Published online Mar 26, 2025. doi: 10.4252/wjsc.v17.i3.98911
Revised: January 3, 2025
Accepted: February 26, 2025
Published online: March 26, 2025
Processing time: 254 Days and 22.7 Hours
Ankylosing spondylitis (AS) is recognized as a long-term inflammatory disorder that leads to inflammation in the spine and joints, alongside abnormal bone growth. In previous studies, we reported that mesenchymal stem cells (MSCs) derived from individuals with AS demonstrated a remarkable inhibition in the formation of osteoclasts compared to those obtained from healthy donors. The mechanism through which MSCs from AS patients achieve this inhibition remains unclear.
To investigate the potential underlying mechanism by which MSCs from indi
We analysed fat mass and obesity-associated (FTO) protein levels in AS-MSCs and MSCs from healthy donors and investigated the effects and mechanism by which FTO in MSCs inhibits osteoclastogenesis by coculturing and measuring the levels of tartrate-resistant acid phosphatase, nuclear factor of activated T cells 1 and cathepsin K.
We found that FTO, an enzyme responsible for removing methyl groups from RNA, was more abundantly expressed in MSCs from AS patients than in those from healthy donors. Reducing FTO levels was shown to diminish the capacity of MSCs to inhibit osteoclast development. Further experimental results revealed that FTO affects the stability of the long non-coding RNA activated by DNA damage (NORAD) by altering its N6-methyladenosine methylation status. Deactivating NORAD in MSCs significantly increased osteoclast formation by affecting miR-4284, which could regulate the MSC-mediated inhibition of osteoclastogenesis reported in our previous research.
This study revealed elevated FTO levels in AS-MSCs and found that FTO regulated the ability of AS-MSCs to inhibit osteoclast formation through the long noncoding RNA NORAD/miR-4284 axis.
Core Tip: This study explores how mesenchymal stem cells (MSCs) from ankylosing spondylitis (AS) patients inhibit osteoclastogenesis. We observed higher expression of fat mass and obesity-associated protein (FTO) in AS-MSCs compared to healthy controls. Reducing FTO expression diminished their osteoclast inhibitory effect. FTO regulates the long non-coding RNA activated by DNA damage (NORAD) by modulating its N6-methyladenosine methylation, and NORAD silencing increased osteoclast formation via miR-4284. These results highlight FTO as a key regulator of AS-MSC function and suggest the long noncoding RNA NORAD/miR-4284 axis as a novel mechanism of osteoclast inhibition in AS.
- Citation: Liu WJ, Wang JX, Li QF, Zhang YH, Ji PF, Jin JH, Zhang YB, Yuan ZH, Feng P, Wu YF, Shen HY, Wang P. Fat mass and obesity-associated protein in mesenchymal stem cells inhibits osteoclastogenesis via lnc NORAD/miR-4284 axis in ankylosing spondylitis. World J Stem Cells 2025; 17(3): 98911
- URL: https://www.wjgnet.com/1948-0210/full/v17/i3/98911.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v17.i3.98911
Ankylosing spondylitis (AS) is an autoimmune disease characterized by chronic inflammation and pathological osteogenesis[1]. Currently, nonsteroidal anti-inflammatory drugs and biologics, such as tumor necrosis factor inhibitors and interleukin-17 inhibitors, have significantly enhanced the management of clinical symptoms related to chronic inflammation[2,3]. However, the molecular mechanisms underlying pathological osteogenesis in AS remain elusive, and the limited therapeutic options pose a significant threat to the quality of life of AS patients.
Mesenchymal stem cells (MSCs), as major sources of osteoblasts, play a crucial role in bone metabolism and homeostasis of the bone microenvironment[4]. Our prior investigation revealed that MSCs derived from individuals with AS (AS-MSCs) exhibited a greater propensity for osteogenic differentiation than MSCs sourced from healthy donors (HD-MSCs)[5]. Additionally, our research revealed that, compared with HD-MSCs, AS-MSCs notably suppressed osteoclastogenesis[6], suggesting that AS-MSCs might play a pivotal role in driving the imbalance between osteogenesis and osteoclastogenesis, thereby potentially contributing to the pathological osteogenesis observed in AS. Nevertheless, the mechanisms through which AS-MSCs promote this imbalance remain elusive.
N6-methyladenosine (m6A) methylation is the most prevalent RNA modification at the post-transcriptional level in eukaryotic cells, and plays a crucial role in various physiological and pathological processes[7]. Research has indicated that m6A modification plays a regulatory role in diverse biological functions of MSCs, including differentiation, senescence, and immune regulatory processes[8]. Moreover, our findings suggest that m6A modification may exacerbate the pathological progression of AS by modulating the direct migration of MSCs[9]. The role of m6A modifications in the ability of MSCs to suppress osteoclastogenesis remains poorly understood.
Long noncoding RNAs (lncRNAs) are a class of endogenous RNAs that exceed 200 nucleotides in length and lack protein-coding potential[10]. They have been implicated in numerous fundamental biological processes as well as various diseases[11]. Mounting evidence shows that lncRNAs can regulate gene expression through various mechanisms, such as binding to proteins and microRNAs, or modulating interactions between proteins and DNA[12]. Furthermore, aberrant expression levels of lncRNAs have been observed in the progression of AS, rendering them promising biomarkers for the diagnosis and treatment of AS[13].
This study aimed to explore the impact of m6A modification on the enhanced ability of AS-MSCs to inhibit osteoclastogenesis compared with that of HD-MSCs. Our findings revealed that FTO, an RNA demethylase, was upregulated in AS-MSCs compared with HD-MSCs. Moreover, we demonstrated that suppressing FTO could attenuate the inhibitory effect of AS-MSCs on osteoclastogenesis by modulating the stability of long non-coding RNA activated by DNA damage (NORAD). This study may offer a novel perspective for understanding the pathologic osteogenesis of AS.
The isolation and expansion of MSCs were carried out according to previously established methods[5], utilizing bone marrow obtained from the posterior superior iliac spine of healthy volunteers and individuals with AS. MSCs were cultured in Dulbecco’s modified Eagle’s medium (Gibco, NY, United States) supplemented with 10% fetal bovine serum (FBS, Gibco, NY, United States) at 37 °C. The culture medium was refreshed every 2 days. Peripheral blood mononuclear cells were isolated via density gradient centrifugation, followed by the further isolation and purification of CD14+ monocytes from peripheral blood mononuclear cells using CD14 MicroBeads (Miltenyi Biotec, Germany). The isolated monocytes were subsequently cultured in Dulbecco’s modified Eagle’s medium containing 10% FBS at 37 °C under 5% CO2.
CD14+ monocytes were seeded at a density of 2.5 × 105 cells/cm2 in the lower chamber of a six-well transwell plate, while MSCs (at a ratio of 10:1, monocyte: MSC) were plated in the upper chamber. The induction medium used for the experiment consisted of 90% alpha minimal essential medium, 10% FBS, 50 ng/mL recombinant human receptor activator of nuclear factor κB ligand, 25 ng/mL recombinant human macrophage-colony stimulating factor, and 50 μg/mL l-ascorbic acid (all from Peprotech, NJ, United States). The culture medium was changed every 3 days to ensure optimal conditions for cell growth and differentiation.
On the 9th day, the cells were washed three times with phosphate buffered saline (PBS). To assess osteoclastogenesis, tartrate resistant acid phosphatase (TRAP) staining was conducted via a leukocyte acid phosphatase assay kit (Sigma, Japan), in accordance with the manufacturer’s guidelines. Cells exhibiting purple staining and possessing three or more nuclei were classified as TRAP-positive osteoclasts. The average number of osteoclasts was determined by evaluating a minimum of 9 fields that covered the entire well.
On the 9th day, the cells were fixed with 4% paraformaldehyde for 5 minutes and thoroughly rinsed with PBS. The samples were subsequently stained with FITC-conjugated phalloidin (Sigma, Japan) and 4’,6-diamidino-2-phenylindole at room temperature for 40 minutes. Following three washes with PBS, the cells were examined via an Axio Observer fluorescence microscope (Carl Zeiss, Germany).
Targeted small interfering RNAs (siRNAs) specific to FTO and lncRNA NORAD, as well as a negative control, were procured from IGEbio (Guangzhou, China). When the cell confluence reached 60%-80%, MSCs were transfected with a mixture containing Opti-MEM reduced serum medium, Lipofectamine™ RNAiMAX (Invitrogen, CA, United States), and siRNA (at a concentration of 1.8 × 106 cells per 1 OD), following the manufacturer’s instructions. After 5 hours of transfection, the transfection medium was removed. The efficiency of siRNA knockdown was evaluated 48 hours later using quantitative polymerase chain reaction (qPCR) and western blotting. The siRNA with the highest efficiency was selected for subsequent experiments.
After the cells were washed three times with PBS, TRIzol (Invitrogen, CA, United States) was added for total RNA extraction. The extracted RNA was then reverse transcribed into complementary DNA using the PrimeScript RT Reagent Kit (TaKaRa, Japan). Subsequently, real-time quantitative reverse transcription-PCR detection was performed using SYBR Green Premix Ex Taq (TaKaRa, Japan) reagent. A real-time fluorescence quantitative PCR system was utilized to measure the gene expression levels. To determine the relative expression levels of the target genes, the 2-ΔΔCt method was employed with GAPDH serving as the internal control. The detailed protocol for these steps can be found in our previously published research. Additionally, the forward and reverse primers for the target genes are below: FTO: Primer#1 5’-ACTTGGCTCCCTTATCTGACC-3’ and primer#2 5’-TGTGCAGTGTGAGAAAGGCTT-3’; lnc NORAD: Primer#1 5’-CTCTCAACTCCACCCCAACC-3’ and primer#2 5’-ACAAACGTGGACGTATCGCT-3’; GAPDH: Primer#1 5’-AAGGTGAAGGTCGGAGTCAA-3’ and primer#2 5’-AATGAAGGGGTCATTGATGG3’; U6: Primer#1 5’-CGCTTCGGCAGCACATATAC-3’ and primer#2 5’-TTCACGAATTTGCGTGTCAT-3’; miR-4284: Primer#1 5’-TCGCCGACGGGCTCACATCA-3’ and primer#2 5’-CTCAACTGGTGTCGTGGAGTCGGC-3’; miR-4284 RT Primer: 5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATGGGGTG-3’.
The cells were washed three times with cold PBS, followed by another wash with RIPA buffer containing a mixture of 1% phosphatase inhibitors and protease inhibitors to extract proteins. The cell lysate was collected and centrifuged at 14000 rpm for 30 minutes at 4 °C. The protein supernatant was collected, and the protein concentration was quantified using a BCA kit. The proteins were subsequently separated by sodium-dodecyl sulfate gel electrophoresis and transferred onto a polyvinylidene fluoride (PVDF) membrane. The PVDF membranes were blocked with 5% milk for 1 hour at room temperature and then incubated overnight with primary antibodies against nuclear factor of activated T cells 1 (NFATc1), TRAP, cathepsin K (CTSK), or beta-actin (dilution 1:1000). After being washed three times with Tris-buffered saline-Tween, the PVDF membrane was incubated with a horseradish peroxidase-conjugated secondary antibody (dilution 1:3000) at room temperature for 1 hour. Following another three washes, the target protein bands were detected via a chemiluminescent HRP substrate, and the density analysis of the bands was performed via ImageJ software.
The m6A RIP assay was conducted via the Magna MeRIP m6A Kit (Merck Millipore) following the protocol provided by the manufacturer. In brief, total RNA was isolated from pretreated MSCs and randomly broken into fragments of 100 or fewer nucleotides. Protein A/G magnetic beads conjugated with either an anti-m6A antibody (ab208577, Abcam, United Kingdon), an anti-FTO antibody (ab126605, Abcam, United Kingdom), or an immunoglobulin G control were added to the fragmented RNA and incubated overnight. Following the collection of the magnetic beads, the immunoprecipitated RNA was further collected to analyze m6A enrichment in target genes via qPCR. Non-immunoprecipitated RNA fragments were used as a control for input.
A mutant (mut) NORAD variant with a mutation in the predicted miR-4284 binding site was constructed and named NORAD mut. The synthesized wild-type (luci-NORAD WT) or luci-NORAD mut sequences were cloned and inserted into the pmirGLO vector, which contains the firefly luciferase reporter gene. For the transfection experiments, 5000 MSCs were seeded per well in 96-well plates. Each construct was co-transfected with NORAD mimics or a negative control via Lipofectamine 3000 transfection reagent (Invitrogen, CA, United States). After 36 hours post-transfection, the cell lysates were collected, and the luciferase activity was measured via the Dual-Luciferase Reporter Assay System (Promega, WI, United States), following the manufacturer’s instructions. The relative luciferase activity was determined by calculating the ratio of firefly to Renilla luciferase activity. This assay helps in assessing the impact of the mutation in the miR-4284 binding site on luciferase expression, shedding light on the regulatory role of miR-4284 in NORAD expression.
Male SKG mice were handled in compliance with the animal care guidelines approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University. Disease induction commenced at 8 weeks of age through the intraperitoneal injection of 3 mg of curdlan. Adenoviruses encoding a specific shRNA targeting FTO (Av-shFTO) or adeno-associated virus of negative control for mouse experiments were procured from OBiO Technology. SKG mice were administered Av-shFTO or adeno-associated virus of negative control via intravenous tail vein injection upon disease onset. At the 8th week post-induction and treatment, the mice were euthanized, and tissue samples were collected for hematoxylin and eosin staining and Safranin O staining.
Each experiment was repeated three times. The data are presented as the mean ± SD. One-way analysis of variance (ANOVA) and Student’s t-test were used for intergroup comparisons with SPSS 20.0 software (Chicago, IL, United States). A P-value < 0.05 was considered statistically significant.
