3618.0] [Reference Citation Analysis (3)] |
| 3. | Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 1440] [Article Influence: 240.0] [Reference Citation Analysis (0)] |
| 4. | Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1150] [Cited by in RCA: 1108] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 5. | Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 637] [Article Influence: 53.1] [Reference Citation Analysis (0)] |
| 6. | Di Fiore R, Suleiman S, Drago-Ferrante R, Subbannayya Y, Pentimalli F, Giordano A, Calleja-Agius J. Cancer Stem Cells and Their Possible Implications in Cervical Cancer: A Short Review. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 7. | Shang R, Lee S, Senavirathne G, Lai EC. microRNAs in action: biogenesis, function and regulation. Nat Rev Genet. 2023;24:816-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 301] [Article Influence: 150.5] [Reference Citation Analysis (0)] |
| 8. | Toden S, Zumwalt TJ, Goel A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 180] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 9. | Xu C, Sun W, Liu J, Pu H, Li Y. MiR-342-3p inhibits LCSC oncogenicity and cell stemness through HDAC7/PTEN axis. Inflamm Res. 2022;71:107-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Komoll RM, Hu Q, Olarewaju O, von Döhlen L, Yuan Q, Xie Y, Tsay HC, Daon J, Qin R, Manns MP, Sharma AD, Goga A, Ott M, Balakrishnan A. MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. J Hepatol. 2021;74:122-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 114] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 11. | Li XR, Chu HJ, Lv T, Wang L, Kong SF, Dai SZ. miR-342-3p suppresses proliferation, migration and invasion by targeting FOXM1 in human cervical cancer. FEBS Lett. 2014;588:3298-3307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 12. | Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391-401. [PubMed] |
| 13. | Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13:877-919. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 341] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 14. | Zhou S, Feng S, Qin W, Wang X, Tang Y, Yuan S. Epigenetic Regulation of Spermatogonial Stem Cell Homeostasis: From DNA Methylation to Histone Modification. Stem Cell Rev Rep. 2021;17:562-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 15. | Wong KK. DNMT1: A key drug target in triple-negative breast cancer. Semin Cancer Biol. 2021;72:198-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 110] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 16. | Pathania R, Ramachandran S, Elangovan S, Padia R, Yang P, Cinghu S, Veeranan-Karmegam R, Arjunan P, Gnana-Prakasam JP, Sadanand F, Pei L, Chang CS, Choi JH, Shi H, Manicassamy S, Prasad PD, Sharma S, Ganapathy V, Jothi R, Thangaraju M. DNMT1 is essential for mammary and cancer stem cell maintenance and tumorigenesis. Nat Commun. 2015;6:6910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 182] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 17. | Lopez-Bertoni H, Lal B, Li A, Caplan M, Guerrero-Cázares H, Eberhart CG, Quiñones-Hinojosa A, Glas M, Scheffler B, Laterra J, Li Y. DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene. 2015;34:3994-4004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 18. | Lee E, Wang J, Yumoto K, Jung Y, Cackowski FC, Decker AM, Li Y, Franceschi RT, Pienta KJ, Taichman RS. DNMT1 Regulates Epithelial-Mesenchymal Transition and Cancer Stem Cells, Which Promotes Prostate Cancer Metastasis. Neoplasia. 2016;18:553-566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 101] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 19. | Wang H, Wu J, Meng X, Ying X, Zuo Y, Liu R, Pan Z, Kang T, Huang W. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis. 2011;32:1033-1042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 20. | Gu L, Liu HM. Forkhead box M1 transcription factor: a novel target for pulmonary arterial hypertension therapy. World J Pediatr. 2020;16:113-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 21. | Kopanja D, Pandey A, Kiefer M, Wang Z, Chandan N, Carr JR, Franks R, Yu DY, Guzman G, Maker A, Raychaudhuri P. Essential roles of FoxM1 in Ras-induced liver cancer progression and in cancer cells with stem cell features. J Hepatol. 2015;63:429-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 22. | Alimardan Z, Abbasi M, Hasanzadeh F, Aghaei M, Khodarahmi G, Kashfi K. Heat shock proteins and cancer: The FoxM1 connection. Biochem Pharmacol. 2023;211:115505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 23. | Luo W, Gao F, Li S, Liu L. FoxM1 Promotes Cell Proliferation, Invasion, and Stem Cell Properties in Nasopharyngeal Carcinoma. Front Oncol. 2018;8:483. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | Wang XL, Cao XZ, Wang DY, Qiu YB, Deng KY, Cao JG, Lin SQ, Xu Y, Ren KQ. Casticin Attenuates Stemness in Cervical Cancer Stem-Like Cells by Regulating Activity and Expression of DNMT1. Chin J Integr Med. 2023;29:224-232. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 25. | Xu P, Wang L, Liu Q, Gao P, Hu F, Xie X, Jiang L, Bi R, Ding F, Yang Q, Xiao H. The abnormal expression of circ-ARAP2 promotes ESCC progression through regulating miR-761/FOXM1 axis-mediated stemness and the endothelial-mesenchymal transition. J Transl Med. 2022;20:318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Wang Q, Liang N, Yang T, Li Y, Li J, Huang Q, Wu C, Sun L, Zhou X, Cheng X, Zhao L, Wang G, Chen Z, He X, Liu C. DNMT1-mediated methylation of BEX1 regulates stemness and tumorigenicity in liver cancer. J Hepatol. 2021;75:1142-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 27. | Sheng Y, Yu C, Liu Y, Hu C, Ma R, Lu X, Ji P, Chen J, Mizukawa B, Huang Y, Licht JD, Qian Z. FOXM1 regulates leukemia stem cell quiescence and survival in MLL-rearranged AML. Nat Commun. 2020;11:928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 28. | Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 Are Promising Prognostic Biomarkers and Novel Targets of Tumor-Suppressive miR-342 in Human Colorectal Cancer. Clin Cancer Res. 2016;22:4947-4957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 29. | Hamurcu Z, Sener EF, Taheri S, Nalbantoglu U, Kokcu ND, Tahtasakal R, Cınar V, Guler A, Ozkul Y, Dönmez-Altuntas H, Ozpolat B. MicroRNA profiling identifies Forkhead box transcription factor M1 (FOXM1) regulated miR-186 and miR-200b alterations in triple negative breast cancer. Cell Signal. 2021;83:109979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y, Jiang G, Lu M, Zhang Z, Yin J, Zeng S, Chen X, Deng M, Jia X, Gu Y, Chen D, Zheng G, He Z. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 2020;27:966-983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 115] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 31. | Gao X, Sheng Y, Yang J, Wang C, Zhang R, Zhu Y, Zhang Z, Zhang K, Yan S, Sun H, Wei J, Wang X, Yu X, Zhang Y, Luo Q, Zheng Y, Qiao P, Zhao Y, Dong Q, Qin L. Osteopontin alters DNA methylation through up-regulating DNMT1 and sensitizes CD133+/CD44+ cancer stem cells to 5 azacytidine in hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 32. | Zhang MY, Calin GA, Yuen KS, Jin DY, Chim CS. Epigenetic silencing of miR-342-3p in B cell lymphoma and its impact on autophagy. Clin Epigenetics. 2020;12:150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 33. | Chang HW, Wang HC, Chen CY, Hung TW, Hou MF, Yuan SS, Huang CJ, Tseng CN. 5-azacytidine induces anoikis, inhibits mammosphere formation and reduces metalloproteinase 9 activity in MCF-7 human breast cancer cells. Molecules. 2014;19:3149-3159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Wang Y, Guo X, Wang L, Xing L, Zhang X, Ren J. miR-342-3p Inhibits Acute Myeloid Leukemia Progression by Targeting SOX12. Oxid Med Cell Longev. 2022;2022:1275141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 35. | Xiong X, Yang M, Yu H, Hu Y, Yang L, Zhu Y, Fei X, Pan B, Xiong Y, Fu W, Li J. MicroRNA-342-3p regulates yak oocyte meiotic maturation by targeting DNA methyltransferase 1. Reprod Domest Anim. 2022;57:761-770. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 36. | Tamizh Selvan G, Venkatachalam P. Ataxia Telengectesia Protein Influences Bleomycin-Induced DNA Damage in Human Fibroblast Cells. Cell Biochem Biophys. 2024;82:1235-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 37. | Selvan TG, Gollapalli P, Kumar SHS, Ghate SD. Early diagnostic and prognostic biomarkers for gastric cancer: systems-level molecular basis of subsequent alterations in gastric mucosa from chronic atrophic gastritis to gastric cancer. J Genet Eng Biotechnol. 2023;21:86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |