Published online Feb 26, 2024. doi: 10.4252/wjsc.v16.i2.114
Peer-review started: October 23, 2023
First decision: December 5, 2023
Revised: December 18, 2023
Accepted: January 29, 2024
Article in press: January 29, 2024
Published online: February 26, 2024
Processing time: 125 Days and 16.9 Hours
Human pluripotent stem cell (hPSC)-derived kidney organoids share similarities with the fetal kidney. However, the current hPSC-derived kidney organoids have some limitations, including the inability to perform nephrogenesis and lack of a corticomedullary definition, uniform vascular system, and coordinated exit path
Core Tip: Pluripotent stem cells (PSCs) are a class of cells with self-renewal and multidirectional differentiation potential, and organoids are a group of tissue analogues induced to form three-dimensional (3D) structures that are structurally and functionally very similar to human organs under specific differentiation conditions. We review research progress on how renal organoid induction protocols can well mimic the human foetal model of renal development and disease. The discovery of human PSCs, and the recent 3D organoid generation methods have opened avenues for in vitro mimicry of human kidney development, disease research, and testing of new drugs directly on human tissue.
- Citation: Long HY, Qian ZP, Lan Q, Xu YJ, Da JJ, Yu FX, Zha Y. Human pluripotent stem cell-derived kidney organoids: Current progress and challenges. World J Stem Cells 2024; 16(2): 114-125
- URL: https://www.wjgnet.com/1948-0210/full/v16/i2/114.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v16.i2.114
The kidney is a complex organ consisting of over 20 cell types and is responsible for producing urine. Additionally, the kidney ensures normal body activity and metabolism through reabsorption and endocrine functions[1]. According to the Global Burden of Disease Alliance, chronic kidney disease (CKD) is predicted to be one of the top five diseases causing loss of life expectancy by 2040[2]. CKD is a major global health problem with increasing incidence and prevalence; it affects approximately 10% of the global population and can lead to end-stage renal failure[3]. Currently, the primary treatments for kidney diseases include medications, dialysis, and kidney transplantation[4]. By 2020, the number of individuals receiving renal replacement therapy exceeded 2.5 million and was expected to double to 5.4 million by 2030[5]. Recently, there has been an increase in the number of people with end-stage renal disease who require dialysis or transplantation[6], presenting a serious and growing public health issue. Therefore, there is an urgent need to develop effective treatments for CKD, and pluripotent stem cells (PSCs) possess potential applications for CKD treatment due to their multi-directional differentiation potential.
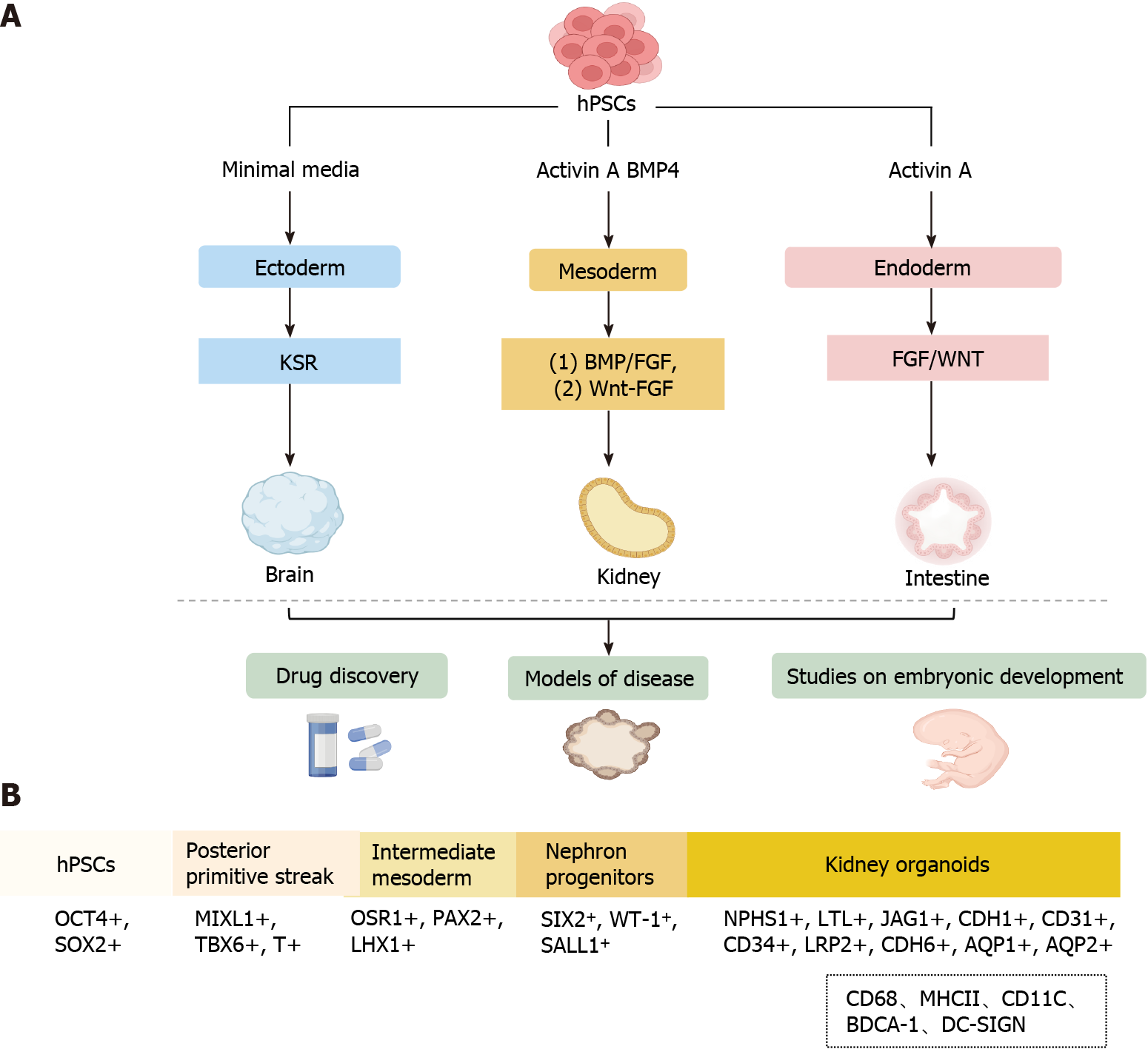
PSCs are a class of cells with the potential for self-renewal and multi-directional differentiation. Organoids are a group of highly PSCs induced into a three-dimensional (3D) structure that closely mimics human organs structurally and functionally under specific differentiation conditions[7-9]. Organoids are physiologically complicated systems for studying organ development, drug screening, and disease modeling because they frequently possess cell types from three germ layers and a structural organization comparable to that of human organs (Figure 1A)[7,10,11].
In this review, we discussed recent advances in the generation of human PSC (hPSC)-derived kidney organoids, with emphasis on the contribution of these advances to the understanding of human kidney development and research in disease modeling. Additionally, we discussed the limitations, future research focus, and applications of hPSC-derived kidney organoids; particularly, methods to produce highly complex hPSC-derived kidney organoids with vascular components were highlighted.
Since the establishment of embryonic stem and induced pluripotent stem cell lines, scientists have employed insights from developmental biology to derive differentiated cell types from these stem cells (Figure 1A), PSCs have high proliferative capacity and can differentiate into all three germ layers: Ectoderm, endoderm, and mesoderm[12-16]. The process of generating kidney organoids exactly mimics the normal development conditions of the kidney in vivo. Specifically, the generation of hPSC-derived organoids is performed using varying growth factors and culture substrates under precise timing to generate primitive streak, intermediate mesoderm (IM), renal unit progenitor cells, and kidney organoids (Figure 1B)[7,8,17-21].
During the embryonic period, the complex kidney develops from the relatively simple posterior kidney, and the embryonic kidney develops from different regions of the mesoderm. Specifically, nephron progenitor cells (NPCs) originate from the posterior IM (PIM) and differentiate into renal unit-like organs through Wnt and fibroblast growth factor (FGF) stimulation. In contrast, the ureteric bud originates from the anterior IM. FGF is a central regulator of the differentiation of the presomitic mesoderm into somatic tissue, which was the initially identified mesoderm-inducing signal[22,23]. Nephron progenitors self-assemble to form multi-compartmentalized kidney organoids composed of different cellular components, including a segmentally patterned epithelium, interstitium, and endothelium, in response to the activation of FGF and/or Wnt signaling[20,21,24]. FGFs are potential mesoderm inducers, while activin A, the first acting factor of transforming growth factor betas, can induce cell transition to the IM stage[25,26]. However, these methods are expensive and time-consuming; therefore, recent efforts have been made to improve scalability and reduce the cost of iPSC-derived renal organoid generation. Morizane and Bonventre[27] induced the differentiation of H9-hESC and HDF-hIPSC cell lines to the primitive streak stage using a Wnt signaling pathway activator (CHIR99021) for 4 d in a planar environment, followed by sequential induction with activin A and FGF9 to bring them to the PIM stage and automatic development of PIM into MM, which further developed towards the renal lineage[27]. Specifically, the cells reached the NPC stage at 9 d, as evidenced by the high expression of four NPC markers (CSIX2, WT1, CPAX2, and SALL1), and the NPCs were capable of subsequently differentiating into foot cells (NPHS1+, WT1+), proximal tubules (LTL+, APQ1+), medullary collaterals (CDH1+, AQP1+), and distal tubules (CDH1+) in the 3D environment[27]. Overall, the main advantage of the protocol developed by Morizane and Bonventre[27] high (90%) efficiency of generating NPCs from H9-hESC and HDF-hIPSC cell lines. Moreover, the culture conditions were modified to allow for the formation of renal organoids in suspension culture, which increased the yield of cellular structures and reduced the overall cost, making the protocol more cost-effective than that developed by Takasato et al[21].
Primitive streak development and subsequent mesoderm formation are highly ordered and spatiotemporally regulated by the interplay of several internal (transcription factors, epigenetic regulators, and chromatin remodeling factors) and external variables. In recent years, numerous protocols have been published on the differentiation of PSCs into kidney organoids (Table 1). Current strategies for the generation of renal organoids include the bone morphogenetic protein (BMP)/FGF pathway and the CHIR99021-FGF pathway. In the BMP/FGF pathway, hPSC differentiation into primitive streak mesoderm (PSM) is induced using BMP4/activin A, followed by stimulation of PSM with FGF9 to differentiate into IM and subsequent stimulation of IM with FGF9/BMP7/RA to differentiate into metanephric mesenchyme (MM) and ureteric bud (UB). Finally, cells in the MM and UB stages can further differentiate into organoids, including collecting ducts, proximal tubules, and foot cells. In the CHIR99021-FGF pathway, PSCs differentiation into PSM is induced using the Wnt agonist CHIR99021, followed by stimulation of PSM with FGF9 to differentiate into IM and subsequent differentiation of IM into MM and UB, both of which can differentiate into kidney organs, including collecting ducts, renal tubules, and glomeruli.
| Posterior primitie streak | Intermediate mesoderm | Nephric progenitor cell | Kidney organoids |
| BMP4 | FGF9 | FGF9 | Without growth factor 6 d |
| Activin A, 2-3 d | 4D | BMP7, RA, 6 d | Kidney organoids |
| SYNPO, NPHS1, WT1, AQP1, SLC3A1, AQP2, SCNNB1[21] | |||
| Activin A, WNT3, 1 d | FGF2, BMP7 | FGF2, BMP7 | HGF 21 d |
| BMP4, FGF2, 2 d | RA, 8 d | RA, 15 d | Kidney organoids E-CADHERIN, ZO1, KRT18, SLC12A3, CD13, ALP, AQP1, MUC1, SYN, NEPHRIN[28] |
| CHIR, 4 d | Without growth factor, 3 d | FGF9, CHIR, 2 d | FGF9, 11 d |
| Kidney organoids | |||
| WT1+, NPHS1+, LTL+, LRP2+, CDH6+, AQP1+, JAG1+, CDH1+, AQP2+, CK8+, CALB1+, CD31+, CD34+[11] | |||
| CHIR, BMP4, 4 d | Activin, 3 d | FGF9, 2 d | FGF9, CHIR 2 d, FGF9, 3d |
| Without growth factor 7 d | |||
| Kidney organoids | |||
| PAX8+, LHX1+, LAM+, PODXL, LTL, CDH1, CDH1+, AQP2+, PDGFRβ+, endomucin+, α-SMA+[27] | |||
| CHIR, 4 d | FGF9, CHIR, 2 d | FGF9, CHIR, 4 d | Without growth factor 18 d |
| Kidney organoids | |||
| NPHS1+, MAFB+, LTL+, CUBN+, LRP2+, HNF4A+, ECAD, GATA3+, SOX17+, PECAM1+[29] | |||
| CHIR, 3 d | Activin, FGF9, 1 d | CHIR, FGF9, 2 d | FGF9 200 ng/mL 4 d, without growth factor 10 d |
| Kidney organoids WT1+, LTL+, NPHS1, SCNN1B, ENDOGLIN, VEGFR[30] | |||
| CHIR, 4 d | FGF9, 3 d | CHIR, 1 h | Without growth factor 15-17 d |
| FGF9, 5 d | Kidney organoids WT1+, LTL+, CDH1+[31] | ||
| CHIR, RA, BMP4, FGF2, 1 d | FGF9, RA, NOGGIN, 2 d | FGF9, CHIR, 3 d | CHIR, FGF2, 2 d, without growth factor 8 d |
| CHIR, BMP7, FGF2, 0.5 d | Kidney organoids, NEPHRIN, WT1, LTL, AQP1, SGLT2, JAGGED-1 | ||
| CHIR, BMP7, FGF2, A83-01, 1.5 d | GED1, CDH6, BRN1, AQP2, AQP3[32] | ||
| CHIR, BMP7, FGF2, Activin A, Y-27632, 3 d |
Recently, the protocols for generating kidney organoids have attracted considerable attention (Table 1). However, the generated kidney organoids are incomplete and lack the complete vascular network structure, and the nephron organoids mainly contain glomeruli and proximal tubules, and protocols to effectively induce distal tubule formation are needed. Taguchi et al[33] developed a new protocol for inducing ureteric bud-like organs from PSCs, and although this class of organs has the properties of the collecting duct principal cells, the generation of intercalary cells remains a challenge.
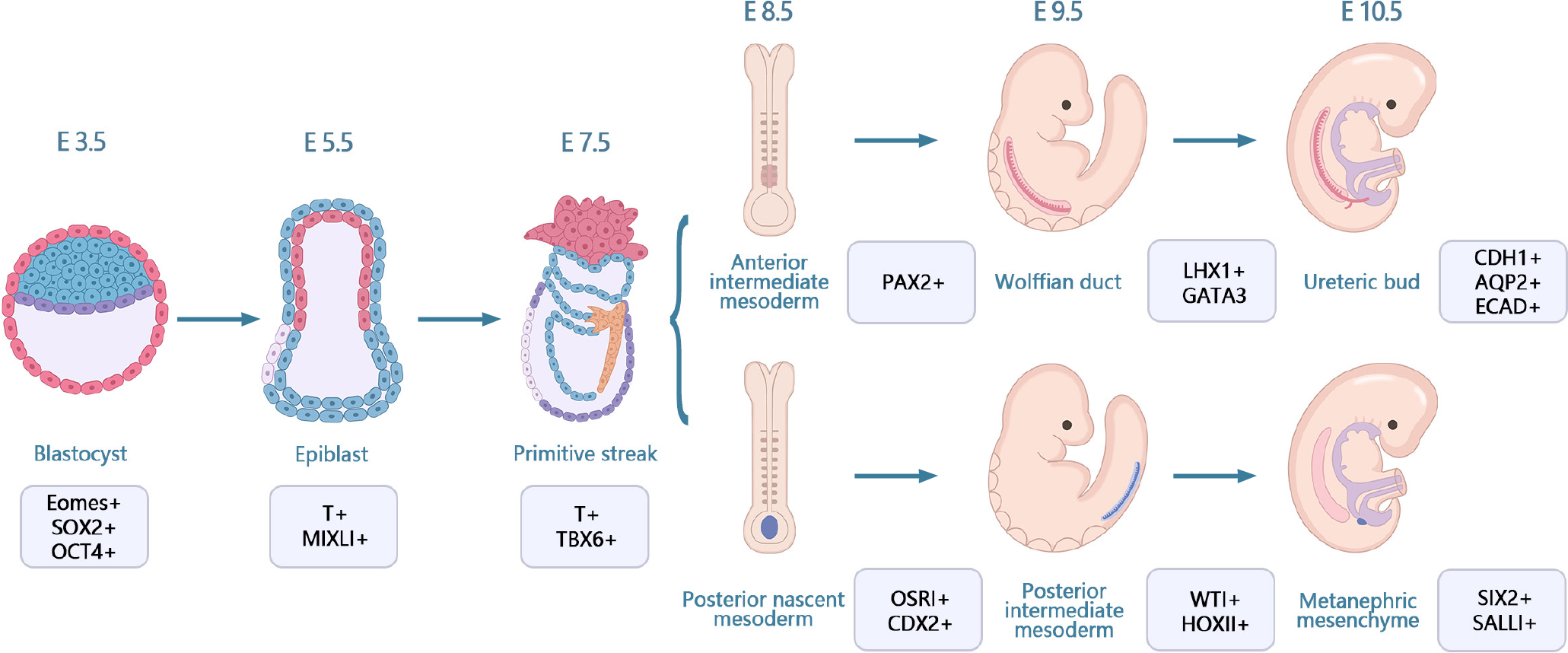
Kidney development originates from the primary stripe of the embryo, which differentiates via the mesoderm into the anterior mesoderm. The anterior mesoderm develops into renal tubes and branching ureteric buds and eventually differentiates into collecting ducts. The posterior renal mesenchyme develops into renal units with renal distal progenitors and collecting duct progenitors that induce and differentiate from each other, ultimately forming a complex renal unit-collecting duct network structure[34]. Kidney development in vivo involves four stages: Stage I, differentiation of PSCs into cells of the posterior primitive streak; stage II, cells of the posterior primitive streak are designated to differentiate into cells of the PIM; stage III, transition of PIM to NPCs of the posterior renal mesenchyme; and stage IV, mesenchyme-to-epithelial transition from the posterior mesenchyme to the renal unit (Figure 2)[17,35]. Organ development events may be studied using hPSC-derived organoids owing to their similarities with fetal tissues[36,37]. Several important steps in human kidney development have been replicated in hPSC-derived kidney organoids.
Recently, kidney organoids have become an important topic in renal development and regenerative medicine[38]. Researchers can use kidney organs derived from hPSCs to model human kidney development and study the biological and molecular mechanisms involved. Kidney organoids are not only structurally similar to the human kidneys but also recapitulate some of the specific functions of the kidneys, including reabsorption[39]. Various cell populations are generated during kidney development, which forms self-assembled tissues through signal transduction[27]. Takasato et al[36] generated kidney organoids from hESC using the CHIR99021-FGF9 induction procedure. Specifically, growth factors were withdrawn from the in vitro culture at 12-18 d, and the elongated ECAD+ ureteral epithelium was surrounded by several clusters of mesenchyme (expressing the molecular hallmarks of the MM, WT1, SIX2, and PAX2) by day 18. The MM formation looked like a renal unit, expressing JAG1 and CDH6 proteins, with a visible connection between the ureteral epithelium and the renal vesicle lumen. The stepwise differentiation protocol for generating human renal tissue from PSCs includes the progression from an initial pattern to a posterior primitive streak, followed by differentiation to IM and then posterior renal mesenchyme. In the Takasato protocol for the differentiation of hPSCs, the mesoderm was induced by the addition of a classical Wnt agonist, followed by the addition of FGF9. Thereafter, the cells were placed on a Transwell filter on day 7 of culture to allow spontaneous tissue formation. A comparative analysis of the single-cell profile of human kidney-like organs and cells of the fetal renal cortex showed that the key renal cell types in the two organs were the same[36].
Notably, it is important to understand the cellular lineage relationships during nephrogenesis to better define them in the human kidney. In the past ten years, sophisticated methods have been developed to track cell fate in mice[40]. Although the genealogical relationships in mouse kidney development have been extensively elucidated, this information cannot be fully applied to humans due to interspecies differences. However, genetic lineage analysis may be utilized in hPSCs to monitor the development of human progenitor cells into kidney organoids[41]. Additionally, the homeodomain transcriptional regulator SIX2 is a crucial element in the kidney mesenchyme[42]. SIX2+ NPCs are pluripotent and can differentiate into multinodular epithelial cells, including the peduncle and wall epithelium of the glomerulus as well as the epithelium of the proximal tubule, Henle’s loop, and distal tubule. Additionally, SIX2+ NPCs can self-renew in the interstitial space in the embryonic kidney[42]. Howden et al[41] investigated whether self-renewing SIX2+ NPCs can give rise to renal unit cell types during human renal organ culture and found that SIX2-expressing cells can differentiate into proximal tubule renal unit segments but not into the collecting duct lineage, which is consistent with findings in mice. Collectively, these results provide proof-of-principle for tracking cell fates in human renal organoids to understand renal development in vivo. The origin of the renal vascular system has remained unclear in previous genetic studies on model organisms. Takasato et al[36] showed that kidney organoids featured PDGFRA+ early mesangial cells invading the glomeruli and PDGFRA+ perivascular cells that lie along the KDR1 endothelia. In the growing kidney, the renal interstitium develops into pericytes and mesangial cells[43]. Low et al[11] reported an increased percentage of vascular progenitor cells, including SIX1+/KDR+/PECAM1- cells, in renal organoids by analyzing the transcriptomes of 62506 single cells on days 10, 12, and 14 of renal differentiation, with the NPC cluster possessing the highest percentage of SIX1+/KDR+/PECAM1- cells. Additionally, there was an increase in the percentage of SIX1+/KDR+/PECAM1+ and SIX1+/KDR-/PECAM1+ mature endothelial cells. These results indicate that NPCs are an unconventional source of renal vasculature, which is consistent with the sequencing data of fetal kidney samples obtained during mid-pregnancy[44]. Additionally, single-cell analysis of vascular endothelial cell subclusters revealed that the vascular endothelial cells of a kidney-like organ form a well-established genetic network and could specifically generate different endothelial subtype structures, such as arterioles and venules.
The establishment of these kidney organoid models has provided novel insights into the process of renal development and may advance the study of renal physiology and pathology. Kidney organoids are used to evaluate developmental and genetic diseases, and despite these promising advances in currently developed protocols, challenges remain. Under physiological conditions, the intrarenal immune system consists mainly of macrophages (CD68, major histocompatibility complex II) and dendritic cells (CD11C, blood dendritic cell antigen 1, DC-SIGN) (Figure 1B)[45,46]. Immune cells have been detected in human tissues, including the kidney, but almost all current in vitro organoid models lack immune cells, although protocols for co-culturing organoids with immune cells have now been developed[47,48], and these protocols provide a new platform for studying complex epithelial-immune cell crosstalk. However, due to the relative novelty of kidney organoids, co-culture with immune cells has not yet been established, and in the future, this could potentially be a new way for kidney organoids to generate immune cells.
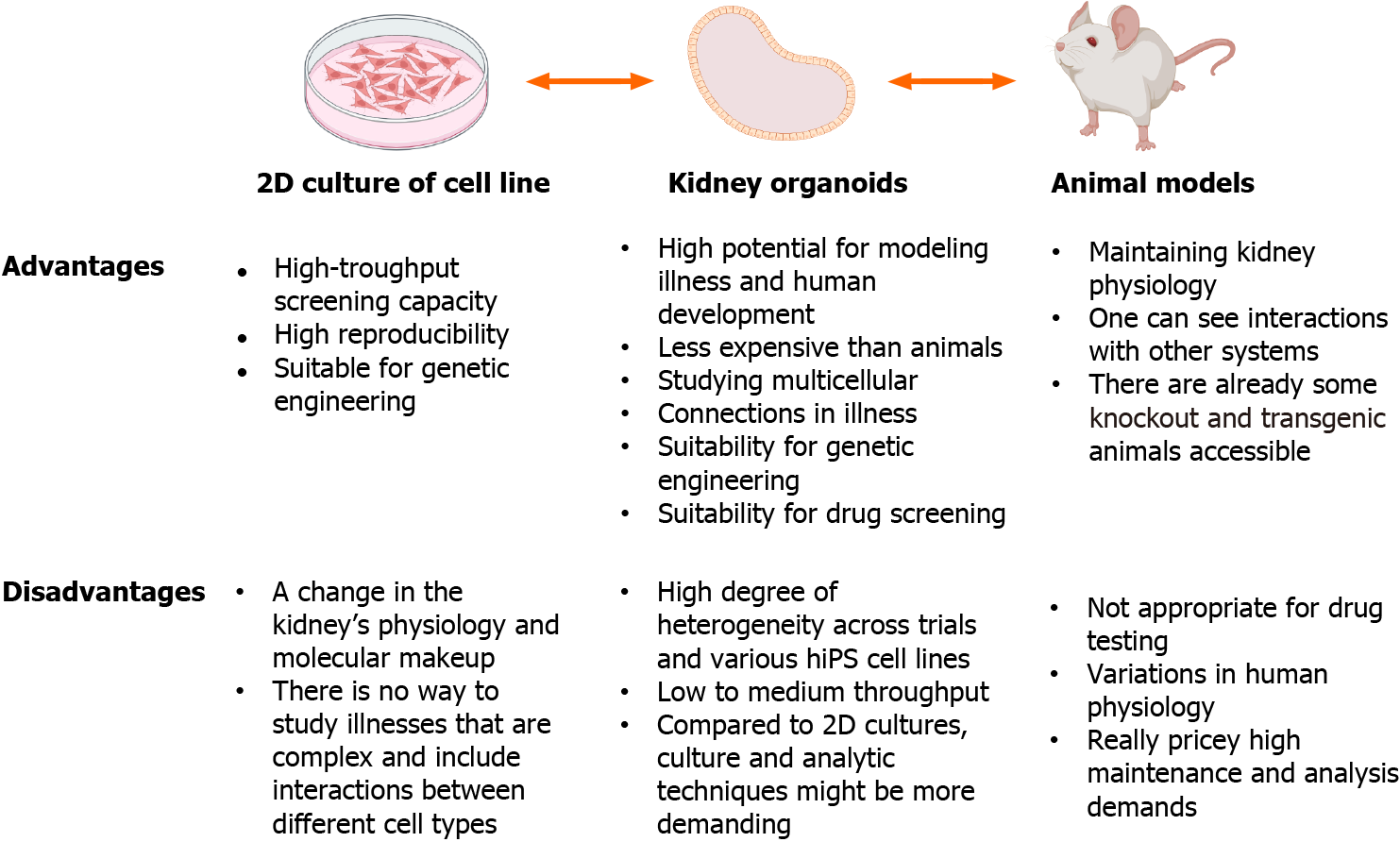
Organoids are superior to 2D cell cultures and animal models for in vitro experiments (Figure 3), and may provide comprehensive insights into development, homeostasis, and pathogenesis. For example, more than 300 genes associated with childhood kidney disease[49] are not present in the mouse genome, and human pathology genes are differentially expressed in mice[50,51]. Targeted differentiation of hPSCs into kidney organoids, combined with single-cell RNA sequencing technology can be used to explore disease mechanisms and identify novel directions for disease modeling[7,52,53]. Additionally, the use of CRISPR/Cas9 to edit hPSCs, introduce specific mutations, and compare them with samples from homogeneous genetic backgrounds is another approach for studying inherited kidney diseases[54,55].
Polycystic kidney disease (PKD) is one of the most prevalent monogenic kidney diseases worldwide and is classified into two types: Autosomal dominant (ADPKD) and autosomal recessive (ARPKD)[56,57]. ADPKD is frequently caused by defects in polycystin-1 (PC1) and PC2, which are encoded by PKD1 and PKD2, respectively[58-60]. Freedman et al[7] knocked down the PKD1 and PKD2 genes in human-induced PSCs (hiPSCs) using CRISPR/Cas9 genome editing technology and utilized these cells to generate kidney organoids, providing new insights into the role of the microenvironment in the PKD process. PKD cysts were visible approximately 35 d after the first plating, and continued to grow during the culture period; moreover, the cysts showed a significant affinity for Lotus tetragonolobus lectin. Importantly, isogenic control hPSCs that were plated and differentiated alongside the edited cells did not develop cysts under these circumstances[7], successfully validating the PKD model for organoid culture. In subsequent studies with improved culture conditions, suspension-cultured organoids promoted the formation of cysts (up to 75% of organoids developed as cysts) on day 21 of differentiation compared with those cultured under apposed conditions. Under these circumstances, control organoids with the same genetic background seldom developed cysts, demonstrating that cystogenesis is a unique result of PKD mutations[61]. In a recent study, gene-edited kidney organoids were used to screen potential therapeutic agents (247 enzyme inhibitors) for polycystic kidneys, and it was observed that nine compounds inhibited cyst growth without hindering the overall growth of the organoids[60]. The establishment of these disease models has improved the understanding of complex molecular and cellular events underlying the pathogenesis of PKD, thus helping to identify new targets for disease regulation and therapy[62].
A rare X-linked hereditary ailment called Fabry disease results in abnormalities in the glycosphingolipid metabolic pathway owing to the absence or low activity of the lysosomal enzyme α-galactosidase A (α-Gal A)[63]. Cell morphology and function are impaired by the accumulation of globular triacyl ceramide (Gb3) and related neutral sphingolipids in lysosomes due to α-Gal A[63,64]. Gb3 accumulates in renal cells, such as podocytes, glomerular endothelial cells, mesangial cells, tubular epithelial cells, and vascular endothelial cells, resulting in Fabry nephropathy[65]. Fabry nephropathy is characterized by microalbuminuria and proteinuria. As the disease progresses, renal function deteriorates leading to end stage renal disease, which is the leading cause of Fabry nephropathy-related deaths[66]. Animal experi
Most cases of congenital nephrotic syndrome (CNS) are caused by mutations in the NPHS1 (NEPHRIN) and NPHS2 (PODOCIN) genes[68], which encode important podocyte proteins. Podocytes in the glomeruli of kidneys maintain the filtration barrier by creating interdigitating foot processes with intervening slit diaphragms, the disruption of which causes proteinuria. Patient-derived organoids have been employed to understand hereditary illnesses that affect the glomerulus[69]. Specifically, patient cell lines have been used to develop 3D organoid-derived human glomeruli (OrgGloms) to simulate CNS. Expectedly, there was a decrease in NEPHRIN protein in OrgGloms, supporting the hypothesized degradation of the shortened protein encoded by the exon 10 NPHS1 variation. NEPHRIN interacts intracellularly with the adaptor protein PODOCIN, facilitating PODOCIN signaling activity and appropriate localization. Additionally, PODOCIN and NEPHRIN proteins were downregulated in CNS OrgGloms generated from patients with CNS[69]. Overall, these findings demonstrate that the use of human kidney organoids may provide new perspectives on the cellular causes of glomerular disease.
Tuberous sclerosis complex (TSC) is a multisystem tumor-forming disease caused by the absence of TSC1 or TSC2[70,71]. Renal manifestations of TSC include mainly cysts and angiomyolipoma[72]. However, the cellular pathogenesis of TSC remains unknown despite the well-described single-gene etiology[73]. Pietrobon et al[74] utilized a genetically engineered human kidney organoid model that encapsulated the pleiotropic features of TSC kidney disease in in vitro and in situ xenografts, and temporal single-cell RNA sequencing indicated that deletion of TSC1 or TSC2 affects multiple developmental processes in stromal, nephron (epithelial) and neural-glial cells. Deletion of TSC1 or TSC2 results in early upregulation of matrix-associated genes; additionally, epithelial cells in TSC1-/- and TSC2-/- organoids exhibit an epithelial-to-mesenchymal transition that is insensitive to rapamycin. Moreover, melanocytic differentiation from MITF+ SCPs was induced by the loss of TSC1 or TSC2. Conclusively, these results illustrate the pleiotropic developmental consequences of TSC1 or TSC2 double-allele inactivation and provide insights into the pathogenesis of TSC[74].
Nephronophthisis (NPHP) is an autosomal recessive interstitial nephritis that manifests as tubular basement membrane damage, tubular dilatation, and interstitial fibrosis, and its pathogenesis is unclear[75]. Forbes et al[76] performed whole-exome sequencing of NPHP1 and its core family line and identified compound heterozygous mutations in the IFT140 gene. Predecessor mutant iPSCs have been generated using reprogramming and gene editing and were induced to form renal organoids. The organoid tubules from the proband had primary cilia that were truncated and shaped like clubs; however, gene repair reversed this feature. Additionally, epithelial cells collected from organoids and subjected to differential expression analysis revealed the downregulation of genes related to apicobasal polarity, cell-cell junctions, and dynein motor assembly. Polarization deficiency was verified in the proband using Matrigel cyst cultures. Tanigawa et al[53] showed that gene repair could restore the deficit in slit diaphragm development in NPHS1 mutant podocytes using a PSC line from a patient with an NPHS1 missense mutation.
In addition to the aforementioned disease models, several hPSC-derived kidney-like organs have been used as disease models in recent years, including BRD4780, a small molecule that clears MUC1-fs from patient cells, knock-in mouse kidneys, and renal organoids. BRD4780 is promising for the treatment of MKD and other toxic protein diseases[77]. Jansen et al[78] assessed the effects of severe acute respiratory syndrome coronavirus 2 on human kidneys using kidney organoids and found that coronavirus disease 2019-induced kidney damage and fibrosis were related to increased collagen-I expression and accumulation. Additionally, Gupta et al[79] used hPSC-derived kidney organoids to simulate the transition from intrinsic to incomplete repair of acute kidney injury. A single exposure to cisplatin resulted in intrinsic repair, preservation of tubular structure, and upregulation of genes associated with homologous-directed repair. Targeted drug screening was used to identify the DNA ligase IV inhibitor SCR7, which increases FANCD2-mediated repair and delays the progression of chronic injury in organoids. Conclusively, these results suggest that targeting the FANCD2/RAD51 pathway may have therapeutic potential for kidney disease, and highlight the important role of kidney organoids in exploring potential treatments.
However, the currently investigated protocols can only produce immature kidney organoids. Therefore, since only diseases that exhibit abnormalities early in embryonic development can be established by kidney organoids, more mature organoids will need to be developed in the future to be able to model and study late-onset diseases.
Despite several remarkable progress in the development of kidney organoids, there are still several challenges. Current challenges hindering the development of kidney organoids and their future applications include immature tissue development, scanty blood vessel formation, inability to form effective collecting tubes, and the emergence of several non-kidney cells (e.g., myocytes and neurons) during the process of induced differentiation. More detailed analyses of the developing kidney at the individual cell level are required. Ultimately, organotypic renal structures need to be equipped with renal elements, collecting ducts, ureters, interstitia, and vascular flow in order to generate transplantable kidneys. However, recent studies have made important breakthroughs in addressing these issues. Regarding vascularization, planting the renal organoid under the peritoneum of the in situ kidney generates abundant blood vessels and matures.
Generally, research on kidney organoids is continuously progressing and better results are expected in the future. Addressing the above limitations would facilitate the development of functional renal organoids and possible artificial kidneys for patients in need of transplants to treat CKD. However, extensive studies are still necessary to address these issues.
We thank Dr. Yong Chen, Central Laboratory of Guizhou Provincial People’s Hospital, for his guidance on the manuscript.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Li SC, United States; Moreno-Gómez-Toledano R, Spain S-Editor: Wang JJ L-Editor: A P-Editor: Yuan YY
| 1. | Liyanage T, Toyama T, Hockham C, Ninomiya T, Perkovic V, Woodward M, Fukagawa M, Matsushita K, Praditpornsilpa K, Hooi LS, Iseki K, Lin MY, Stirnadel-Farrant HA, Jha V, Jun M. Prevalence of chronic kidney disease in Asia: a systematic review and analysis. BMJ Glob Health. 2022;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 146] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 2. | Foreman KJ, Marquez N, Dolgert A, Fukutaki K, Fullman N, McGaughey M, Pletcher MA, Smith AE, Tang K, Yuan CW, Brown JC, Friedman J, He J, Heuton KR, Holmberg M, Patel DJ, Reidy P, Carter A, Cercy K, Chapin A, Douwes-Schultz D, Frank T, Goettsch F, Liu PY, Nandakumar V, Reitsma MB, Reuter V, Sadat N, Sorensen RJD, Srinivasan V, Updike RL, York H, Lopez AD, Lozano R, Lim SS, Mokdad AH, Vollset SE, Murray CJL. Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet. 2018;392:2052-2090. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1036] [Cited by in RCA: 1569] [Article Influence: 224.1] [Reference Citation Analysis (0)] |
| 3. | Kovesdy CP. Epidemiology of chronic kidney disease: an update 2022. Kidney Int Suppl (2011). 2022;12:7-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 1308] [Article Influence: 436.0] [Reference Citation Analysis (0)] |
| 4. | Evans M, Lewis RD, Morgan AR, Whyte MB, Hanif W, Bain SC, Davies S, Dashora U, Yousef Z, Patel DC, Strain WD. A Narrative Review of Chronic Kidney Disease in Clinical Practice: Current Challenges and Future Perspectives. Adv Ther. 2022;39:33-43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 133] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 5. | Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, Li X, Wang L, Liu Y, Liu J, Zhang M, Qi J, Yu S, Afshin A, Gakidou E, Glenn S, Krish VS, Miller-Petrie MK, Mountjoy-Venning WC, Mullany EC, Redford SB, Liu H, Naghavi M, Hay SI, Murray CJL, Liang X. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394:1145-1158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2594] [Cited by in RCA: 2386] [Article Influence: 397.7] [Reference Citation Analysis (1)] |
| 6. | Fletcher BR, Damery S, Aiyegbusi OL, Anderson N, Calvert M, Cockwell P, Ferguson J, Horton M, Paap MCS, Sidey-Gibbons C, Slade A, Turner N, Kyte D. Symptom burden and health-related quality of life in chronic kidney disease: A global systematic review and meta-analysis. PLoS Med. 2022;19:e1003954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 160] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 7. | Freedman BS, Brooks CR, Lam AQ, Fu H, Morizane R, Agrawal V, Saad AF, Li MK, Hughes MR, Werff RV, Peters DT, Lu J, Baccei A, Siedlecki AM, Valerius MT, Musunuru K, McNagny KM, Steinman TI, Zhou J, Lerou PH, Bonventre JV. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun. 2015;6:8715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 451] [Cited by in RCA: 553] [Article Influence: 55.3] [Reference Citation Analysis (0)] |
| 8. | van den Berg CW, Ritsma L, Avramut MC, Wiersma LE, van den Berg BM, Leuning DG, Lievers E, Koning M, Vanslambrouck JM, Koster AJ, Howden SE, Takasato M, Little MH, Rabelink TJ. Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo. Stem Cell Reports. 2018;10:751-765. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 231] [Cited by in RCA: 311] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 9. | Kim JW, Nam SA, Yi J, Kim JY, Lee JY, Park SY, Sen T, Choi YM, Kim HL, Kim HW, Park J, Cho DW, Kim YK. Kidney Decellularized Extracellular Matrix Enhanced the Vascularization and Maturation of Human Kidney Organoids. Adv Sci (Weinh). 2022;9:e2103526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 80] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 10. | Zeng Z, Huang B, Parvez RK, Li Y, Chen J, Vonk AC, Thornton ME, Patel T, Rutledge EA, Kim AD, Yu J, Grubbs BH, McMahon JA, Pastor-Soler NM, Hallows KR, McMahon AP, Li Z. Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors. Nat Commun. 2021;12:3641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 11. | Low JH, Li P, Chew EGY, Zhou B, Suzuki K, Zhang T, Lian MM, Liu M, Aizawa E, Rodriguez Esteban C, Yong KSM, Chen Q, Campistol JM, Fang M, Khor CC, Foo JN, Izpisua Belmonte JC, Xia Y. Generation of Human PSC-Derived Kidney Organoids with Patterned Nephron Segments and a De Novo Vascular Network. Cell Stem Cell. 2019;25:373-387.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 242] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 12. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11399] [Cited by in RCA: 10427] [Article Influence: 386.2] [Reference Citation Analysis (0)] |
| 13. | Schwartzentruber J, Foskolou S, Kilpinen H, Rodrigues J, Alasoo K, Knights AJ, Patel M, Goncalves A, Ferreira R, Benn CL, Wilbrey A, Bictash M, Impey E, Cao L, Lainez S, Loucif AJ, Whiting PJ; HIPSCI Consortium, Gutteridge A, Gaffney DJ. Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet. 2018;50:54-61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 165] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 14. | Drakhlis L, Biswanath S, Farr CM, Lupanow V, Teske J, Ritzenhoff K, Franke A, Manstein F, Bolesani E, Kempf H, Liebscher S, Schenke-Layland K, Hegermann J, Nolte L, Meyer H, de la Roche J, Thiemann S, Wahl-Schott C, Martin U, Zweigerdt R. Human heart-forming organoids recapitulate early heart and foregut development. Nat Biotechnol. 2021;39:737-746. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 240] [Article Influence: 60.0] [Reference Citation Analysis (0)] |
| 15. | Rostovskaya M, Bredenkamp N, Smith A. Towards consistent generation of pancreatic lineage progenitors from human pluripotent stem cells. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Günther C, Winner B, Neurath MF, Stappenbeck TS. Organoids in gastrointestinal diseases: from experimental models to clinical translation. Gut. 2022;71:1892-1908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 17. | Taguchi A, Nishinakamura R. Nephron reconstitution from pluripotent stem cells. Kidney Int. 2015;87:894-900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Hariharan K, Stachelscheid H, Rossbach B, Oh SJ, Mah N, Schmidt-Ott K, Kurtz A, Reinke P. Parallel generation of easily selectable multiple nephronal cell types from human pluripotent stem cells. Cell Mol Life Sci. 2019;76:179-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 19. | Yu X, Jiang S, Li K, Yang X, Zhang D, Du X, Feng K, Zhao S. Maturation of Nephrons by Implanting hPSC-derived Kidney Progenitors Under Kidney Capsules of Unilaterally Nephrectomized Mice. Curr Stem Cell Res Ther. 2023;18:551-559. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 20. | Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol. 2015;33:1193-1200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 531] [Cited by in RCA: 671] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 21. | Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG, Little MH. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 2014;16:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 517] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 22. | Griffin K, Patient R, Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983-2994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 159] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 529] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 24. | Li Z, Araoka T, Wu J, Liao HK, Li M, Lazo M, Zhou B, Sui Y, Wu MZ, Tamura I, Xia Y, Beyret E, Matsusaka T, Pastan I, Rodriguez Esteban C, Guillen I, Guillen P, Campistol JM, Izpisua Belmonte JC. 3D Culture Supports Long-Term Expansion of Mouse and Human Nephrogenic Progenitors. Cell Stem Cell. 2016;19:516-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 146] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 25. | Kimelman D, Kirschner M. Synergistic induction of mesoderm by FGF and TGF-beta and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell. 1987;51:869-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 639] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 26. | Smith JC, Price BM, Van Nimmen K, Huylebroeck D. Identification of a potent Xenopus mesoderm-inducing factor as a homologue of activin A. Nature. 1990;345:729-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 491] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 27. | Morizane R, Bonventre JV. Generation of nephron progenitor cells and kidney organoids from human pluripotent stem cells. Nat Protoc. 2017;12:195-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 159] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 28. | Kang M, Han YM. Differentiation of human pluripotent stem cells into nephron progenitor cells in a serum and feeder free system. PLoS One. 2014;9:e94888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Kumar SV, Er PX, Lawlor KT, Motazedian A, Scurr M, Ghobrial I, Combes AN, Zappia L, Oshlack A, Stanley EG, Little MH. Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells. Development. 2019;146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 30. | Garreta E, Prado P, Tarantino C, Oria R, Fanlo L, Martí E, Zalvidea D, Trepat X, Roca-Cusachs P, Gavaldà-Navarro A, Cozzuto L, Campistol JM, Izpisúa Belmonte JC, Hurtado Del Pozo C, Montserrat N. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat Mater. 2019;18:397-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 198] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 31. | Yoshimura Y, Muto Y, Ledru N, Wu H, Omachi K, Miner JH, Humphreys BD. A single-cell multiomic analysis of kidney organoid differentiation. Proc Natl Acad Sci U S A. 2023;120:e2219699120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 32. | Tsujimoto H, Kasahara T, Sueta SI, Araoka T, Sakamoto S, Okada C, Mae SI, Nakajima T, Okamoto N, Taura D, Nasu M, Shimizu T, Ryosaka M, Li Z, Sone M, Ikeya M, Watanabe A, Osafune K. A Modular Differentiation System Maps Multiple Human Kidney Lineages from Pluripotent Stem Cells. Cell Rep. 2020;31:107476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 33. | Taguchi A, Nishinakamura R. Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell. 2017;21:730-746.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 305] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 34. | Little MH, McMahon AP. Mammalian kidney development: principles, progress, and projections. Cold Spring Harb Perspect Biol. 2012;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 310] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 35. | Peng G, Suo S, Cui G, Yu F, Wang R, Chen J, Chen S, Liu Z, Chen G, Qian Y, Tam PPL, Han JJ, Jing N. Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature. 2019;572:528-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 36. | Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, Little MH. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 904] [Cited by in RCA: 1074] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 37. | Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, Humphreys BD. Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell. 2018;23:869-881.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 413] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 38. | Nishikawa M, Kimura H, Yanagawa N, Hamon M, Hauser P, Zhao L, Jo OD. An optimal serum-free defined condition for in vitro culture of kidney organoids. Biochem Biophys Res Commun. 2018;501:996-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 39. | Shi M, McCracken KW, Patel AB, Zhang W, Ester L, Valerius MT, Bonventre JV. Human ureteric bud organoids recapitulate branching morphogenesis and differentiate into functional collecting duct cell types. Nat Biotechnol. 2023;41:252-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 48] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 40. | Humphreys BD, DiRocco DP. Lineage-tracing methods and the kidney. Kidney Int. 2014;86:481-488. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 41. | Howden SE, Vanslambrouck JM, Wilson SB, Tan KS, Little MH. Reporter-based fate mapping in human kidney organoids confirms nephron lineage relationships and reveals synchronous nephron formation. EMBO Rep. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 42. | Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214-5228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 372] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 43. | Kobayashi A, Mugford JW, Krautzberger AM, Naiman N, Liao J, McMahon AP. Identification of a multipotent self-renewing stromal progenitor population during mammalian kidney organogenesis. Stem Cell Reports. 2014;3:650-662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 186] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 44. | Hochane M, van den Berg PR, Fan X, Bérenger-Currias N, Adegeest E, Bialecka M, Nieveen M, Menschaart M, Chuva de Sousa Lopes SM, Semrau S. Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development. PLoS Biol. 2019;17:e3000152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 45. | Hume DA, Gordon S. Mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Identification of resident macrophages in renal medullary and cortical interstitium and the juxtaglomerular complex. J Exp Med. 1983;157:1704-1709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 152] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 46. | Soos TJ, Sims TN, Barisoni L, Lin K, Littman DR, Dustin ML, Nelson PJ. CX3CR1+ interstitial dendritic cells form a contiguous network throughout the entire kidney. Kidney Int. 2006;70:591-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 232] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 47. | Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A, Ashenberg O, Su CW, Smillie C, Shekhar K, Chen Z, Wu C, Ordovas-Montanes J, Alvarez D, Herbst RH, Zhang M, Tirosh I, Dionne D, Nguyen LT, Xifaras ME, Shalek AK, von Andrian UH, Graham DB, Rozenblatt-Rosen O, Shi HN, Kuchroo V, Yilmaz OH, Regev A, Xavier RJ. T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell. 2018;175:1307-1320.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 426] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 48. | Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, van Rooij N, van Leerdam ME, Depla A, Smit EF, Hartemink KJ, de Groot R, Wolkers MC, Sachs N, Snaebjornsson P, Monkhorst K, Haanen J, Clevers H, Schumacher TN, Voest EE. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell. 2018;174:1586-1598.e12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 667] [Cited by in RCA: 738] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 49. | Little MH, Quinlan C. Advances in our understanding of genetic kidney disease using kidney organoids. Pediatr Nephrol. 2020;35:915-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 50. | Weber S, Taylor JC, Winyard P, Baker KF, Sullivan-Brown J, Schild R, Knüppel T, Zurowska AM, Caldas-Alfonso A, Litwin M, Emre S, Ghiggeri GM, Bakkaloglu A, Mehls O, Antignac C, Network E, Schaefer F, Burdine RD. SIX2 and BMP4 mutations associate with anomalous kidney development. J Am Soc Nephrol. 2008;19:891-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 160] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 51. | Combes AN, Wilson S, Phipson B, Binnie BB, Ju A, Lawlor KT, Cebrian C, Walton SL, Smyth IM, Moritz KM, Kopan R, Oshlack A, Little MH. Haploinsufficiency for the Six2 gene increases nephron progenitor proliferation promoting branching and nephron number. Kidney Int. 2018;93:589-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 52. | Freedman BS, Lam AQ, Sundsbak JL, Iatrino R, Su X, Koon SJ, Wu M, Daheron L, Harris PC, Zhou J, Bonventre JV. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J Am Soc Nephrol. 2013;24:1571-1586. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 53. | Tanigawa S, Islam M, Sharmin S, Naganuma H, Yoshimura Y, Haque F, Era T, Nakazato H, Nakanishi K, Sakuma T, Yamamoto T, Kurihara H, Taguchi A, Nishinakamura R. Organoids from Nephrotic Disease-Derived iPSCs Identify Impaired NEPHRIN Localization and Slit Diaphragm Formation in Kidney Podocytes. Stem Cell Reports. 2018;11:727-740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 54. | Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10336] [Cited by in RCA: 11145] [Article Influence: 928.8] [Reference Citation Analysis (0)] |
| 55. | Sterneckert JL, Reinhardt P, Schöler HR. Investigating human disease using stem cell models. Nat Rev Genet. 2014;15:625-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 198] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 56. | Zhang Z, Bai H, Blumenfeld J, Ramnauth AB, Barash I, Prince M, Tan AY, Michaeel A, Liu G, Chicos I, Rennert L, Giannakopoulos S, Larbi K, Hughes S, Salvatore SP, Robinson BD, Kapur S, Rennert H. Detection of PKD1 and PKD2 Somatic Variants in Autosomal Dominant Polycystic Kidney Cyst Epithelial Cells by Whole-Genome Sequencing. J Am Soc Nephrol. 2021;32:3114-3129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 57. | Tan AY, Zhang T, Michaeel A, Blumenfeld J, Liu G, Zhang W, Zhang Z, Zhu Y, Rennert L, Martin C, Xiang J, Salvatore SP, Robinson BD, Kapur S, Donahue S, Bobb WO, Rennert H. Somatic Mutations in Renal Cyst Epithelium in Autosomal Dominant Polycystic Kidney Disease. J Am Soc Nephrol. 2018;29:2139-2156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 58. | Qian F, Watnick TJ, Onuchic LF, Germino GG. The molecular basis of focal cyst formation in human autosomal dominant polycystic kidney disease type I. Cell. 1996;87:979-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 418] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 59. | Ding H, Li LX, Harris PC, Yang J, Li X. Extracellular vesicles and exosomes generated from cystic renal epithelial cells promote cyst growth in autosomal dominant polycystic kidney disease. Nat Commun. 2021;12:4548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 55] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 60. | Tran T, Song CJ, Nguyen T, Cheng SY, McMahon JA, Yang R, Guo Q, Der B, Lindström NO, Lin DC, McMahon AP. A scalable organoid model of human autosomal dominant polycystic kidney disease for disease mechanism and drug discovery. Cell Stem Cell. 2022;29:1083-1101.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 73] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 61. | Cruz NM, Song X, Czerniecki SM, Gulieva RE, Churchill AJ, Kim YK, Winston K, Tran LM, Diaz MA, Fu H, Finn LS, Pei Y, Himmelfarb J, Freedman BS. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat Mater. 2017;16:1112-1119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 223] [Article Influence: 27.9] [Reference Citation Analysis (1)] |
| 62. | Li SR, Gulieva RE, Helms L, Cruz NM, Vincent T, Fu H, Himmelfarb J, Freedman BS. Glucose absorption drives cystogenesis in a human organoid-on-chip model of polycystic kidney disease. Nat Commun. 2022;13:7918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 63. | Brady RO, Gal AE, Bradley RM, Martensson E, Warshaw AL, Laster L. Enzymatic defect in Fabry's disease. Ceramidetrihexosidase deficiency. N Engl J Med. 1967;276:1163-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 779] [Cited by in RCA: 750] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 64. | Hopkin RJ, Cabrera G, Charrow J, Lemay R, Martins AM, Mauer M, Ortiz A, Patel MR, Sims K, Waldek S, Warnock DG, Wilcox WR. Risk factors for severe clinical events in male and female patients with Fabry disease treated with agalsidase beta enzyme replacement therapy: Data from the Fabry Registry. Mol Genet Metab. 2016;119:151-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 65. | de Menezes Neves PDM, Machado JR, Custódio FB, Dos Reis Monteiro MLG, Iwamoto S, Freire M, Ferreira MF, Dos Reis MA. Ultrastructural deposits appearing as "zebra bodies" in renal biopsy: Fabry disease?- comparative case reports. BMC Nephrol. 2017;18:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 66. | Branton MH, Schiffmann R, Sabnis SG, Murray GJ, Quirk JM, Altarescu G, Goldfarb L, Brady RO, Balow JE, Austin Iii HA, Kopp JB. Natural history of Fabry renal disease: influence of alpha-galactosidase A activity and genetic mutations on clinical course. Medicine (Baltimore). 2002;81:122-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 319] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 67. | Kim JW, Kim HW, Nam SA, Lee JY, Cho HJ, Kim TM, Kim YK. Human kidney organoids reveal the role of glutathione in Fabry disease. Exp Mol Med. 2021;53:1580-1591. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 68. | Trautmann A, Bodria M, Ozaltin F, Gheisari A, Melk A, Azocar M, Anarat A, Caliskan S, Emma F, Gellermann J, Oh J, Baskin E, Ksiazek J, Remuzzi G, Erdogan O, Akman S, Dusek J, Davitaia T, Özkaya O, Papachristou F, Firszt-Adamczyk A, Urasinski T, Testa S, Krmar RT, Hyla-Klekot L, Pasini A, Özcakar ZB, Sallay P, Cakar N, Galanti M, Terzic J, Aoun B, Caldas Afonso A, Szymanik-Grzelak H, Lipska BS, Schnaidt S, Schaefer F; PodoNet Consortium. Spectrum of steroid-resistant and congenital nephrotic syndrome in children: the PodoNet registry cohort. Clin J Am Soc Nephrol. 2015;10:592-600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 202] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 69. | Hale LJ, Howden SE, Phipson B, Lonsdale A, Er PX, Ghobrial I, Hosawi S, Wilson S, Lawlor KT, Khan S, Oshlack A, Quinlan C, Lennon R, Little MH. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat Commun. 2018;9:5167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 167] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 70. | Northrup H, Krueger DA; International Tuberous Sclerosis Complex Consensus Group. Tuberous sclerosis complex diagnostic criteria update: recommendations of the 2012 Iinternational Tuberous Sclerosis Complex Consensus Conference. Pediatr Neurol. 2013;49:243-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 933] [Cited by in RCA: 1001] [Article Influence: 83.4] [Reference Citation Analysis (0)] |
| 71. | van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, Burley M, Jeremiah S, Woodward K, Nahmias J, Fox M, Ekong R, Osborne J, Wolfe J, Povey S, Snell RG, Cheadle JP, Jones AC, Tachataki M, Ravine D, Sampson JR, Reeve MP, Richardson P, Wilmer F, Munro C, Hawkins TL, Sepp T, Ali JB, Ward S, Green AJ, Yates JR, Kwiatkowska J, Henske EP, Short MP, Haines JH, Jozwiak S, Kwiatkowski DJ. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1226] [Cited by in RCA: 1134] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 72. | Eijkemans MJ, van der Wal W, Reijnders LJ, Roes KC, van Waalwijk van Doorn-Khosrovani SB, Pelletier C, Magestro M, Zonnenberg B. Long-term Follow-up Assessing Renal Angiomyolipoma Treatment Patterns, Morbidity, and Mortality: An Observational Study in Tuberous Sclerosis Complex Patients in the Netherlands. Am J Kidney Dis. 2015;66:638-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 92] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 73. | Kwiatkowski DJ. Animal models of lymphangioleiomyomatosis (LAM) and tuberous sclerosis complex (TSC). Lymphat Res Biol. 2010;8:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 74. | Pietrobon A, Yockell-Lelièvre J, Flood TA, Stanford WL. Renal organoid modeling of tuberous sclerosis complex reveals lesion features arise from diverse developmental processes. Cell Rep. 2022;40:111048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 75. | König J, Kranz B, König S, Schlingmann KP, Titieni A, Tönshoff B, Habbig S, Pape L, Häffner K, Hansen M, Büscher A, Bald M, Billing H, Schild R, Walden U, Hampel T, Staude H, Riedl M, Gretz N, Lablans M, Bergmann C, Hildebrandt F, Omran H, Konrad M; Gesellschaft für Pädiatrische Nephrologie (GPN). Phenotypic Spectrum of Children with Nephronophthisis and Related Ciliopathies. Clin J Am Soc Nephrol. 2017;12:1974-1983. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 76. | Forbes TA, Howden SE, Lawlor K, Phipson B, Maksimovic J, Hale L, Wilson S, Quinlan C, Ho G, Holman K, Bennetts B, Crawford J, Trnka P, Oshlack A, Patel C, Mallett A, Simons C, Little MH. Patient-iPSC-Derived Kidney Organoids Show Functional Validation of a Ciliopathic Renal Phenotype and Reveal Underlying Pathogenetic Mechanisms. Am J Hum Genet. 2018;102:816-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 77. | Dvela-Levitt M, Kost-Alimova M, Emani M, Kohnert E, Thompson R, Sidhom EH, Rivadeneira A, Sahakian N, Roignot J, Papagregoriou G, Montesinos MS, Clark AR, McKinney D, Gutierrez J, Roth M, Ronco L, Elonga E, Carter TA, Gnirke A, Melanson M, Hartland K, Wieder N, Hsu JC, Deltas C, Hughey R, Bleyer AJ, Kmoch S, Živná M, Barešova V, Kota S, Schlondorff J, Heiman M, Alper SL, Wagner F, Weins A, Golub TR, Lander ES, Greka A. Small Molecule Targets TMED9 and Promotes Lysosomal Degradation to Reverse Proteinopathy. Cell. 2019;178:521-535.e23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 78. | Jansen J, Reimer KC, Nagai JS, Varghese FS, Overheul GJ, de Beer M, Roverts R, Daviran D, Fermin LAS, Willemsen B, Beukenboom M, Djudjaj S, von Stillfried S, van Eijk LE, Mastik M, Bulthuis M, Dunnen WD, van Goor H, Hillebrands JL, Triana SH, Alexandrov T, Timm MC, van den Berge BT, van den Broek M, Nlandu Q, Heijnert J, Bindels EMJ, Hoogenboezem RM, Mooren F, Kuppe C, Miesen P, Grünberg K, Ijzermans T, Steenbergen EJ, Czogalla J, Schreuder MF, Sommerdijk N, Akiva A, Boor P, Puelles VG, Floege J, Huber TB; COVID Moonshot consortium, van Rij RP, Costa IG, Schneider RK, Smeets B, Kramann R. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell. 2022;29:217-231.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 164] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 79. | Gupta N, Matsumoto T, Hiratsuka K, Garcia Saiz E, Galichon P, Miyoshi T, Susa K, Tatsumoto N, Yamashita M, Morizane R. Modeling injury and repair in kidney organoids reveals that homologous recombination governs tubular intrinsic repair. Sci Transl Med. 2022;14:eabj4772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 66] [Article Influence: 22.0] [Reference Citation Analysis (0)] |