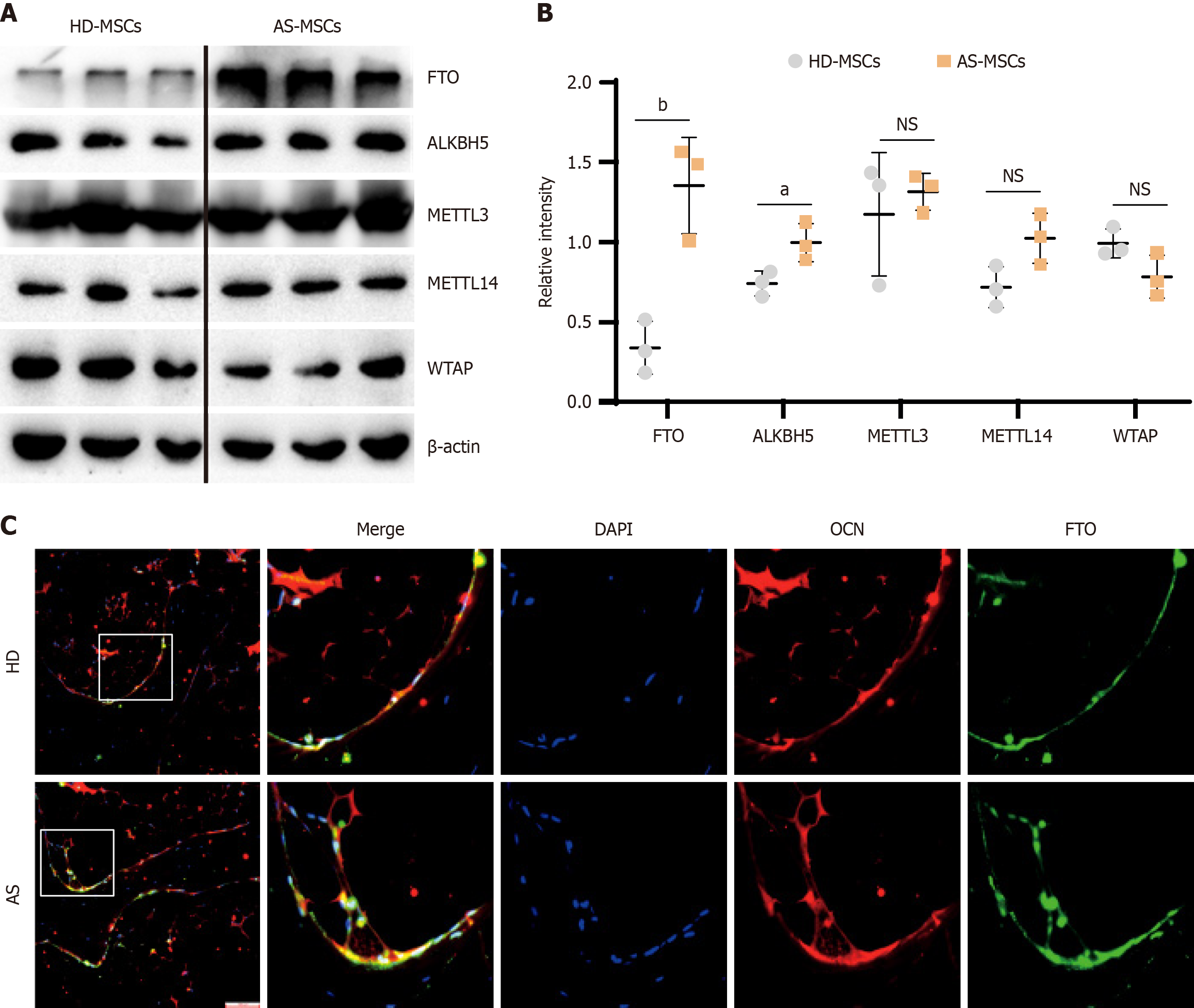
To investigate the involvement of m6A modifications in MSCs in AS, we first confirmed the protein-level differences in the expression of m6A-related methyltransferases and demethylases through western blot analysis. The results revealed a significant increase in the expression level of FTO in AS-MSCs compared with that in normal MSCs (Figure 1A and B). Furthermore, we extended this analysis to bone tissue samples from AS patients and normal controls. Immunofluorescence staining revealed a notably greater level of FTO expression in AS-MSCs marked with osteocalcin than in normal controls (Figure 1C). These findings suggest that FTO in MSCs may play a crucial role in the development of AS, indicating the potential involvement of FTO-mediated m6A modifications in the pathogenesis of AS.
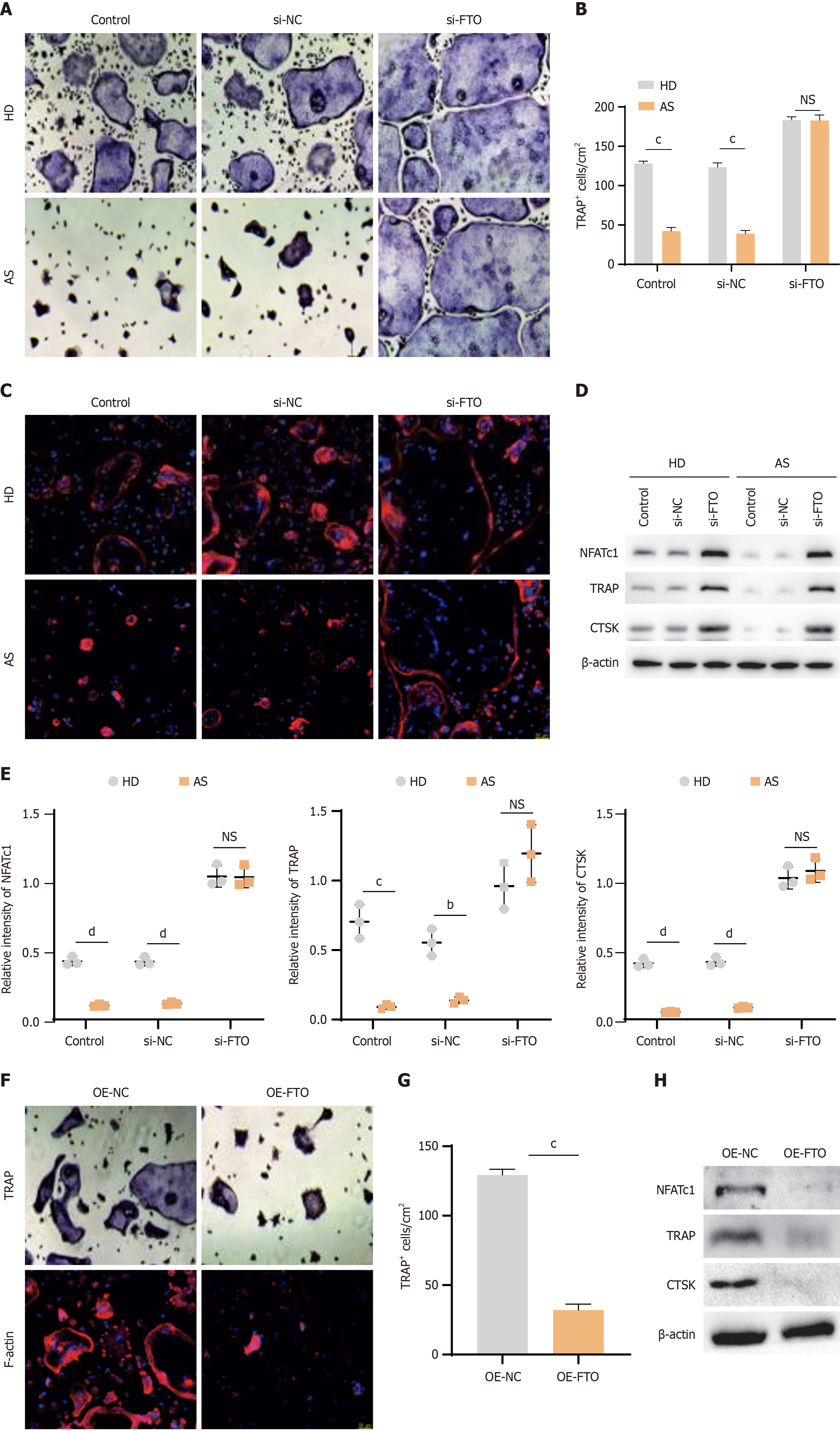
To investigate the effect of FTO upregulation in AS-MSCs, an siRNA was used to silence FTO in MSCs. Then, the MSCs were cocultured with CD14+ monocytes in medium supplemented with macrophage colony-stimulating factor and receptor activator of nuclear factor κB ligand to induce osteoclast differentiation. TRAP staining and F-actin staining were employed to assess osteoclast formation. The results revealed that the number of osteoclasts in the HD-MSC group was greater than that in the AS-MSC group (Figure 2A-C). Following FTO knockdown, the number of osteoclasts increased significantly in both groups. Interestingly, the number of osteoclasts cultured with AS-MSCs recovered to the level observed in those cultured with HD-MSCs. Furthermore, the expression levels of osteoclast formation markers (NFATc1, TRAP, and CTSK) were evaluated via western blot analysis. The results revealed lower levels of NFATc1, TRAP, and CTSK in AS-MSCs and higher levels in the FTO-knockdown group (Figure 2D and E). To further confirm the influence of FTO in MSCs, we used lentiviruses to overexpress FTO. As shown by TRAP and F-actin staining and western blotting, overexpressing FTO in MSCs led to reduced osteoclast formation (Figure 2F-H). In summary, these findings suggest that FTO in MSCs positively regulates the ability of MSCs to inhibit osteoclast formation, highlighting the potential role of FTO in modulating osteoclastogenesis.
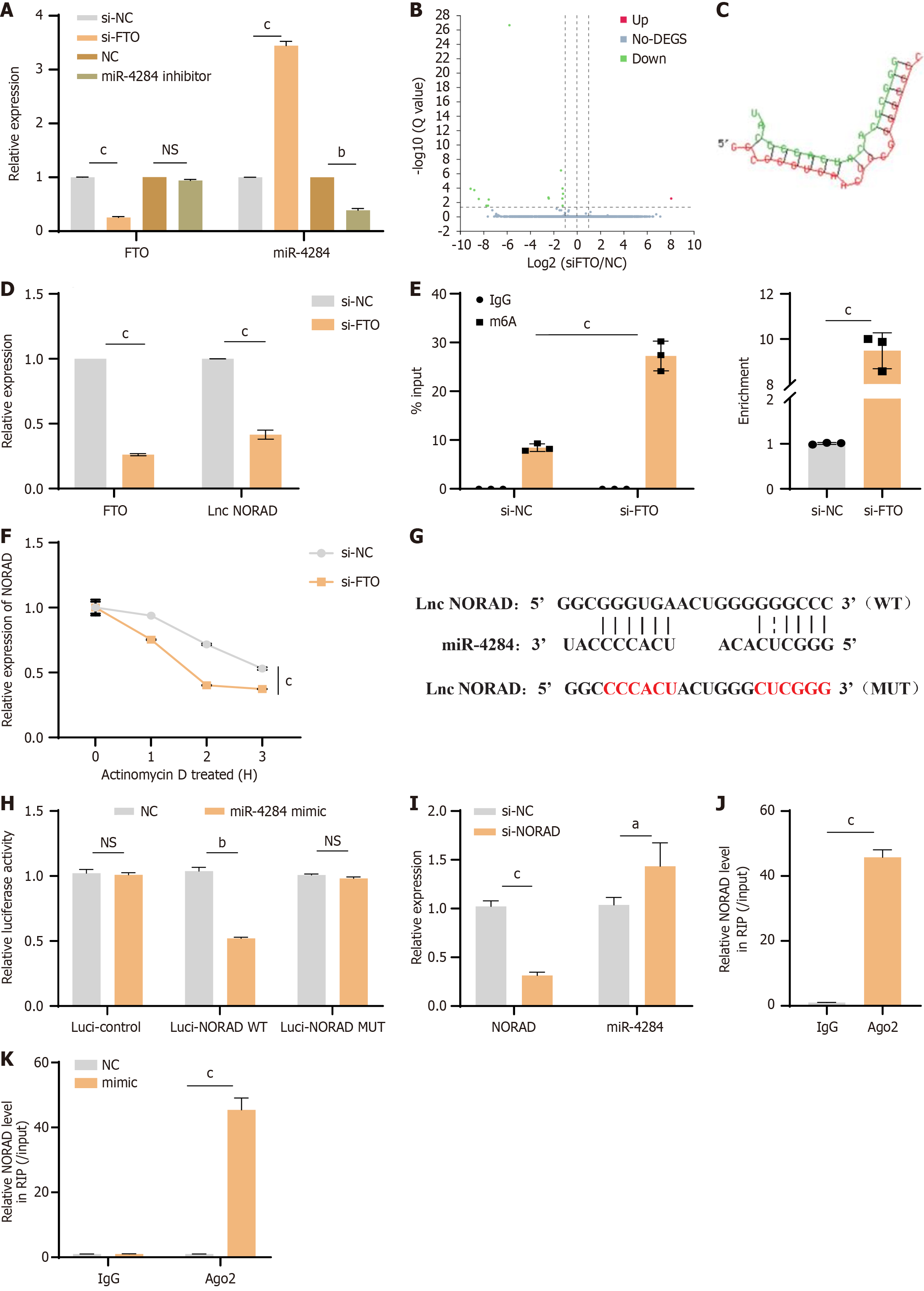
Previously, we demonstrated that miR-4284 could modulate the MSC-mediated inhibition of osteoclast formation[6]. We first examined the correlation between FTO and miR-4284 through qPCR. Our results revealed that the expression of miR-4284 was elevated after siFTO. Interestingly, suppressing miR-4284 did not affect the expression of FTO (Figure 3A). We thus wondered whether lncRNAs participate in the regulatory network between FTO and miR-4284. Next, we performed high-throughput sequencing to compare the lncRNA expression profiles of MSCs with or without FTO knockdown. The results revealed significant differential expression of the lncRNA NORAD in the profiles (Figure 3B), which further validated the interaction between miR-4284 and the lncRNA NORAD via DIANA TOOLS (Figure 3C). The qPCR results confirmed the downregulation of NORAD in the FTO-knockdown group (Figure 3D). Additionally, previous reports suggested that m6A modification could regulate the expression of the lncRNA NORAD[14]. To investigate this, we analysed the m6A levels on the lncRNA NORAD via m6A RNA immunoprecipitation-qPCR analysis. The results indicated that the m6A levels of the lncRNA NORAD in the FTO knockdown group were significantly greater than those in the control group, suggesting that FTO could reduce the m6A modification levels of the lncRNA NORAD (Figure 3E). To further understand how FTO regulates NORAD levels via m6A modification, RNA stability assays were performed using actinomycin D to inhibit RNA transcription. The results demonstrated that the half-life of NORAD in the control group was significantly longer than that in the FTO-knockdown group (Figure 3F). These findings collectively suggest that FTO regulates NORAD stability through m6A modification.
Next, we investigated the relationship between lnc NORAD and miR-4284. The results revealed that miR-4284 contains complementary binding sites with the lncRNA NORAD (Figure 3G). Luciferase assays revealed that the luciferase activity of WT NORAD was significantly increased when NORAD was cotransfected with the miR-4284 mimic, whereas there was no significant difference in the activity of the NORAD MUT (Figure 3H). Further analysis via qPCR demonstrated that lnc NORAD was upregulated after transfection with the miR-4284 mimic (Figure 3I). Additionally, the results from the RNA binding protein immunoprecipitation assay confirmed the interaction relationship between NORAD and miR-4284 (Figure 3J and K). Overall, these findings suggest that the lncRNA NORAD can bind with miR-4284 and potentially downregulate its expression. This finding indicates a regulatory mechanism in which the lncRNA NORAD may modulate the expression or activity of miR-4284, highlighting a potential regulatory axis in the context of MSC-mediated osteoclast formation.
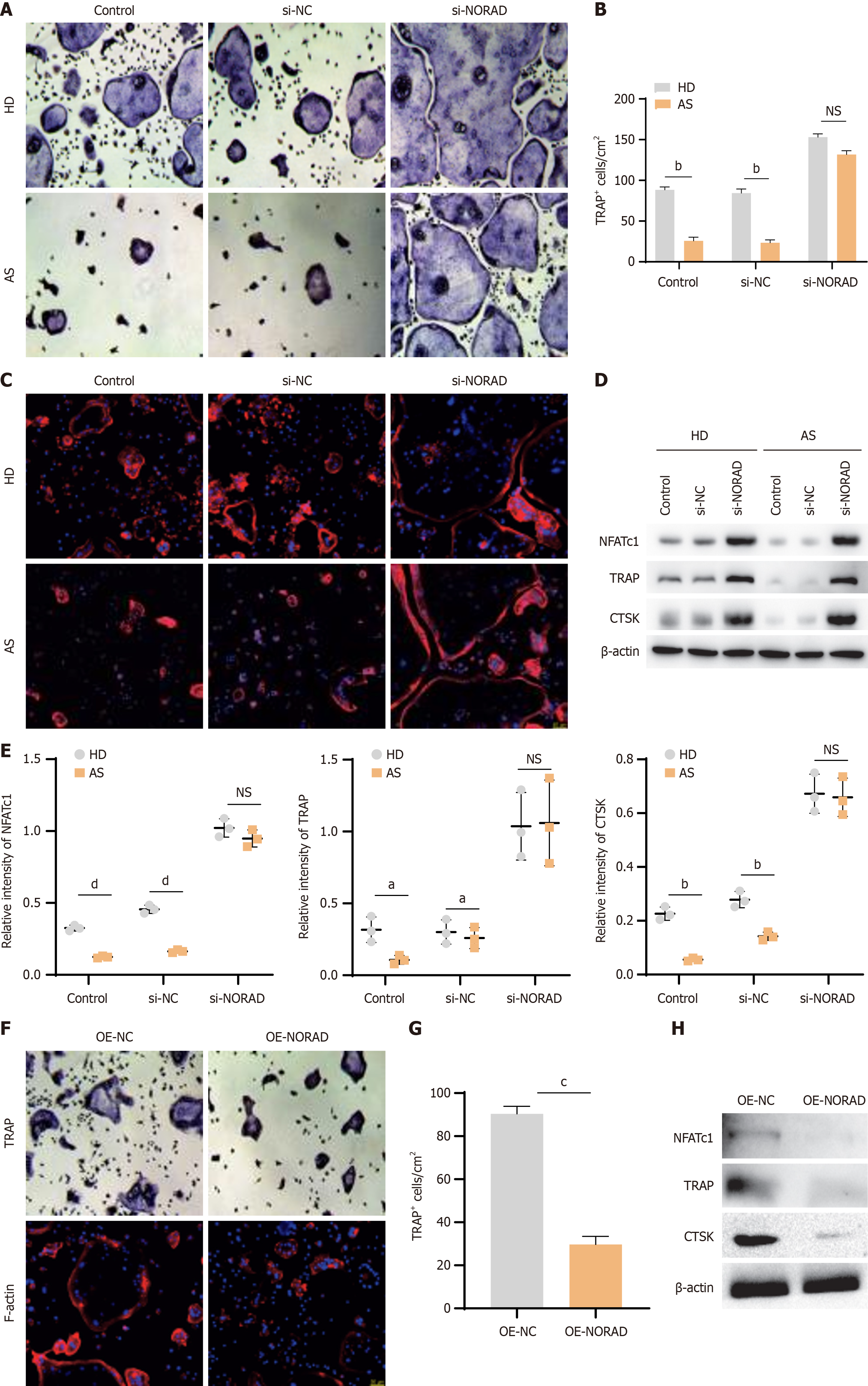
To investigate the effect of NORAD on MSC-mediated osteoclast formation, we cocultured CD14+ monocytes with AS-MSCs or HD-MSCs with or without NORAD knockdown. TRAP staining revealed an increased number of TRAP+ cells in the NORAD-knockdown group, and there was no significant difference in osteoclast formation between AS-MSCs and HD-MSCs after NORAD knockdown (Figure 4A and B). F-actin staining was consistent with the TRAP staining results (Figure 4C). Furthermore, western blot analysis was performed to evaluate the expression levels of osteoclast differentiation markers (NFATc1, TRAP, and CTSK). The results indicated that the lnc NORAD knockdown group presented higher expression levels of these markers than the control group. Additionally, NORAD knockdown abolished the differences in osteoclast differentiation marker expression between AS-MSCs and HD-MSCs (Figure 4D and E). To validate the impact of lnc NORAD on MSCs, we employed lentiviral vectors to increase the expression of lnc NORAD. TRAP, F-actin staining, and western blotting revealed that the overexpression of NORAD in MSCs resulted in decreased osteoclast formation (Figure 4F-H). These findings collectively suggest that the lncRNA NORAD in MSCs facilitates MSC-mediated osteoclast formation, indicating a potential regulatory role for the lncRNA NORAD in modulating osteoclast differentiation processes.
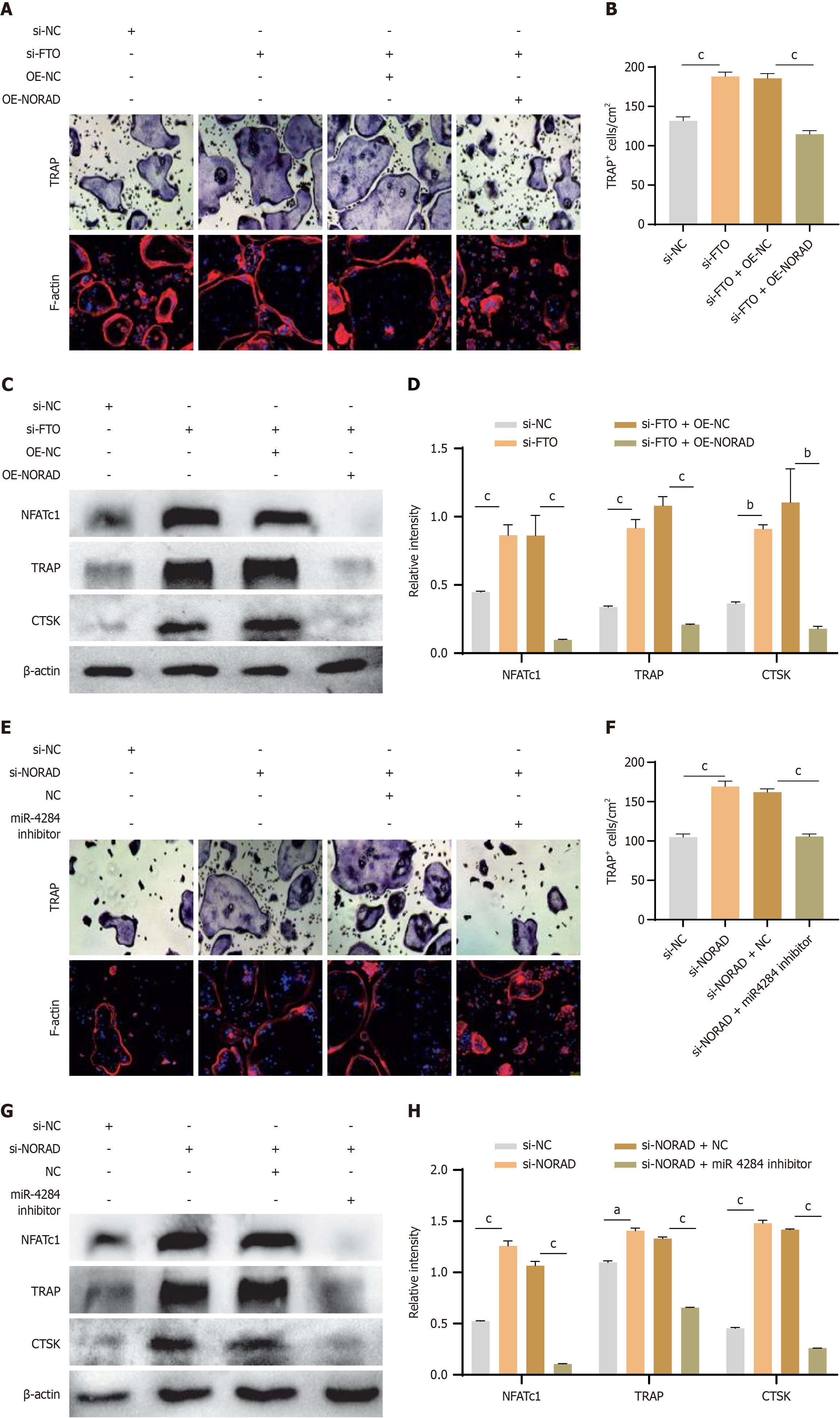
To further verify the effect of the FTO/lnc NORAD interaction in MSCs on osteoclast differentiation, we used siRNAs and lentiviruses to silence FTO while overexpressing NORAD in MSCs. The results of TRAP, F-actin staining, and western blotting demonstrated that overexpressing NORAD significantly attenuated the increased osteoclastogenesis induced by FTO knockdown (Figure 5A-D). Additionally, by downregulating NORAD and inhibiting miR-4284 in MSCs, we observed a marked reduction in osteoclastogenesis resulting from NORAD knockdown, as demonstrated by TRAP assays, F-actin staining, and western blotting (Figure 5E-H). Taken together, these results suggest that FTO positively regulates osteoclast differentiation in MSCs through the NORAD/miR-4284 pathway.
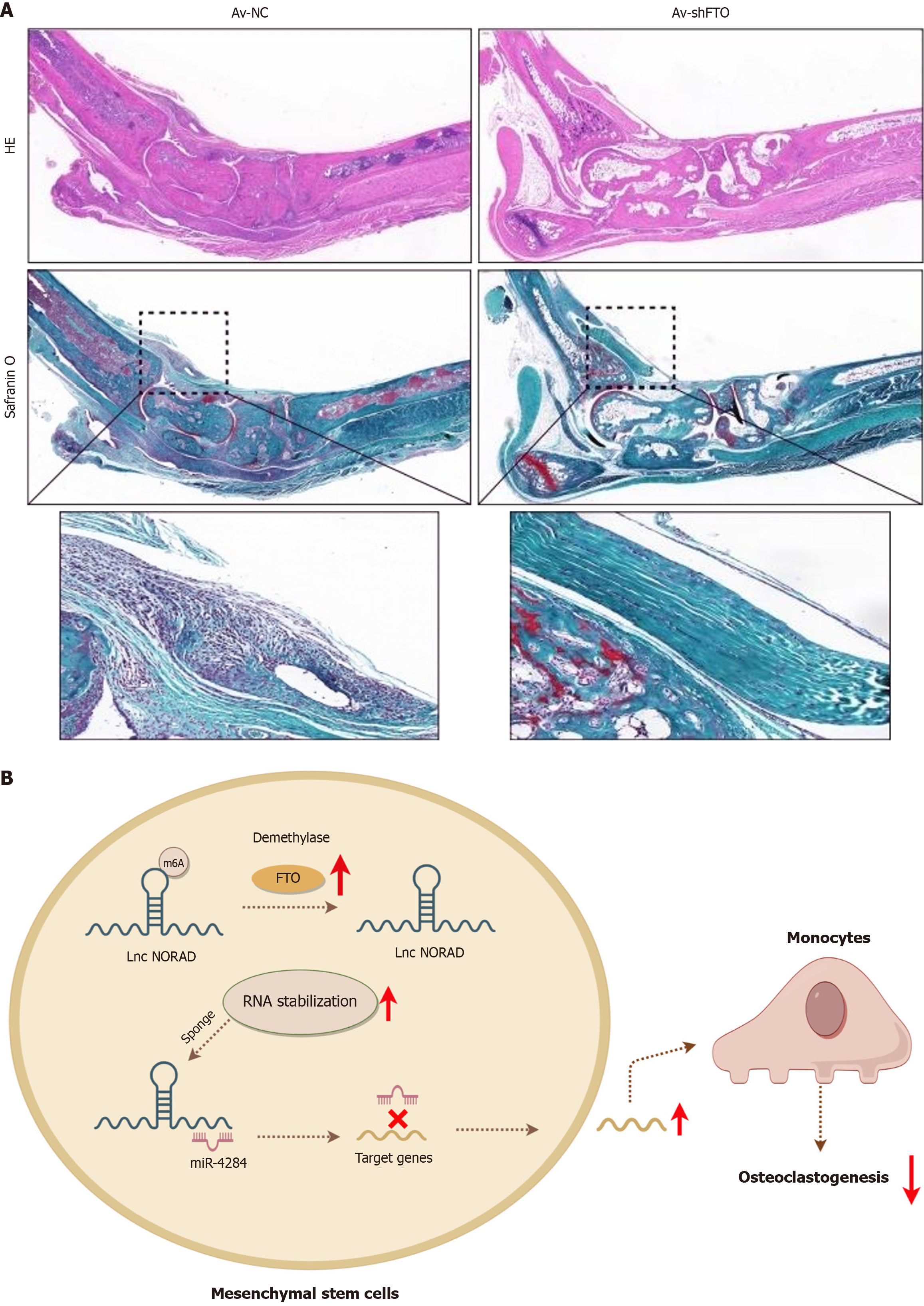
To assess the effect of FTO knockdown in vivo, we established an AS animal model by administering 3 mg of curdlan via intraperitoneal injection to SKG mice to induce AS features. We subsequently administered Av-shFTO via intravenous tail vein injection. Our findings revealed that the suppression of FTO markedly reduced the degree of enthesopathy, as well as the size and thickness of inflamed ankles. These results suggest that FTO could be a potential target for the treatment of AS (Figure 6A).
In this study, we revealed notable upregulation of FTO in AS-MSCs compared with HD-MSCs. Inhibiting FTO diminished the suppressive effects of AS-MSCs on osteoclastogenesis by influencing the stability of the lncRNA NORAD, which subsequently targeted miR-4284 (Figure 6B). This investigation examined the FTO/lnc NORAD/miR-4284 axis to understand the pathological osteogenesis associated with AS.
Pathologic osteogenesis is a significant concern in AS, as it can lead to the development of spinal ankylosis in severe cases, posing a serious threat to their lives[15]. The balance between osteoblasts (bone-forming cells) and osteoclasts (bone-resorbing cells) is crucial for bone homeostasis and remodelling. Dysfunction or abnormal regulation of these cell types can contribute to various bone diseases, potentially leading to abnormal osteogenesis in conditions such as AS[16]. Previous research has indicated that, compared with HD-MSCs, AS-MSCs exhibit a greater capacity for osteogenic differentiation[5]. Additionally, AS-MSCs have been shown to more effectively inhibit osteoclastogenesis[6]. These findings suggest that AS-MSCs may play a crucial role in the pathologic osteogenesis observed in AS. By demonstrating the enhanced osteogenic potential and regulatory effects on osteoclastogenesis of AS-MSCs, this research provides valuable insights into the mechanisms underlying pathologic osteogenesis in AS. Understanding the unique properties of AS-MSCs in bone metabolism may offer new avenues for developing targeted therapies to address abnormal osteogenesis and associated complications in AS patients.
m6A methylation is a crucial RNA modification that impacts the stability, splicing, and translation of RNA molecules, playing a significant role in various biological processes such as cell differentiation, immune responses, and tumor development[17,18]. Studies have shown that m6A modification can influence the biological functions of MSCs. Research by Wu et al[19] revealed that conditional knockout of methyltransferase 3, an m6A methyltransferase, in MSCs could accelerate bone loss and modulate the differentiation potential of MSCs. Similarly, other m6A-related enzymes such as the demethylases FTO have been found to be essential for osteogenesis[20]. These findings underscore the importance of m6A modification in regulating MSC function. In this study, we found that the expression of FTO was elevated in AS-MSCs. Furthermore, silencing FTO in both HD-MSCs and AS-MSCs promoted osteoclastogenesis in vitro when MSCs were co-cultured with monocytes. These results suggest that the upregulation of FTO in AS-MSCs may contribute to the abnormal osteogenesis observed in AS. However, further research is needed to fully understand the role of FTO in AS-MSCs, particularly in an in vivo setting.
The interaction between lncRNAs and microRNAs, which act as molecular sponges is a well-established mechanism for regulating gene expression[21]. Building upon previous research that identified miR-4284 as a key molecule enhancing the ability of AS-MSCs to inhibit osteoclastogenesis, this study aimed to explore the potential role of lncRNAs in mediating the relationship between FTO, miR-4284, and osteoclastogenesis. In this study, global profiling of lncRNA expression in FTO-silenced MSCs revealed lnc NORAD as a crucial link connecting FTO and miR-4284. Further investigations demonstrated that FTO could stabilize lnc NORAD by removing m6A methylation, and in turn, lnc NORAD acted as a sponge to negatively regulate the activity of miR-4284. Importantly, interference with lnc NORAD was found to increase osteoclastogenesis in coculture assays, suggesting that lnc NORAD may contribute to the inhibited osteoclastogenesis exhibited by AS-MSCs. These findings shed light on a potential regulatory axis involving FTO, lnc NORAD, miR-4284, and osteoclastogenesis in the context of AS-MSCs. By elucidating the intricate interplay between these molecules, this study provides valuable insights into the molecular mechanisms underlying the altered osteoclastogenesis observed in AS. Targeting this regulatory network involving lncRNAs, microRNAs, and m6A-modifying enzymes like FTO may offer novel therapeutic strategies for managing bone-related complications in AS.
Indeed, lnc NORAD, which was originally identified as a regulator of genome integrity in response to DNA damage[22-24], has been implicated in various human cancers as an oncogene[25,26]. It has been shown to play a role in manipulating cancer cell development[27,28]. Interestingly, NORAD has also been found to undergo m6A modification, and Li et al[14] reported that the m6A modification of NORAD by WTAP negatively regulates its stability, which aligns with our findings. While recent studies have primarily focused on the role of NORAD as a competitive endogenous RNA that regulates downstream microRNA and mRNA expression[29-31], the specific mechanism through which NORAD targets miR-4284 in the context of AS-MSCs remains unclear in this study. However, whether other target molecular of NORAD mediate the function of FTO-NORAD axis in AS-MSCs remains unknown and broader insights into this process should be further investigated.
In this work, we focused on the role of m6A modification in the enhanced inhibition of osteoclastogenesis by AS-MSCs. Our results revealed that FTO regulated the function of AS-MSCs through the NORAD/miR-4284 axis, which may contribute to elucidating the mechanism of pathological osteogenesis and provide potential treatment targets for AS.
| 1. | Sieper J, Poddubnyy D. Axial spondyloarthritis. Lancet. 2017;390:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 896] [Article Influence: 112.0] [Reference Citation Analysis (0)] |
| 2. | Ritchlin C, Adamopoulos IE. Axial spondyloarthritis: new advances in diagnosis and management. BMJ. 2021;372:m4447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 3. | Sieper J. How to define remission in ankylosing spondylitis? Ann Rheum Dis. 2012;71 Suppl 2:i93-i95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 4. | Yue R, Zhou BO, Shimada IS, Zhao Z, Morrison SJ. Leptin Receptor Promotes Adipogenesis and Reduces Osteogenesis by Regulating Mesenchymal Stromal Cells in Adult Bone Marrow. Cell Stem Cell. 2016;18:782-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 342] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 5. | Xie Z, Wang P, Li Y, Deng W, Zhang X, Su H, Li D, Wu Y, Shen H. Imbalance Between Bone Morphogenetic Protein 2 and Noggin Induces Abnormal Osteogenic Differentiation of Mesenchymal Stem Cells in Ankylosing Spondylitis. Arthritis Rheumatol. 2016;68:430-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 6. | Liu W, Wang P, Xie Z, Wang S, Ma M, Li J, Li M, Cen S, Tang S, Zheng G, Ye G, Wu X, Wu Y, Shen H. Abnormal inhibition of osteoclastogenesis by mesenchymal stem cells through the miR-4284/CXCL5 axis in ankylosing spondylitis. Cell Death Dis. 2019;10:188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 7. | Huang H, Weng H, Chen J. The Biogenesis and Precise Control of RNA m(6)A Methylation. Trends Genet. 2020;36:44-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 8. | Chen X, Hua W, Huang X, Chen Y, Zhang J, Li G. Regulatory Role of RNA N(6)-Methyladenosine Modification in Bone Biology and Osteoporosis. Front Endocrinol (Lausanne). 2019;10:911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 73] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 9. | Xie Z, Yu W, Zheng G, Li J, Cen S, Ye G, Li Z, Liu W, Li M, Lin J, Su Z, Che Y, Ye F, Wang P, Wu Y, Shen H. TNF-α-mediated m(6)A modification of ELMO1 triggers directional migration of mesenchymal stem cell in ankylosing spondylitis. Nat Commun. 2021;12:5373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 10. | Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393-407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1719] [Cited by in RCA: 2605] [Article Influence: 434.2] [Reference Citation Analysis (0)] |
| 11. | Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1788] [Cited by in RCA: 2126] [Article Influence: 212.6] [Reference Citation Analysis (0)] |
| 12. | Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol Rev. 2016;96:1297-1325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1378] [Cited by in RCA: 1324] [Article Influence: 147.1] [Reference Citation Analysis (1)] |
| 13. | Sun R, Wang X, Sun X, Zhao B, Zhang X, Gong X, Wong SH, Chan MTV, Wu WKK. Emerging Roles of Long Non-Coding RNAs in Ankylosing Spondylitis. Front Immunol. 2022;13:790924. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 14. | Li G, Ma L, He S, Luo R, Wang B, Zhang W, Song Y, Liao Z, Ke W, Xiang Q, Feng X, Wu X, Zhang Y, Wang K, Yang C. WTAP-mediated m(6)A modification of lncRNA NORAD promotes intervertebral disc degeneration. Nat Commun. 2022;13:1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 91] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 15. | Braun J, Sieper J. Ankylosing spondylitis. Lancet. 2007;369:1379-1390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1266] [Cited by in RCA: 1374] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 16. | Rosen CJ. Bone remodeling, energy metabolism, and the molecular clock. Cell Metab. 2008;7:7-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49:7239-7255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 315] [Article Influence: 78.8] [Reference Citation Analysis (0)] |
| 18. | Sendinc E, Shi Y. RNA m6A methylation across the transcriptome. Mol Cell. 2023;83:428-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 193] [Reference Citation Analysis (0)] |
| 19. | Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y, Li J, Sheng R, Deng P, Wang Y, Zheng R, Jiang Y, Ye L, Chen Q, Zhou X, Lin S, Yuan Q. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 326] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 20. | Zhang Q, Riddle RC, Yang Q, Rosen CR, Guttridge DC, Dirckx N, Faugere MC, Farber CR, Clemens TL. The RNA demethylase FTO is required for maintenance of bone mass and functions to protect osteoblasts from genotoxic damage. Proc Natl Acad Sci U S A. 2019;116:17980-17989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 21. | Ghafouri-Fard S, Abak A, Tavakkoli Avval S, Rahmani S, Shoorei H, Taheri M, Samadian M. Contribution of miRNAs and lncRNAs in osteogenesis and related disorders. Biomed Pharmacother. 2021;142:111942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 673] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 23. | Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP, Subramanian V, Chen J, Schenone M, Guttman M, Carr SA, Lander ES. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561:132-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 278] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 24. | Ventura A. NORAD: Defender of the Genome. Trends Genet. 2016;32:390-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Soghli N, Yousefi T, Abolghasemi M, Qujeq D. NORAD, a critical long non-coding RNA in human cancers. Life Sci. 2021;264:118665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 26. | Yang Z, Zhao Y, Lin G, Zhou X, Jiang X, Zhao H. Noncoding RNA activated by DNA damage (NORAD): Biologic function and mechanisms in human cancers. Clin Chim Acta. 2019;489:5-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 27. | Han T, Wu Y, Hu X, Chen Y, Jia W, He Q, Bian Y, Wang M, Guo X, Kang J, Wan X. NORAD orchestrates endometrial cancer progression by sequestering FUBP1 nuclear localization to promote cell apoptosis. Cell Death Dis. 2020;11:473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 28. | Sun Y, Wang J, Ma Y, Li J, Sun X, Zhao X, Shi X, Hu Y, Qu F, Zhang X. Radiation induces NORAD expression to promote ESCC radiotherapy resistance via EEPD1/ATR/Chk1 signalling and by inhibiting pri-miR-199a1 processing and the exosomal transfer of miR-199a-5p. J Exp Clin Cancer Res. 2021;40:306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 29. | Jia Y, Tian C, Wang H, Yu F, Lv W, Duan Y, Cheng Z, Wang X, Wang Y, Liu T, Wang J, Liu L. Long non-coding RNA NORAD/miR-224-3p/MTDH axis contributes to CDDP resistance of esophageal squamous cell carcinoma by promoting nuclear accumulation of β-catenin. Mol Cancer. 2021;20:162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 30. | Argoetti A, Shalev D, Polyak G, Shima N, Biran H, Lahav T, Hashimshony T, Mandel-Gutfreund Y. lncRNA NORAD modulates STAT3/STAT1 balance and innate immune responses in human cells via interaction with STAT3. Nat Commun. 2025;16:571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 31. | Zhang X, Chen T, Li Z, Wan L, Zhou Z, Xu Y, Yan D, Zhao W, Chen H. NORAD exacerbates metabolic dysfunction-associated steatotic liver disease development via the miR-511-3p/Rock2 axis and inhibits ubiquitin-mediated degradation of ROCK2. Metabolism. 2025;164:156111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |