Published online Mar 26, 2024. doi: 10.4252/wjsc.v16.i3.305
Peer-review started: December 13, 2023
First decision: January 24, 2024
Revised: January 31, 2024
Accepted: February 28, 2024
Article in press: February 28, 2024
Published online: March 26, 2024
Processing time: 102 Days and 13 Hours
Mesenchymal stem cells (MSCs) modulated by various exogenous signals have been applied extensively in regenerative medicine research. Notably, nanosecond pulsed electric fields (nsPEFs), characterized by short duration and high strength, significantly influence cell phenotypes and regulate MSCs differentiation via multiple pathways. Consequently, we used transcriptomics to study changes in messenger RNA (mRNA), long noncoding RNA (lncRNA), microRNA (miRNA), and circular RNA expression during nsPEFs application.
To explore gene expression profiles and potential transcriptional regulatory mechanisms in MSCs pretreated with nsPEFs.
The impact of nsPEFs on the MSCs transcriptome was investigated through whole transcriptome sequencing. MSCs were pretreated with 5-pulse nsPEFs (100 ns at 10 kV/cm, 1 Hz), followed by total RNA isolation. Each transcript was normalized by fragments per kilobase per million. Fold change and difference significance were applied to screen the differentially expressed genes (DEGs). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were performed to elucidate gene functions, complemented by quantitative polymerase chain reaction verification.
In total, 263 DEGs were discovered, with 92 upregulated and 171 downregulated. DEGs were predominantly enriched in epithelial cell proliferation, osteoblast differentiation, mesenchymal cell differentiation, nuclear division, and wound healing. Regarding cellular components, DEGs are primarily involved in condensed chromosome, chromosomal region, actin cytoskeleton, and kinetochore. From aspect of molecular functions, DEGs are mainly involved in glycosaminoglycan binding, integrin binding, nuclear steroid receptor activity, cytoskeletal motor activity, and steroid binding. Quantitative real-time polymerase chain reaction confirmed targeted transcript regulation.
Our systematic investigation of the wide-ranging transcriptional pattern modulated by nsPEFs revealed the differential expression of 263 mRNAs, 2 miRNAs, and 65 lncRNAs. Our study demonstrates that nsPEFs may affect stem cells through several signaling pathways, which are involved in vesicular transport, calcium ion transport, cytoskeleton, and cell differentiation.
Core Tip: Nanosecond pulsed electric fields (nsPEFs) have been found to regulate the osteogenic, chondrogenic, and adipogenic differentiation of mesenchymal stem cells (MSCs). We hypothesized that several key factors may be regulated by nsPEFs, thereby influencing the biological functions of MSCs. Following exposure of MSCs to nsPEFs, we identified the differential expression of 263 messenger RNAs, 65 long noncoding RNAs, and 2 microRNAs. Verification by quantitative polymerase chain reaction and Gene Ontology and Kyoko Encyclopedia of Genes and Genomes enrichment analyses demonstrated the involvement of chromosome, cytoskeleton, and calcium signaling pathways following nsPEFs pretreatment. These results may be very meaningful for the further application of nsPEFs in MSCs.
- Citation: Lin JJ, Ning T, Jia SC, Li KJ, Huang YC, Liu Q, Lin JH, Zhang XT. Evaluation of genetic response of mesenchymal stem cells to nanosecond pulsed electric fields by whole transcriptome sequencing. World J Stem Cells 2024; 16(3): 305-323
- URL: https://www.wjgnet.com/1948-0210/full/v16/i3/305.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v16.i3.305
Mesenchymal stem cells (MSCs), as seed cells in regenerative repair, have been extensively applied in preclinical and clinical research in regenerative medicine, such as osteoarthritis[1], cartilage defects[2], and bone defects[3]. The differentiation and function of MSCs can be modulated by various exogenous signals, including biological factors[4], drug formulations[5], and physical signals[6]. The quest for an appropriate exogenous signal to regulate the functions and differentiation of stem cells remains a dynamic area of investigation for numerous researchers.
Pulsed electric fields (PEFs), as a crucial biophysical signal, can induce changes in cell membranes and alterations in intracellular calcium ion concentrations. Under specific conditions, PEFs can significantly influence cell phenotypes and regulate stem cell differentiation through multiple pathways[7,8]. However, the biological effects of traditional PEFs are relatively weak, and the time required for the emergence of a differentiating response can often range from hours to days[9]. This may be attributed to the fact that the pulse width of traditional PEFs is in the microsecond range or higher, exceeding the intrinsic charging and discharging time of cell membranes (in the range of hundreds of nanoseconds). As a result, traditional PEFs face difficulties in deeply penetrating the cell interior due to the shielding effect of the cell membrane[10]. In contrast, nanosecond PEFs (nsPEFs) represent nanosecond-duration, high-strength electric fields, with a shorter pulse width than the charging and discharging time of the cell membrane. Furthermore, nsPEFs can deeply penetrate into cellular organelles and exhibit significant biological effects[11]. In our previous research, it was found that nsPEFs can influence the osteogenic, adipogenic, and chondrogenic differentiation of MSCs by regulating DNA methylation and the MAPK signaling pathway[12]. Although nsPEFs show strong regulatory effects on MSCs differentiation, previous studies have mainly focused on specific molecules or pathways, and a comprehensive exploration of the mechanisms by which nsPEFs regulate MSCs has not been conducted.
Transcriptomics analysis, by examining messenger RNA (mRNA), long noncoding RNA (lncRNA), microRNA (miRNA), and circular RNA (circRNA), allows for a comprehensive understanding of changes in gene expression. It also holds significant importance in unraveling alterations in biological processes. In this study, we first utilized high-throughput transcriptomics sequencing to detect the changes of mRNA, miRNA, lncRNA, and circRNA expression in MSCs after nsPEFs treatment. Additionally, we carried out Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses to explore the biological processes and signaling pathways associated with differentially expressed target genes. Furthermore, we validated their expression levels using quantitative real-time polymerase chain reaction, providing further support for the application of nsPEFs in MSCs.
All animal experiments were approved by the Institutional Animal Care and Use Committee of Peking University (COE-GeZ-7). Rat bone marrow MSCs (rMSCs) were harvested from 8-wk-old Sprague-Dawley rats according to our previous study[13]. MSCs were cultured in expansion medium composed of Dulbecco’s modified Eagle’s medium (DMEM, Hyclone) supplemented with 10% (v/v) fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Amresco), in a humidified incubator at 37 °C with 5% CO2. Cells were trypsinized with 0.25% (w/v) trypsin (Invitrogen, Carlsbad, CA, United States) upon reaching 85% confluence. MSCs at passage 5 were used for subsequent experiments.
We previously found that nsPEFs (100 ns, 10 kV/cm, 1 Hz, 5 pulses) can improve the stemness of porcine bone marrow MSCs, human bone marrow MSCs, and rMSCs, and promote osteochondral defect repair in rats[12-14]. In this study, nsPEFs with the same parameters were applied to regulate MSCs performance. According to our previous study[12-14], one million MSCs were suspended in 1 mL of DMEM within a 0.4-cm gap cuvette (Bio-Rad, 165-2088, United States) and stimulated by 5 pulses of nsPEFs (100 ns at 10 kV/cm, 1 Hz), and the time interval between two pulses was 1 s. The cells were then subjected to nsPEFs with a duration of 100 ns, as previously described. Five pulses were applied at 1-s intervals between each pulse. MSCs without nsPEFs stimulation served as the control group.
Total RNA was isolated from the cells 24 h after exposure with the miRNA extraction kit (Cat#TR205-200, Tanmo). Qualified total RNA was further purified with the RNAClean XP Kit (Cat#A63987, Beckman Coulter, Inc.Kraemer Boulevard Brea, CA, United States) and the RNase-Free DNase Set (Cat#79254, QIAGEN, GmBH, Germany). RNA quantity was assessed by UV spectrometry at 260 nm/280 nm absorbance on a spectropchotometer (NanoDrop Technologies, Wilmington, DE, United States).
The filtered clean reads were mapped to the reference genome database. Each transcript was normalized by fragments per kilobase per million to eliminate the influence of gene length and sequencing depth. The counts of each sample were mapped to the annotated genome after standardization and normalization. Finally, fold change (FC) and difference significance were used to screen the differentially expressed genes (DEGs). Each group of cells was sequenced with three independent biological replicates.
GO term and KEGG pathway enrichment analyses were performed using the tool for Function Annotation in DAVID (https://david.ncifcrf.gov/). The KEGG pathway maps were obtained from the KEGG database (http://www.kegg.jp/).
Total RNA was extracted from 1 × 106 cells treated with 1 mL TRIzol. RNA purity and concentration were determined by a NanoDrop 2000 spectropchotometer (Thermo Fisher Scientific). cDNA was synthesized with the ReverTra Ace qPCR Kit (TOYOBO, FSQ-101) and then subjected to quantitative polymerase chain reaction using Power SYBR Green PCR Master Mix (ABI, 4368708). The mRNA levels were determined using 50 ng of cDNA on an Applied Biosystems 7300. All template amplifications were conducted in triplicate with a three-step polymerase chain reaction process. Using Actin expression as a normalization control, the relative expression was calculated using the 2-ΔΔCt method. The primer sequences are provided in Table 1.
| Primer | Primer sequence (5’ to 3’) |
| Actg2-F | CTCTCTCCACCTTCCAGCAAA |
| Actg2-R | AGGGCCCCGCTTCATC |
| Nek2-F | TGGAGGGCCTGACAATCTG |
| Nek2-R | CCACCACACTGAGTTTCTGGTTT |
| Cenpf-F | GGAGAGCCTGTGTGCATGTG |
| Cenpf-R | ACGTGAGCAGGAGGTTATGAAAC |
| Scin-F | TGGCCGAAGATGATGTCATG |
| Scin-R | CTTTGCCAATCCAAATGAAGATC |
| Kif20b-F | TGTGCCACACCAGTCACAATT |
| Kif20b-R | ACCTCGCCACTCTTCCTCTTC |
| Nog-F | CGAGGGTTTTCAATGAACTTTTTT |
| Nog-R | AGTGCATTACATGAACCAGAAAGC |
| Ereg-F | GGGTTGCCACAAGTCTGAACA |
| Ereg-R | GCATGCTGCACATCCTTGTC |
| Asic3-F | GCCTGCTTACCATCCTTGAGA |
| Asic3-R | CCCCAGGACTCTGTCTTGGA |
| Aldh3a1-F | TCCCACCGCCGCTCTT |
| Aldh3a1-R | GCCTTGTGAGCTTCTTCATTCA |
| Tubb2b-F | GGCGAGGATGAGGCTTGA |
| Tubb2b-R | TTCACCTCAGCTTTCCCTAACC |
| Cryba4-F | CGTGCTGGAGAGCGATCA |
| Cryba4-R | AGCCCCACTCCCTGAAGTG |
| Nr3c2-F | CGCACAGCAATATGAAAACCA |
| Nr3c2-R | GCCCCTTTCCCCCAGAA |
| Stxbp5l-F | AAGCCTCAGCAGGAAAAGCA |
| Stxbp5l-R | TGCCCGGTCCAGGAATG |
| Actin-F | TCTGTGTGGATTGGTGGCTCTA |
| Actin-R | CTGCTTGCTGATCCACATCTG |
All numerical data from quantitative polymerase chain reaction are presented as the mean ± SD. Comparisons between groups were performed by the independent sample t-test. Results are presented as the mean ± SD. The Student’s t-test was used to evaluate the difference between the two groups using Prism 8.21 software (GraphPad). The statistical significance level was set at P < 0.05.
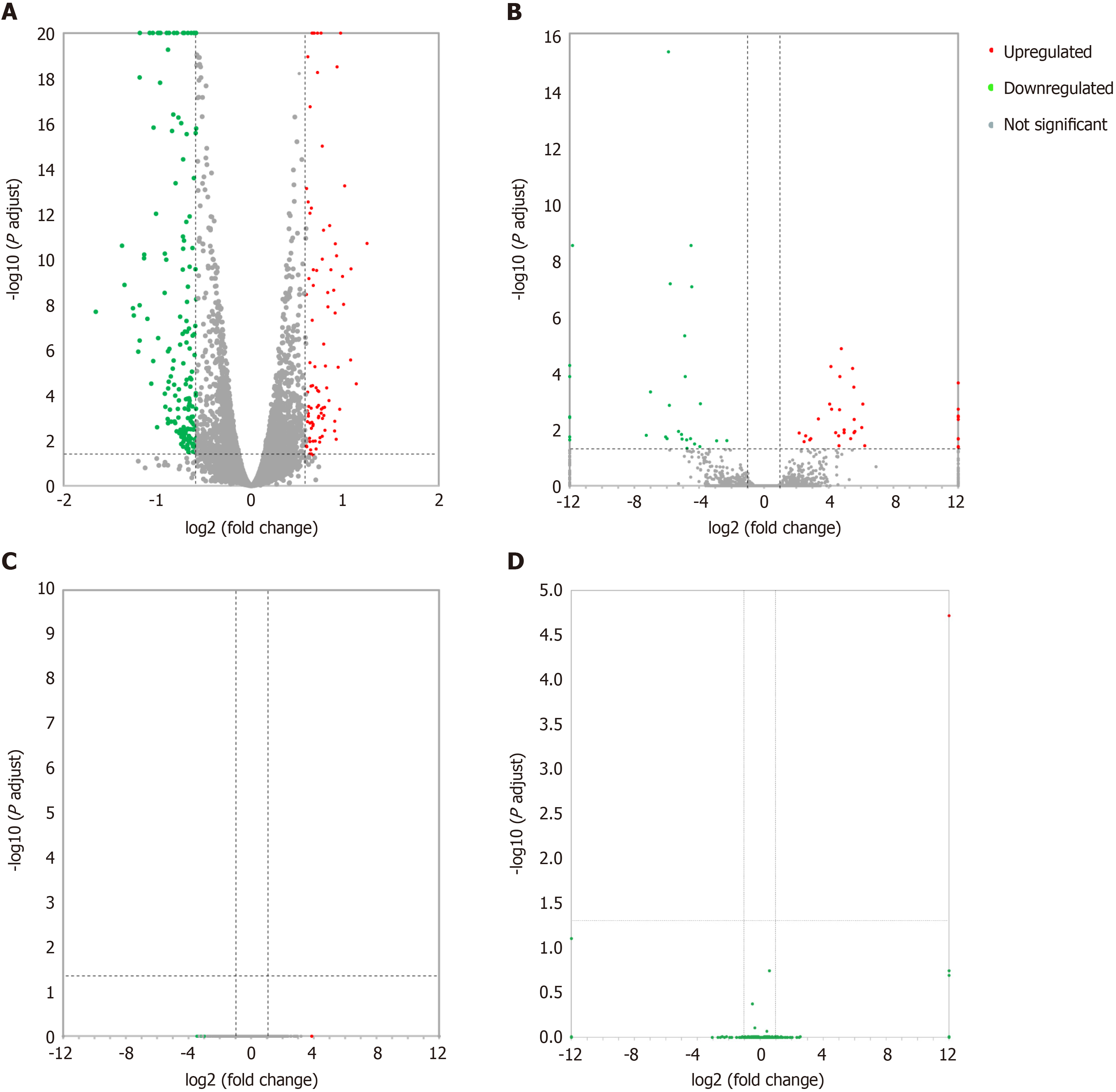
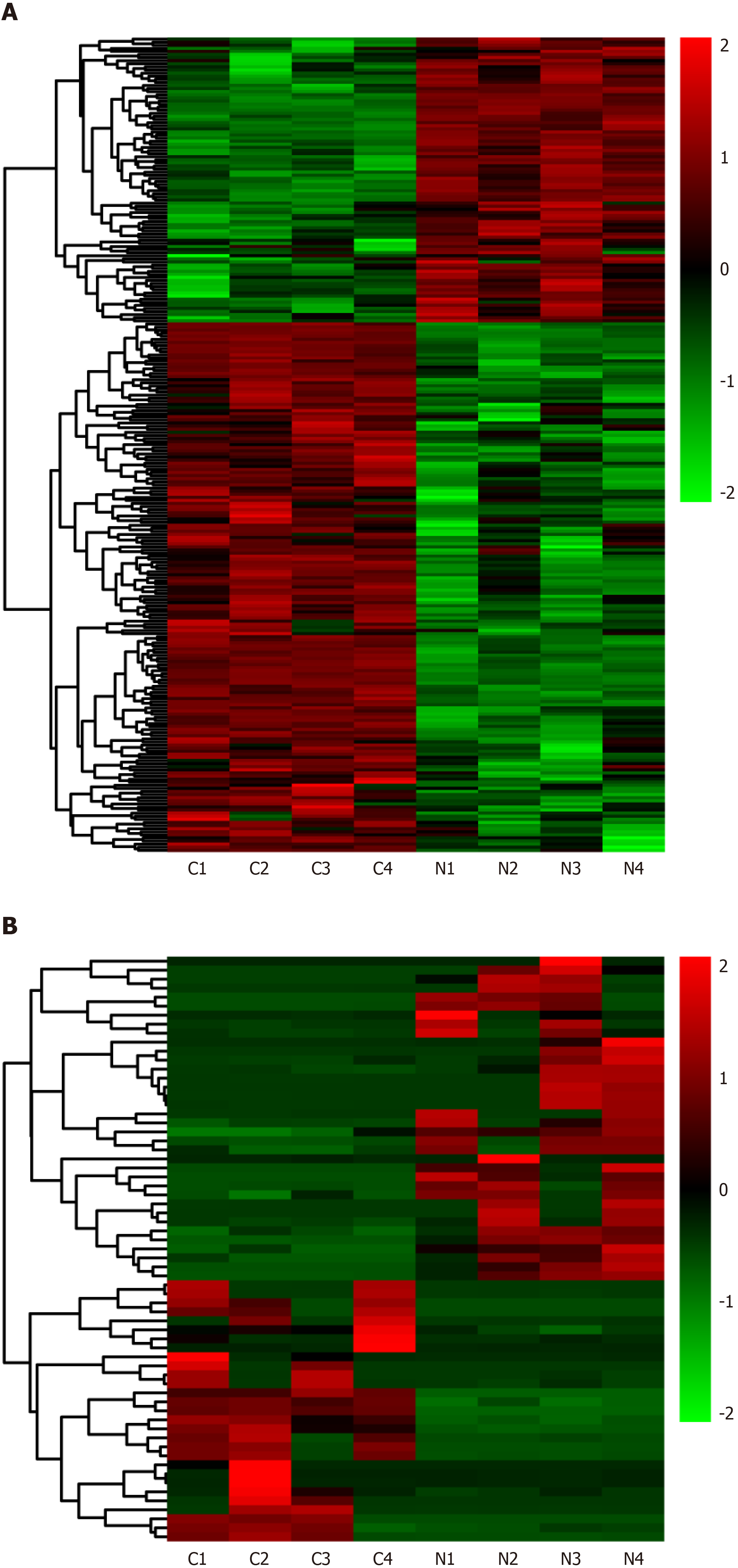
Differentially expressed lncRNAs and mRNAs (n = 4) are displayed using Volcano plots (Figure 1) and heat maps (Figure 2). The top 20 differentially expressed lncRNAs and mRNAs in the nsPEFs-treated group compared to the control group are listed in Tables 2 and 3, respectively.
| LncRNA_ID | Locus | Log2FC | Q value |
| NONRATT027355.2 | 7:143840739-143841394 | 6.223946 | 0.038374 |
| NONRATT014734.2 | 19:44137444-44139463 | 6.103516 | 0.001285 |
| NONRATT000391.2 | 1:78818388-78818948 | 6.035452 | 0.008784 |
| NONRATT030693.2 | X:115351001-115352258 | 5.611966 | 0.011922 |
| MSTRG.1686.44 | 1:180852586-181353774 | 5.574714 | 0.004533 |
| NONRATT029722.2 | 9:113936538-113936864 | 5.563687 | 0.013263 |
| MSTRG.1686.24 | 1:180804226-181295177 | 5.546189 | 0.000326 |
| NONRATT024954.2 | 6:50889024-50923726 | 5.468331 | 7.04E-05 |
| NONRATT010080.2 | 14:83648401-83660792 | 5.356187 | 0.021489 |
| NONRATT021215.2 | 4:29083076-29092661 | -5.06373 | 0.022244 |
| NONRATT015697.2 | 2:211078318-211078606 | -5.08835 | 0.01522 |
| NONRATT002358.2 | 1:80335470-80336633 | -5.27782 | 0.011922 |
| NONRATT015446.2 | 2:188562082-188565332 | -5.7896 | 7.11E-08 |
| NONRATT026659.2 | 7:30291089-30293287 | -5.84278 | 0.001419 |
| NONRATT017622.2 | 20:7219444-7220293 | -5.89986 | 4.44E-16 |
| NONRATT031234.2 | X:116752819-116754309 | -5.9906 | 0.021489 |
| NONRATT024688.2 | 6:136358084-136363672 | -6.06997 | 0.018677 |
| NONRATT024848.2 | 6:25912732-26051235 | -7.01177 | 0.000482 |
| NONRATT005942.2 | 10:84682448-84688899 | -7.27093 | 0.016558 |
| NONRATT002900.2 | X:157319040-157323878 | -11.8256 | 3.16E-09 |
| Gene ID | Gene name | Log2FC | Q value |
| ENSRNOG00000017609 | Cnga4 | 4.049502 | 0.041927 |
| ENSRNOG00000005883 | Nek10 | 3.164517 | 0.001515 |
| ENSRNOG00000051612 | AABR07044570.1 | 3.006014 | 0.020392 |
| ENSRNOG00000003891 | Porf1 | 2.826433 | 0.000369 |
| ENSRNOG00000042070 | Ticam2 | 2.821013 | 0.00501 |
| ENSRNOG00000046566 | Tub | 2.803217 | 0.006364 |
| ENSRNOG00000032973 | Il13ra2 | 2.661856 | 1.68E-08 |
| ENSRNOG00000025261 | AABR07050407.1 | 2.232324 | 0.00943 |
| ENSRNOG00000001959 | Mx1 | 2.073926 | 0.049836 |
| ENSRNOG00000031598 | Atp8b4 | 2.055746 | 0.024695 |
| ENSRNOG00000003283 | Rcsd1 | -2.09387 | 0.018919 |
| ENSRNOG00000010454 | Ccno | -2.1785 | 0.011229 |
| ENSRNOG00000002456 | Hlf | -2.44194 | 0.021066 |
| ENSRNOG00000052129 | Nwd1 | -2.45693 | 0.032717 |
| ENSRNOG00000014424 | RGD1563354 | -2.46926 | 0.016841 |
| ENSRNOG00000049115 | Ccr5 | -2.73203 | 0.01083 |
| ENSRNOG00000055401 | Kcnc1 | -3.13004 | 0.021817 |
| ENSRNOG00000055318 | AABR07068030.1 | -3.34625 | 0.000321 |
| ENSRNOG00000014556 | Cdh20 | -3.47543 | 0.025307 |
| ENSRNOG00000054723 | AABR07058174.1 | -3.5912 | 0.047547 |
| ENSRNOG00000011946 | Ptn | -3.6805 | 0.000634 |
In total, 263 DEGs were identified in the PRJNA931816 dataset, of which 92 and 171 were significantly (|log2FC| > 0.585 and q < 0.05) upregulated and downregulated, respectively (Figures 1A and 2A). Of these DEGs, 65 were lncRNAs, of which 36 and 19 were significantly (|log2FC| > 1 and q < 0.05) upregulated and downregulated, respectively (Figures 1B and 2B); 0 were circRNAs (Figure 1C); and 2 were miRNAs, both of which were significantly upregulated (Figure 1D).
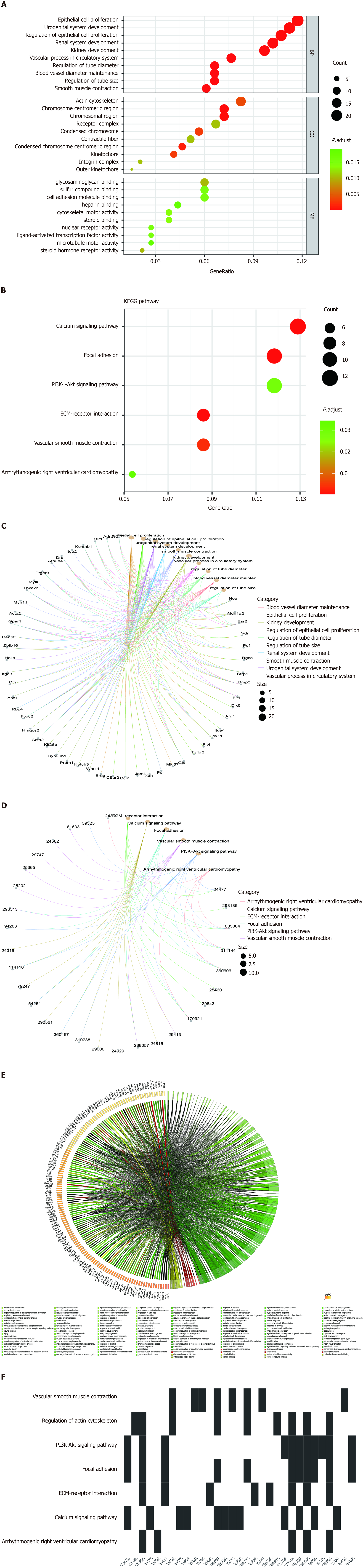
To investigate the biological functions and pathways of DEGs, KEGG and GO analyses were conducted for the 263 DEGs. Figure 3 demonstrates the mainly enriched functional annotations from three aspects, including biological processes, cellular components (CC), and molecular functions (MF). From the perspective of biological processes, DEGs were mainly enriched in epithelial cell proliferation, osteoblast differentiation, mesenchymal cell differentiation, nuclear division, and wound healing. From the perspective of CC, DEGs are mainly involved in condensed chromosome, chromosomal region, actin cytoskeleton, and the kinetochore. From the perspective of MF, DEGs are mainly involved in glycosaminoglycan binding, integrin binding, nuclear steroid receptor activity, cytoskeletal motor activity, and steroid binding. When the upregulated mRNAs were enriched, 12 mRNAs were found to be involved in chromosome segregation (biological process). For example, among the 12 mRNAs, Top2a is a conserved regulator of chromatin topology which plays an important role in catalyzing reversible DNA double-strand breaks[15]. From the perspective of CC, upregulated mRNAs were mainly enriched in the chromosome, centromeric region, kinetochore, and midbody. From the perspective of MF, upregulated mRNAs were mainly enriched in integrin binding. When the downregulated mRNAs were enriched, they are involved in the extracellular space (CC). Among the 12 mRNAs, Wnt11 encodes a protein that plays an important role in regulating extracellular matrix (ECM) organization[16], and Smoc1 encodes an extracellular glycoprotein that is a critical regulator of cell attachment to the ECM by binding to calcium[17]. From the perspective of GO, downregulated mRNAs were mainly enriched in the positive regulation of gene expression, cell differentiation, and ventricular septum morphogenesis (Figures 3A and C). The detailed relationship between DEGs and GO are shown by the Chord diagram of GO (Figure 3E). GO analysis classified genes heavily involved in epithelial cell proliferation and osteoblast differentiation, among others.
Moreover, KEGG analysis shown in Figures 3B, D, and F demonstrated that DEGs mainly participate in the calcium signaling pathway, ECM-receptor interaction, focal adhesion, and vascular smooth muscle contraction. A Waterfall plot was generated to reveal the potential effects of nsPEFs on signaling pathways (Figure 3F). Other related signaling pathways such as the regulation of actin cytoskeleton, PI3K-Akt signaling pathway, Rap1 signaling pathway, cGMP-PKG signaling pathway, and Hippo signaling pathway-multiple species may also contribute to completing the reaction process of MSCs to nsPEFs. The term cluster showed that nsPEFs may stimulate the cells through the calcium signaling pathway, etc.
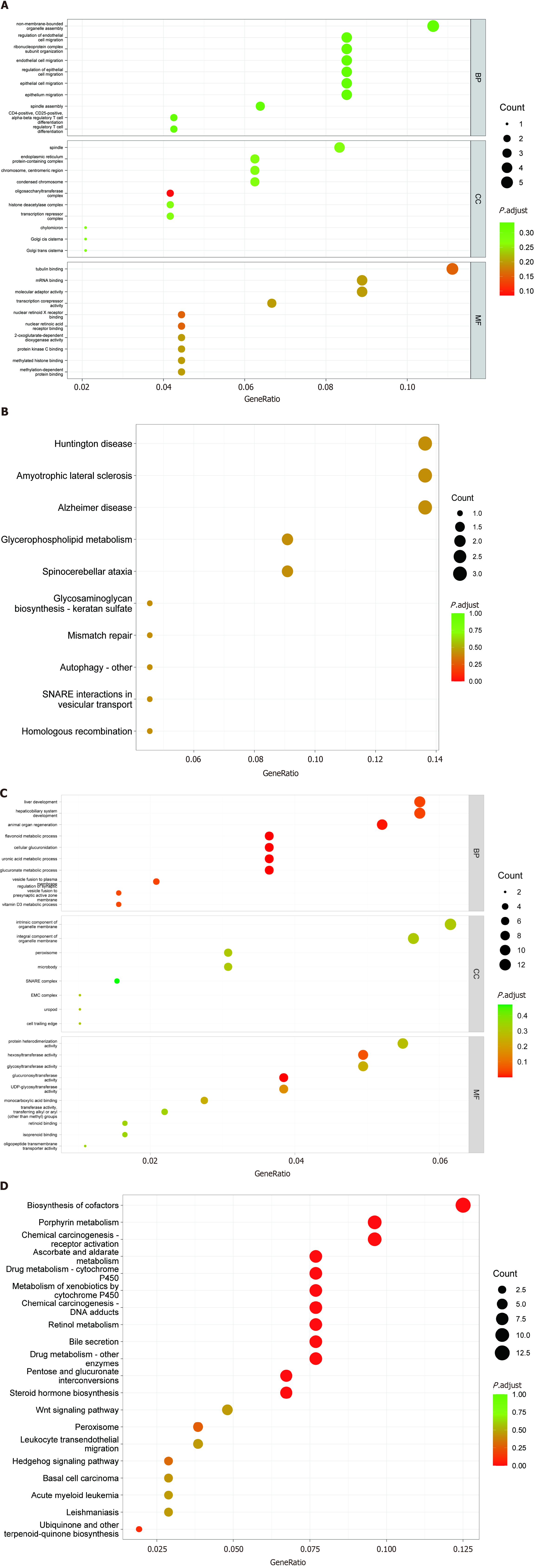
The GO enrichment analysis results for the differentially expressed lncRNAs, miRNAs, and mRNAs are shown in Figure 4. Based on the target genes of differentially expressed lncRNAs, the most significantly enriched biological processes were regulation of endothelial cell migration, and ribonucleoprotein complex subunit organization. The most significantly enriched CC were the oligosaccharyltransferase complex, endoplasmic reticulum protein-containing complex, chromosome, and centromeric region. The most significantly enriched MF were tubulin binding, nuclear retinoid X receptor binding, and nuclear retinoic acid receptor binding (Figure 4A). Moreover, in Figure 4B, KEGG analysis shows that differentially expressed lncRNAs mainly participate in the glycerophospholipid metabolism signaling pathway.
Based on the target genes of differentially expressed miRNAs, the most significantly enriched biological processes were the flavonoid metabolic process, cellular glucuronidation, vesicle fusion to plasma membrane, and animal organ regeneration. The most significantly enriched CC were the intrinsic component of organelle membrane, the integral component of organelle membrane, and the peroxisome. The most significantly enriched MF were glucuronosyltransferase activity, hexosyltransferase activity, and MAP kinase activity (Figure 4C). In addition, KEGG analysis in Figure 4D shows that differentially expressed miRNAs mainly participate in porphyrin metabolism, ascorbate and aldarate metabolism, biosynthesis of cofactors, and the pentose and glucuronate interconversions signaling pathway.
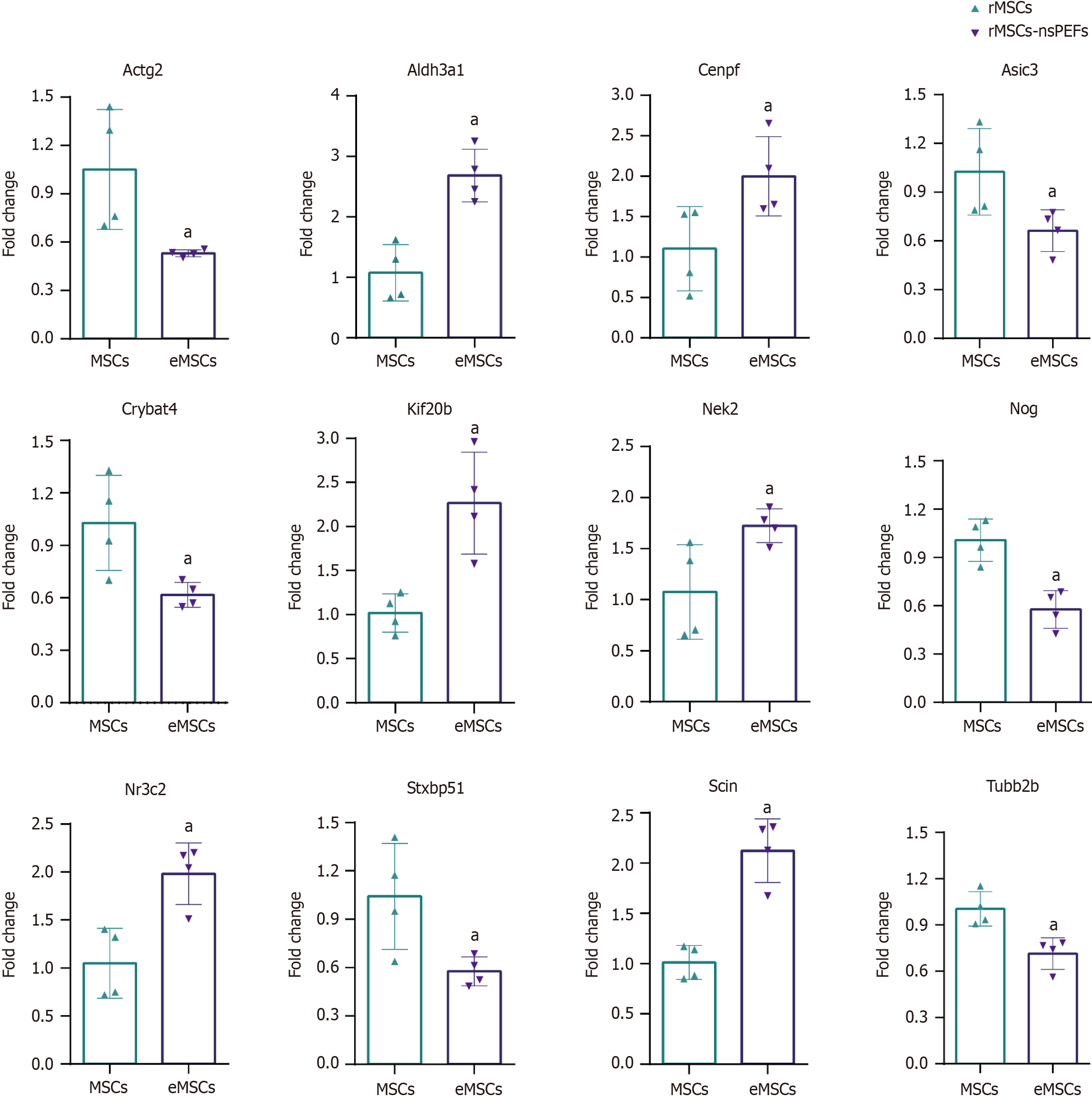
We confirmed the accuracy of sequencing data for selected mRNAs, and the mRNA validation results were consistent with the RNA-seq data (Figure 5). According to the related signaling pathway, we selected seven upregulated mRNAs, aldehyde dehydrogenase 3 family member A1 (Aldh3a1), centromere protein F (Cenpf), kinesin family member 20B
In this study, nsPEFs-treated rMSCs were evaluated by whole transcriptome sequencing in terms of mRNA, lncRNA, circRNA, and miRNA expression. Previous studies have shown that nsPEFs can regulate the expression levels of some mRNAs in MSCs and upregulate the differentiation potential of MSCs[12]. nsPEFs can regulate the chondrogenic differentiation of MSCs through phosphorylation of molecules of the MAPK signaling pathways[14]. nsPEFs can also regulate the differentiation potential of MSCs through demethylation of stemness genes[12], and can induce nodule formation in osteoblasts[18]. However, the lack of a systematic study hinders further application of nsPEFs in stem cell differentiation.
Using whole transcriptome sequencing, 263 differentially expressed mRNAs (q < 0.05, |log2FC| > 0.585), 2 differentially expressed miRNAs, and 65 differentially expressed lncRNAs were identified, which are involved in stem cell differentiation, calcium ions, plasma membrane, cell skeleton, chromatin, cell adhesion, etc. Our previous study found that nsPEFs (100 ns, 10kV/cm) with specific parameter combinations could promote the chondrogenic, osteoblastic, and adipogenic differentiation of stem cells, but did not induce apoptosis under these parameters[13,15]. Therefore, a combination of 100 ns and 10 kV/cm was selected to conduct electrical stimulation treatment on rMSCs to determine their effects on the gene expression profile. mRNA analysis indicated that nsPEFs may affect cell differentiation, calcium ions, plasma membrane, and other aspects. The expression levels of Scin, Ereg, Kif20b, Aldh3a1, Nr3c2, and Cenpf were upregulated. To better understand the universal effects of nsPEFs on mammalian cells, we compared the gene expression profiles between rMSCs (our data), TM3 cells, Jurkat cells, and U937 cells based on publicly available data[19,20]. We found that the gene expression levels of 23 genes were co-upregulated in rMSCs and TM3 cells, including Kif20b, which indicated that nsPEFs may cause common effects (Supplementary Figure 1). Compared with rMSCs, we found more common DEGs in TM3 cells (23 genes) than in Jurkat cells (3 genes) and U937 cells (6 genes). The reason for this may be that TM3 and rMSCs are adherent cells, and they are from mice or rats, while U937 and Jurkat cells are suspension cells, and are from humans. The 23 common DEGs are involved in the ECM-receptor interaction signaling pathway. Electric fields were reported to regulate ECM structure[21] and synthesis[22]. On the other hand, the ECM was reported to participate in regeneration[23] and play an important role in stem cell fate[24]. The ECM may be involved in the regulation of cell fate by nsPEFs, which could be further investigated focusing on ECM-receptor interaction. KIF20B was reported to be involved in cell proliferation[25]. Scin belongs to the gelolysin protein superfamily, which is involved in the regulation of cytoskeleton and transport in cell vesicles. It has an important regulatory role in the release of intracellular calcium ions, behaving as a filamentous actin-severing and capping protein[26]. The actin filament network, which in turn leads to the release of secretory vesicles[27], plays an important role in actin-dependent membrane fusion[26]. Ereg belongs to the epidermal growth factor family and is separated from stem cells. It has been reported that Ereg can promote the migration and chemotactic ability of adipose stem cells through the MAPK signaling pathway[28]. Cenpf plays an important role in the microtubule network, which may be related to SNARE proteins which are involved in plasma membrane circulation[29]. These genes remain to be explored in nsPEFs-treated stem cells.
MiRNAs play an important biological function by regulating downstream gene translation. We found that nsPEFs had few effects on miRNAs, and the expression levels of novel.118 and novel.106 were significantly upregulated. GO/KEGG analyses of the target genes of miRNAs showed that nsPEFs may affect vesicle fusion to the plasma membrane, MAPK, etc. In addition, nsPEFs were reported to affect the MAPK signaling pathway by phosphorylation of p38, JNK, and ERK[30]. Bone regeneration can be regulated via the MAPK signaling pathway under specific hydrogel treatment[31]. High-voltage PEFs with short durations, can permeabilize cell membranes with a duration ranging from microseconds to nanoseconds[32]. nsPEFs can also regulate membrane pore formation and upregulate the release of exosomes[33], and vesicle fusion was related to exosome formation[34]. In addition, studies have shown that electroporation can increase the production of exosomes by increasing intracellular calcium ions[35]. Following nsPEFs treatment, exosome release from tumor cells was also significantly increased[33]. Exosomes, released from stem cells, could stimulate wound regeneration and bone regeneration[36,37]. Thus, nsPEFs may affect exosome formation through vesicle fusion to the plasma membrane in stem cells, which requires further investigation.
Based on the target genes of differentially expressed lncRNAs, the most significant MF involved tubulin binding after nsPEFs treatment and the most significant signaling pathways involved glycerophospholipid metabolism and mismatch repair. In addition, studies have shown that nsPEFs with certain parameters can be applied to regulate the level of cell differentiation[12], promote the release of the intracellular calcium pool[38], and trigger reversible perforation of the cell membrane[39]. A previous study showed that nsPEFs could affect chromosome structure by inducing extracellular release of chromosomal DNA in a calcium-dependent manner[40]. nsPEFs with high intensity (60 kV/cm) can induce damage to the cytoskeleton and nuclear membrane[41]. Chromosome structure is sensitive to physical stimulation. Extremely-low-frequency magnetic fields could stabilize active chromatin, partially depending upon chromatin status[42]. Chromosomes can be oriented, aligned, and translated by high-frequency electric fields, in a frequency-dependent manner[43]. Chromatin accessibility played an important role in gene expression and cell fate[44]. Chromatin undergoes a binary off/on switch during cell fate transitions[45]. Electric fields were reported to change cell fate partially through regulation of calcium and modulation of electrically charged cell-surface receptors in response to the electric field[46]. Thus, nsPEFs may affect the chromatin accessibility and fate of stem cells, which remains to be explored.
The calcium signaling pathway may play an important role in the process of reaction of MSCs to nsPEFs. A previous study showed that nsPEFs could induce calcium flux in osteoblasts[47]. In our study, DEGs and the target genes of lncRNAs and miRNAs were enriched in the calcium signaling pathway as shown by GO/KEGG analyses. Calcium release caused by nsPEFs may be due to nanopore formation in the endoplasmic reticulum[48]. Furthermore, nsPEFs may activate TMEM16F (or anoctamin 6), a protein functioning as a calcium-dependent scramblase, which contributes to the reaction of calcium release due to nsPEFs[49]. BAPTA-AM, a calcium chelator, could attenuate the upregulated phosphorylation level of JNK caused by nsPEFs[14]. Calcium may contribute to apoptosis in hair follicle stem cells through Piezo1[50]. Calcium uptake was reported to control mitochondrial calcium homeostasis and hematopoietic stem cell differentiation, two important determinants in stem cell fate[51]. Calcium-activated potassium channel activity can influence MSC differentiation through membrane potential and intracellular calcium oscillations[52]. Consequently, the calcium signaling pathway may play an important role in the effects caused by nsPEFs in stem cells.
There are few studies on the effect of nsPEFs on stem cells at the whole transcriptomic level. The effects of nsPEFs on stem cells were systematically studied, and 263 differentially expressed mRNAs, 2 differentially expressed miRNAs, and 65 differentially expressed lncRNAs were identified. It was shown that nsPEFs may affect stem cells via several signaling pathways and may involve vesicular transport, calcium ion transport, the cytoskeleton, and cell differentiation. Our study is the first to investigate the expression profile of the whole transcriptome in nsPEFs-treated stem cells. This study provides a certain basis for the application of nsPEFs in stem cell differentiation and tissue regeneration.
Mesenchymal stem cells (MSCs) have been extensively applied in preclinical and clinical research in regenerative medicine. Their differentiation and function are modulated by various exogenous signals, which provide potential strategies for researchers to explore appropriate exogenous signals to regulate the functions and differentiation of stem cells. Nanosecond pulsed electric fields (nsPEFs) represent nanosecond-duration, high-strength electric fields to significantly influence cell phenotypes and regulate stem cell differentiation through multiple pathways. Thus, we used transcriptomics analysis to analyze messenger RNA (mRNA), long noncoding RNA (lncRNA), microRNA (miRNA), and circular RNA (circRNA) expression to identify changes in gene expression following treatment with nsPEFs.
The differentiation and function of MSCs are regulated by nsPEFs. However, the mechanism, especially changes in gene expression after nsPEFs treatment, remains unclear.
To reveal gene expression in MSCs pretreated with nsPEFs and explore the potential gene regulatory mechanism.
We used whole transcriptome sequencing to investigate the effects of nsPEFs on MSC transcriptome. Five pulses of nsPEFs (100 ns at 10 kV/cm, 1 Hz) were applied to pretreat MSCs. Total RNA was isolated after pretreatment of MSCs; each transcript was normalized by fragments per kilobase per million. Fold change and difference significance were used to screen the differentially expressed genes (DEGs). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses were conducted to identify gene function, and the results were verified by quantitative polymerase chain reaction.
The top 20 differentially expressed lncRNAs and mRNAs were revealed. Two hundred and sixty-three DEGs were identified in the PRJNA931816 dataset, of which 92 were upregulated and 171 were significantly downregulated, respectively. DEGs were mainly enriched in epithelial cell proliferation, osteoblast differentiation, mesenchymal cell differentiation, nuclear division, and wound healing. As for cellular components, DEGs were mainly involved in condensed chromosome, chromosomal region, actin cytoskeleton, and the kinetochore. With regard to molecular functions, DEGs are mainly involved in glycosaminoglycan binding, integrin binding, nuclear steroid receptor activity, cytoskeletal motor activity, and steroid binding. Quantitative real-time polymerase chain reaction was used to verify the seven upregulated mRNAs, Aldh3a1, Cenpf, Kif20b, Ereg, Nek2, Nr3c2, and Scin, and six downregulated mRNAs, Actg2, Asic3, Crybat4, Nog, Stxbp5 L, and Tubb2b.
Our systematic investigation of the wide-ranging transcriptional pattern modulated by nsPEFs revealed the differential expression of 263 mRNAs, 2 miRNAs, and 65 lncRNAs. We showed that nsPEFs may affect stem cells via several signaling pathways and involve vesicular transport, calcium ion transport, the cytoskeleton, and cell differentiation.
This study is the first to investigate the expression profile of the whole transcriptome in nsPEFs-treated stem cells. The findings provide a certain basis for the application of nsPEFs in stem cell differentiation and tissue regeneration.
The authors are grateful to Peking University, Peking University People Hospital, and Peking University Shenzhen Hospital for providing the research instruments and workplace.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Delben PB, Brazil; Mohammadzadeh I, Iran; Soltani R, Iran S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Zhao YQ
| 1. | McKinney JM, Pucha KA, Doan TN, Wang L, Weinstock LD, Tignor BT, Fowle KL, Levit RD, Wood LB, Willett NJ. Sodium alginate microencapsulation of human mesenchymal stromal cells modulates paracrine signaling response and enhances efficacy for treatment of established osteoarthritis. Acta Biomater. 2022;141:315-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 2. | Zhang J, Zhang M, Lin R, Du Y, Wang L, Yao Q, Zannettino A, Zhang H. Chondrogenic preconditioning of mesenchymal stem/stromal cells within a magnetic scaffold for osteochondral repair. Biofabrication. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 3. | Choi DH, Lee KE, Oh SY, Lee SM, Jo BS, Lee JY, Park JC, Park YJ, Park KD, Jo I, Park YS. Tonsil-derived mesenchymal stem cells incorporated in reactive oxygen species-releasing hydrogel promote bone formation by increasing the translocation of cell surface GRP78. Biomaterials. 2021;278:121156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Fan L, Chen J, Tao Y, Heng BC, Yu J, Yang Z, Ge Z. Enhancement of the chondrogenic differentiation of mesenchymal stem cells and cartilage repair by ghrelin. J Orthop Res. 2019;37:1387-1397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 5. | Chen Y, An X, Wang Z, Guan S, An H, Huang Q, Zhang H, Liang L, Huang B, Wang H, Lu M, Nie H, Wang J, Dai X, Lu X. Transcriptome and lipidome profile of human mesenchymal stem cells with reduced senescence and increased trilineage differentiation ability upon drug treatment. Aging (Albany NY). 2021;13:9991-10014. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 6. | Kusuyama J, Bandow K, Shamoto M, Kakimoto K, Ohnishi T, Matsuguchi T. Low intensity pulsed ultrasound (LIPUS) influences the multilineage differentiation of mesenchymal stem and progenitor cell lines through ROCK-Cot/Tpl2-MEK-ERK signaling pathway. J Biol Chem. 2014;289:10330-10344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Hess R, Jaeschke A, Neubert H, Hintze V, Moeller S, Schnabelrauch M, Wiesmann HP, Hart DA, Scharnweber D. Synergistic effect of defined artificial extracellular matrices and pulsed electric fields on osteogenic differentiation of human MSCs. Biomaterials. 2012;33:8975-8985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Guo W, Zhang X, Yu X, Wang S, Qiu J, Tang W, Li L, Liu H, Wang ZL. Self-Powered Electrical Stimulation for Enhancing Neural Differentiation of Mesenchymal Stem Cells on Graphene-Poly(3,4-ethylenedioxythiophene) Hybrid Microfibers. ACS Nano. 2016;10:5086-5095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 166] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 9. | Bicer M, Sheard J, Iandolo D, Boateng SY, Cottrell GS, Widera D. Electrical Stimulation of Adipose-Derived Stem Cells in 3D Nanofibrillar Cellulose Increases Their Osteogenic Potential. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Yao C, Hu X, Mi Y, Li C, Sun C. Window effect of pulsed electric field on biological cells. IEEE Trans Dielectr Electr Insul. 2009;16:1259-1266. [DOI] [Full Text] |
| 11. | Ning T, Zhang K, Heng BC, Ge Z. Diverse effects of pulsed electrical stimulation on cells - with a focus on chondrocytes and cartilage regeneration. Eur Cell Mater. 2019;38:79-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 12. | Li K, Ning T, Wang H, Jiang Y, Zhang J, Ge Z. Nanosecond pulsed electric fields enhance mesenchymal stem cells differentiation via DNMT1-regulated OCT4/NANOG gene expression. Stem Cell Res Ther. 2020;11:308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Li K, Fan L, Lin J, Heng BC, Deng Z, Zheng Q, Zhang J, Jiang Y, Ge Z. Nanosecond pulsed electric fields prime mesenchymal stem cells to peptide ghrelin and enhance chondrogenesis and osteochondral defect repair in vivo. Sci China Life Sci. 2022;65:927-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Ning T, Guo J, Zhang K, Li K, Zhang J, Yang Z, Ge Z. Nanosecond pulsed electric fields enhanced chondrogenic potential of mesenchymal stem cells via JNK/CREB-STAT3 signaling pathway. Stem Cell Res Ther. 2019;10:45. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 15. | Uusküla-Reimand L, Wilson MD. Untangling the roles of TOP2A and TOP2B in transcription and cancer. Sci Adv. 2022;8:eadd4920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 100] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 16. | Heilig AK, Nakamura R, Shimada A, Hashimoto Y, Nakamura Y, Wittbrodt J, Takeda H, Kawanishi T. Wnt11 acts on dermomyotome cells to guide epaxial myotome morphogenesis. Elife. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | Kim JA, Choi YA, Yun HS, Bae YC, Shin HI, Park EK. Extracellular calcium-binding peptide-modified ceramics stimulate regeneration of calvarial bone defects. Tissue Eng Regen Med. 2016;13:57-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Vadlamani RA, Nie Y, Detwiler DA, Dhanabal A, Kraft AM, Kuang S, Gavin TP, Garner AL. Nanosecond pulsed electric field induced proliferation and differentiation of osteoblasts and myoblasts. J R Soc Interface. 2019;16:20190079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Kasprzycka W, Trębińska-Stryjewska A, Lewandowski RB, Stępińska M, Osuchowska PN, Dobrzyńska M, Achour Y, Osuchowski ŁP, Starzyński J, Mierczyk Z, Trafny EA. Nanosecond Pulsed Electric Field Only Transiently Affects the Cellular and Molecular Processes of Leydig Cells. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Roth CC, Glickman RD, Tolstykh GP, Estlack LE, Moen EK, Echchgadda I, Beier HT, Barnes RA Jr, Ibey BL. Evaluation of the Genetic Response of U937 and Jurkat Cells to 10-Nanosecond Electrical Pulses (nsEP). PLoS One. 2016;11:e0154555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 21. | Nguyen HT, Wei C, Chow JK, Nguy L, Nguyen HK, Schmidt CE. Electric field stimulation through a substrate influences Schwann cell and extracellular matrix structure. J Neural Eng. 2013;10:046011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Aaron RK, Ciombor DM, Simon BJ. Treatment of nonunions with electric and electromagnetic fields. Clin Orthop Relat Res. 2004;21-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 163] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 23. | Yang Y, Lin H, Shen H, Wang B, Lei G, Tuan RS. Mesenchymal stem cell-derived extracellular matrix enhances chondrogenic phenotype of and cartilage formation by encapsulated chondrocytes in vitro and in vivo. Acta Biomater. 2018;69:71-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 24. | Chermnykh E, Kalabusheva E, Vorotelyak E. Extracellular Matrix as a Regulator of Epidermal Stem Cell Fate. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 25. | Chen J, Zhao CC, Chen FR, Feng GW, Luo F, Jiang T. KIF20B Promotes Cell Proliferation and May Be a Potential Therapeutic Target in Pancreatic Cancer. J Oncol. 2021;2021:5572402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Wang X, Shelton SD, Bordieanu B, Frank AR, Yi Y, Venigalla SSK, Gu Z, Lenser NP, Glogauer M, Chandel NS, Zhao H, Zhao Z, McFadden DG, Mishra P. Scinderin promotes fusion of electron transport chain dysfunctional muscle stem cells with myofibers. Nat Aging. 2022;2:155-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Chumnarnsilpa S, Robinson RC, Grimes JM, Leyrat C. Calcium-controlled conformational choreography in the N-terminal half of adseverin. Nat Commun. 2015;6:8254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 28. | Cao Y, Wang L, Yang H, Lin X, Li G, Han N, Du J, Fan Z. Epiregulin promotes the migration and chemotaxis ability of adipose-derived mesenchymal stem cells via mitogen-activated protein kinase signaling pathways. J Cell Biochem. 2018;119:8450-8459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 29. | Pooley RD, Moynihan KL, Soukoulis V, Reddy S, Francis R, Lo C, Ma LJ, Bader DM. Murine CENPF interacts with syntaxin 4 in the regulation of vesicular transport. J Cell Sci. 2008;121:3413-3421. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Morotomi-Yano K, Akiyama H, Yano K. Nanosecond pulsed electric fields activate MAPK pathways in human cells. Arch Biochem Biophys. 2011;515:99-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 31. | Liu Y, Zhang Y, Zheng Z, Zhong W, Wang H, Lin Z, Li L, Wu G. Incorporation of NGR1 promotes bone regeneration of injectable HA/nHAp hydrogels by anti-inflammation regulation via a MAPK/ERK signaling pathway. Front Bioeng Biotechnol. 2022;10:992961. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 32. | Safaei Z, Thompson GL. Histone deacetylase 4 and 5 translocation elicited by microsecond pulsed electric field exposure is mediated by kinase activity. Front Bioeng Biotechnol. 2022;10:1047851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 33. | Qian J, Chen T, Wu Q, Zhou L, Zhou W, Wu L, Wang S, Lu J, Wang W, Li D, Xie H, Su R, Guo D, Liu Z, He N, Yin S, Zheng S. Blocking exposed PD-L1 elicited by nanosecond pulsed electric field reverses dysfunction of CD8(+) T cells in liver cancer. Cancer Lett. 2020;495:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3060] [Cited by in RCA: 5611] [Article Influence: 801.6] [Reference Citation Analysis (0)] |
| 35. | Yang Z, Shi J, Xie J, Wang Y, Sun J, Liu T, Zhao Y, Zhao X, Wang X, Ma Y, Malkoc V, Chiang C, Deng W, Chen Y, Fu Y, Kwak KJ, Fan Y, Kang C, Yin C, Rhee J, Bertani P, Otero J, Lu W, Yun K, Lee AS, Jiang W, Teng L, Kim BYS, Lee LJ. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat Biomed Eng. 2020;4:69-83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 506] [Article Influence: 84.3] [Reference Citation Analysis (0)] |
| 36. | Zhang Y, Shi L, Li X, Liu Y, Zhang G, Wang Y. Placental stem cells-derived exosomes stimulate cutaneous wound regeneration via engrailed-1 inhibition. Front Bioeng Biotechnol. 2022;10:1044773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 37. | Ma S, Zhang Y, Li S, Li A, Li Y, Pei D. Engineering exosomes for bone defect repair. Front Bioeng Biotechnol. 2022;10:1091360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 38. | Mao Z, Zhang Y, Lu N, Cheng S, Hong R, Liu QH. Carbon Nanotubes Enabling Highly Efficient Cell Apoptosis by Low-Intensity Nanosecond Electric Pulses via Perturbing Calcium Handling. Small. 2020;16:e1904047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Pakhomov AG, Gianulis E, Vernier PT, Semenov I, Xiao S, Pakhomova ON. Multiple nanosecond electric pulses increase the number but not the size of long-lived nanopores in the cell membrane. Biochim Biophys Acta. 2015;1848:958-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 40. | Koga T, Morotomi-Yano K, Sakugawa T, Saitoh H, Yano KI. Nanosecond pulsed electric fields induce extracellular release of chromosomal DNA and histone citrullination in neutrophil-differentiated HL-60 cells. Sci Rep. 2019;9:8451. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 41. | Stacey M, Fox P, Buescher S, Kolb J. Nanosecond pulsed electric field induced cytoskeleton, nuclear membrane and telomere damage adversely impact cell survival. Bioelectrochemistry. 2011;82:131-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 42. | Manser M, Sater MR, Schmid CD, Noreen F, Murbach M, Kuster N, Schuermann D, Schär P. ELF-MF exposure affects the robustness of epigenetic programming during granulopoiesis. Sci Rep. 2017;7:43345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Andrews MJ, McClure JA. Effects of high frequency electric fields on mammalian chromosomes in vitro. J Biol Phys. 1978;6:69-86. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 44. | Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet. 2019;20:207-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 668] [Cited by in RCA: 1045] [Article Influence: 174.2] [Reference Citation Analysis (0)] |
| 45. | Li D, Shu X, Zhu P, Pei D. Chromatin accessibility dynamics during cell fate reprogramming. EMBO Rep. 2021;22:e51644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 46. | Thrivikraman G, Boda SK, Basu B. Unraveling the mechanistic effects of electric field stimulation towards directing stem cell fate and function: A tissue engineering perspective. Biomaterials. 2018;150:60-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 236] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 47. | Zhou P, He F, Han Y, Liu B, Wei S. Nanosecond pulsed electric field induces calcium mobilization in osteoblasts. Bioelectrochemistry. 2018;124:7-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 48. | Scarlett SS, White JA, Blackmore PF, Schoenbach KH, Kolb JF. Regulation of intracellular calcium concentration by nanosecond pulsed electric fields. Biochim Biophys Acta. 2009;1788:1168-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 59] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 49. | Muratori C, Pakhomov AG, Gianulis E, Meads J, Casciola M, Mollica PA, Pakhomova ON. Activation of the phospholipid scramblase TMEM16F by nanosecond pulsed electric fields (nsPEF) facilitates its diverse cytophysiological effects. J Biol Chem. 2017;292:19381-19391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 50. | Xie Y, Chen D, Jiang K, Song L, Qian N, Du Y, Yang Y, Wang F, Chen T. Hair shaft miniaturization causes stem cell depletion through mechanosensory signals mediated by a Piezo1-calcium-TNF-α axis. Cell Stem Cell. 2022;29:70-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 51. | Bonora M, Kahsay A, Pinton P. Mitochondrial calcium homeostasis in hematopoietic stem cell: Molecular regulation of quiescence, function, and differentiation. Int Rev Cell Mol Biol. 2021;362:111-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Pchelintseva E, Djamgoz MBA. Mesenchymal stem cell differentiation: Control by calcium-activated potassium channels. J Cell Physiol. 2018;233:3755-3768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |