Published online Dec 26, 2024. doi: 10.4252/wjsc.v16.i12.990
Revised: November 5, 2024
Accepted: December 2, 2024
Published online: December 26, 2024
Processing time: 98 Days and 19.6 Hours
In this editorial, we have taken an in-depth look at the article published by Wan et al. The study showed that preconditioning mesenchymal stem cells (MSCs) pro
Core Tip: Reducing the programmed death of pre-conditioned bone marrow-derived mesenchymal stem cells can significantly enhance their engraftment, survival, and differentiation potential, and modulate the immune microenvironment of the recipient tissue. Autophagy, an important complementary pathway of programmed cell death, plays a crucial role in cellular homeostasis, self-renewal, and functional regulation. Regulating autophagy in mesenchymal stem cells provides a new perspective and strategy for stem cell therapy.
- Citation: Chai M, Zhang CY, Chen S, Xu DH. Application of autophagy in mesenchymal stem cells. World J Stem Cells 2024; 16(12): 990-1001
- URL: https://www.wjgnet.com/1948-0210/full/v16/i12/990.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v16.i12.990
Programmed cell death (PCD), a genetically controlled mode of active cell death, plays a crucial role in the development and homeostatic maintenance of organisms[1]. Autophagy is an important complementary pathway to PCD, and has gained attention in recent years for its role in cellular homeostasis, self-renewal, and functional regulation. The mesenchymal stem cells (MSCs) are known to attenuate neuronal damage and dysfunction by regulating autophagy, thus improving the prognosis of neurodegenerative diseases[2]. Furthermore, MSCs and their exosomes can alleviate hepatic injury by promoting the autophagic flux in liver cells[3]. Autophagy may also mediate the therapeutic effects of MSCs in cardiovascular diseases, diabetes, and autoimmune diseases[4]. Taken together, MSCs can improve the microenvironment of damaged tissues and promote tissue repair and regeneration by regulating autophagy.
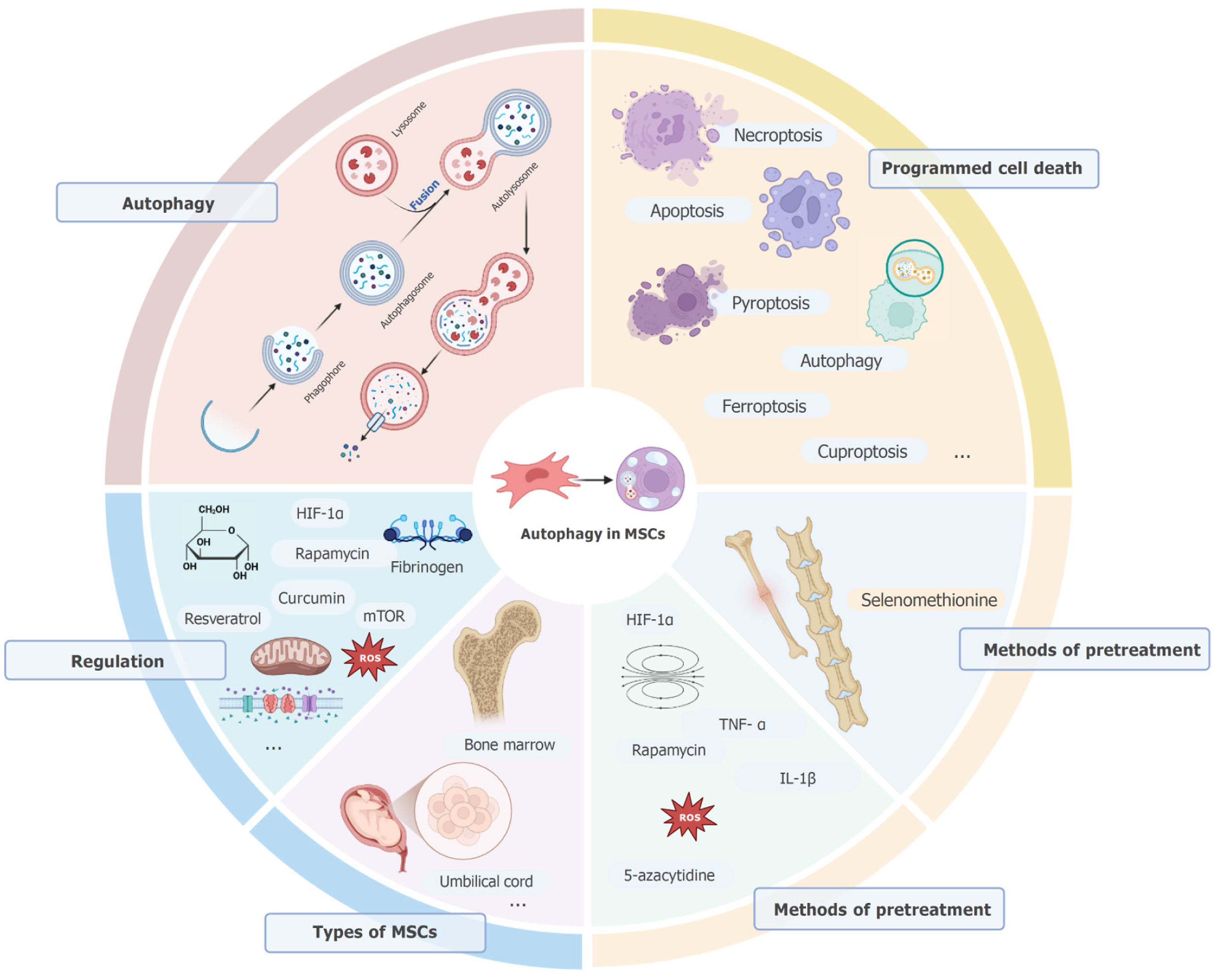
As studies increasingly delve into the functional link between autophagy and MSCs, and discover new mechanisms and targets of autophagy in MSCs, we can expect more effective strategies for the clinical application of MSCs. At the same time, the complexity and diversity of autophagy regulation in the MSCs need to be elucidated to avoid potential adverse effects (Figure 1).
PCD is a genetically controlled, active form of cell death as opposed to necrosis, which is a passive process caused by external physical, chemical or biological factors. Necrosis is often accompanied by the rupture of cell membranes and the release of cellular contents, which can trigger an inflammatory response. In contrast, the cell membrane remains intact during PCD, no inflammatory response is elicited, and the entire death process requires energy expenditure. Various types of PCD have been discovered so far, including apoptosis, necroptosis, pyroptosis, autophagy, ferroptosis and cuproptosis.
Apoptosis, the most prevalent and extensively researched type of PCD, is characterized by chromatin condensation and margination, cellular shrinkage, phosphatidylserine externalization on the inner leaflet of the cell membrane, and the formation of apoptotic bodies through cellular exocytosis[5]. It is triggered by multiple stimuli, and is essential during embryonic development and aging to maintain cellular homeostasis. In addition, apoptosis also plays a pivotal role in defending against autoimmune responses and in the elimination of damaged cells[6]. The apoptotic cascade is driven by the endogenous (mitochondrial) and the exogenous (death receptor-mediated) pathways, which are primarily controlled by members of the Bcl-2 family[7] and caspase family[8] of genes. Dysregulated apoptosis is a key contributor to the onset and progression of cancer. Furthermore, neurodegenerative disorders such as Alzheimer’s disease and Parkinson’s disease, as well as autoimmune disorders, are closely linked to aberrant apoptosis. The study titled “Pretreatment can alleviate programmed cell death in mesenchymal stem cells” which was published by Wan et al[9] has shown that preconditioning MSCs with prostaglandin E1 upregulates hypoxia-inducible factor-1α (HIF-1α)[10]. This finding offers significant insights into the therapeutic advantages of MSCs in combating pulmonary hypertension. Necroptosis is a mode of PCD that is triggered by death receptor signaling via the receptor interacting protein kinase 1-receptor interacting protein kinase 3-mixed lineage kinase domain-like pathway[11]. Unlike apoptosis, necroptosis culminates in the rupture of cell membrane, resulting in the discharge of intracellular factors that can initiate an inflammatory reaction.
Pyroptosis, also known as cellular inflammatory necrosis, is a newly discovered mode of PCD. It is triggered by activation of the inflammasome complex, and manifests as continuous swelling of the cell until the plasma membrane ruptures and releases pro-inflammatory cellular contents. Six primary inflammasomes have been identified so far: Nucleotide-binding domain, leucine-rich repeat, and pyrin domain-containing protein 3, nucleotide-binding domain, leucine-rich repeat, and pyrin domain-containing protein 1, NLR family CARD domain-containing protein 7, ice protease-activating factor, NLR family CARD domain-containing protein 4, and absent in melanoma 2[12]. During pyroptosis, several members of the caspase family are activated, which cleave the pore-forming gasdermin family proteins[13]. The gasdermins then perforate the cell membrane, thereby facilitating the release of inflammatory mediators. Pyroptosis is pivotal in the pathology of various diseases, including cancer[14], inflammatory disorders, and neurodegenerative diseases. Modulating the initiation and progression of pyroptosis could potentially mitigate these conditions.
Ferroptosis represents a unique form of iron-dependent, regulated cell death, which is initiated by the buildup of lipid peroxides in cellular membranes[15]. Iron ions facilitate peroxidation of polyunsaturated fatty acid phospholipids through the Fenton reaction, resulting in cellular membrane damage and subsequent cell death. This process is tightly regulated by intracellular antioxidant systems, particularly glutathione peroxidase 4, which is able to reduce phospholipid peroxides and inhibit iron-mediated death[16]. The induction and inhibition of ferroptosis in MSCs has emerged as a potential therapeutic strategy for anti-tumor treatment.
Cuproptosis is a copper-mediated form of regulatory cell death that is caused by the inhibition of the tricarboxylic acid cycle, proteotoxic stress and mitochondrial dysfunction[17]. It is regulated by various signaling pathways, including those involved in the generation of reactive oxygen species (ROS) and the breakdown of solute carrier family 7 member 11[18]. Copper chelators, such as D-penicillamine and tetrathiomolybdate ammonium[19], can potentially inhibit the growth of cancer cells by modulating cuproptosis.
Autophagy is a self-catabolic process wherein the cytoplasmic contents or organelles are enveloped by a bilayer membrane into an autophagosome[20], which subsequently fuses with lysosomes to form an autolysosome, resulting in the degradation of the encapsulated contents[21]. While autophagy is essential for recycling cellular components and maintaining homeostasis, excessive autophagy can lead to type II cell death[22]. Based on the mechanism of action, autophagy can be categorized into three types - macroautophagy, microautophagy, and chaperone-mediated autophagy. Macroautophagy is the most common type, and involves the encapsulation and degradation of macromolecules and organelles in the autophagosomes[23]. In contrast, microautophagy does not require formation of autophagosomes as the lysosomes or vacuoles directly engulf and degrade substances in the cytoplasm[24]. During chaperone-mediated autophagy, molecular chaperones such as HSC70 selectively recognize and bind to soluble proteins in the cytoplasm, and transport them to lysosomes for degradation[25].
Autophagy is triggered in response to starvation, hypoxia, endoplasmic reticulum stress, and other stressors. These signals lead to the activation of autophagy-related genes (Atg) via the mammalian target of rapamycin (mTOR)[26] pathway. The Atg-mediated formation of autophagosomes is orchestrated by multiple autophagy-related proteins, including but not limited to Beclin-1[27], ATG14, and VPS34. The autophagosome membrane continuously extends, enveloping large molecules and organelles in the cytoplasm, ultimately self-closing to form mature autophagosomes. The fusion of autophagosomes with lysosomes results in autolysosomes, wherein the cellular contents are digested by the lysosomal enzymes. The degradation products, including amino acids and fatty acids, are then released back into the cytoplasm for cellular recycling[21]. Autophagy not only breaks down intracellular macromolecules to provide energy and raw materials for the cell during starvation or nutritional deficiency, but also contributes to cell renewal and repair by degrading aberrant or aging organelles. In addition, autophagy can remove intracellular pathogens like bacteria and viruses, as well as abnormal protein aggregates, thereby safeguarding cells against infection and damage[28].
Autophagy and apoptosis exhibit a synergistic relationship in the event of cellular damage or stress. In fact, autophagy can serve as an upstream regulator of apoptosis by breaking down damaged organelles or proteins, thus supplying the essential signals or raw materials for the apoptotic process. For instance, tumor necrosis factor-α (TNF-α) and other signaling molecules that are upregulated during autophagy may initiate the apoptotic cascade by activating critical proteins involved in the pathways[29]. Furthermore, excessive accumulation of autophagosomes or a saturation in their degradation capacity can also induce a shift in the cell death pathway. However, the metabolic regulation of autophagy is often at odds with apoptosis. For example, under conditions of starvation or oxidative stress, autophagy can inhibit apoptosis by reducing the abundance of pro-apoptotic proteins in the cytoplasm.
MSCs are adult stem cells that primarily originate from the mesoderm, and exhibit self-renewal and multipotent differentiation abilities. The MSCs have been detected in various fetal and adult tissues, including but not limited to bone marrow, placenta, umbilical cord, adipose tissue, mucous membranes, bones, muscles, lungs, liver, pancreas, and even amniotic fluid and umbilical cord blood[30]. Under suitable in vivo or in vitro conditions, MSCs can differentiate into various cell types, such as osteoblasts, chondrocytes, adipocytes, myoblasts, neuronal cells, hepatocytes, endothelial cells, etc. Depending on their source, MSCs are broadly classified as bone marrow MSCs (BM-MSCs), umbilical cord MSCs (UC-MSCs), adipose MSCs, and amniotic membrane MSCs[31].
Both intrinsic and extrinsic factors affect autophagy in the MSCs. Intrinsic factors include cell signaling pathways and mitochondrial function, and the extrinsic factors include nutrients, hypoxia, extracellular matrix, drugs, and chemicals. Multiple signaling pathways are involved in the regulation of autophagy in MSCs. For example, the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (Akt)/mTOR signaling pathway is a major negative regulatory pathway of autophagy. Hair follicle MSC-derived exosomes inhibit the PI3K/AKT/mTOR signaling pathway via miR-214-3p, which in turn maintains mitochondrial dynamic stability and enhances mitochondrial autophagy[32]. Furthermore, the AMP-activated protein kinase (AMPK) signaling pathway can activate autophagy by inhibiting the mechanistic target of rapamycin complex 1 activity[33]. ROS produced by mitochondria act as signaling molecules to activate autophagy in order to scavenge damaged mitochondria and maintain intracellular redox homeostasis[34]. In addition, changes in the membrane potential of mitochondria can affect autophagy[35].
MSCs acquire energy and nutrients during starvation by activating autophagy. For example, glucose deficiency induces the onset of autophagy in MSCs, whereas glucose supplementation inhibits autophagy[36]. Furthermore, MSCs can adapt to hypoxic conditions by activating autophagy, and maintain survival and function. HIF-1α plays an important role in hypoxia-induced autophagy in MSCs[37]. Components of the extracellular matrix, such as fibrinogen, can influence autophagy in MSCs by activating cell surface receptors, which in turn regulate intracellular signaling pathways[38]. Several drugs and chemicals are known to modulate autophagy in MSCs. For example, rapamycin is an mTOR inhibitor that inhibits autophagy by activating mTOR activity[39]. In addition, natural compounds such as resveratrol[40] and curcumin[41] have been shown to regulate autophagy in MSCs.
The UC-MSCs are most promising for clinical application on account of their high quality, purity, and quantity, as well as the convenience of collection and lack of ethical concerns[42]. Nevertheless, as MSCs were first discovered in the bone marrow, BM-MSCs are the more widely used in cell therapy and tissue engineering[43]. Recent studies suggest that autophagy may play a key role in the biology of BM-MSCs and UC-MSCs.
BM-MSCs: BM-MSCs mainly exist in the bone marrow cavity, and in connective tissues such as trabecular bone, skeletal muscle, and periosteum. Autophagy plays a role in the proliferation, differentiation, and therapeutic potential of BM-MSCs. Pierrefite-Carle et al[44] hypothesized that autophagy provides BM-MSCs with energy substrates during their differentiation. The authors observed significant accumulation of autophagic vacuoles in the undifferentiated BM-MSCs, along with stagnant autophagic flow. In addition, the autophagosome marker LC 3-II was downregulated in the BM-MSCs during the early stages of osteogenic differentiation (within 12 hours of culture), suggesting that these autophagic vacuoles may serve as a source of energy substrates for the differentiation process. The significance of autophagy in BM-MSCs is further underscored by its role in the progression of bone degenerative diseases, such as disc degeneration and osteoarthritis, as well as in bone metabolic disorders like osteoporosis, metaplastic osteitis, and osteosclerosis. Autophagy modulators, such as mechanically targeted rapamycin kinase inhibitors, AMPK activators, and phytochemicals, have been shown to promote bone regeneration[45]. Wu et al[46] showed that hypoxic conditions induced autophagy in the BM-MSCs in vitro through the activation of the extracellular signal-regulated kinase-1/2 pathway. Furthermore, Zheng et al[47] found that senescent BM-MSCs have enhanced autophagic activity as well as increased levels of inflammatory factors such as interleukin (IL)-6 and IL-8, which upregulate intracellular FoxO3a protein levels. Modulating autophagy and FoxO3a expression could delay BM-MSC senescence and improve its therapeutic efficacy. Li and Qu[48] showed that the PI3K/Akt/nuclear factor-kappaB, mitogen-activated protein kinase/extracellular signal-regulated kinase, and stem cell factor/c-kit pathways are involved in the interaction between autophagy and apoptosis, and that modulation of autophagic activity could potentially improve the therapeutic efficacy of BM-MSCs against myocardial infarction.
UC-MSCs: UC-MSCs are versatile stem cells that exist in the newborn’s umbilical cord, predominantly within Wharton’s Jelly[49], and can differentiate into bone, cartilage, fat, muscle, tendon, nerve, liver, cardiac muscle, and other cell types in vitro in response to suitable induction factors. UC-MSCs can express surface markers such as CD105, CD73, and CD90[50].
UC-MSCs can reduce immune rejection to allografts by suppressing the activation of immune cells. Additionally, they can facilitate hematopoietic recovery[51] and enhance the engraftment of hematopoietic stem cells. The biological function and therapeutic efficacy of the UC-MSCs can be enhanced by regulating autophagic activity. Moreover, UC-MSCs themselves can be used as autophagy modulators. For example, in inflammatory diseases, the transplantation of UC-MSCs can inhibit the production and release of inflammatory factors, thereby reducing inflammatory responses and tissue damage.
He et al[52] showed that autophagy is crucial for the function of UC-MSCs-derived exosomes (UC-MDEs), and detected high expression of the autophagy markers BECN1 and MAP 1 LC3B in UC-MDEs by transmission electron microscopy. Furthermore, the autophagy inhibitor 3-methyladenine significantly reduced the ameliorative effect of UC-MDEs on glycolipid metabolism in type 2 diabetic rats. In a study conducted by Ma et al[53] exosomes from TNF-α-preconditioned UC-MSCs inhibited autophagy in the acinar cells of severe acute pancreatitis by shuttling 3,4-dihydroxyphenylglycol and suppressing the mTOR pathway, and alleviated the symptoms of severe acute pancreatitis. Furthermore, Han et al[54] showed that UC-MSCs can promote diabetic wound healing by inducing autophagy, which may have potential clinical applications. Wang et al[55] showed that 100 nM to 10 μM rapamycin induced autophagy in UC-MSCs, and the optimal dose was 100 nM. Enhancing autophagy in the UC-MSCs improved the pro-angiogenic activity of the conditioned medium, which in turn promoted wound healing and tissue repair. Ma et al[56] showed that the combination of UC-MDEs and autophagy activators significantly enhanced the function of human corneal epithelial cells and attenuated corneal defects, apoptosis, and inflammation through the activation of the AMPK/mTOR/ULK1 pathway, thereby providing a new therapeutic strategy for corneal wound healing and ocular surface regeneration. Yin et al[57] were able to restore ovarian function in mice and increase the circulation of CD8+CD28- T cells using UC-MSCs. Mechanistically, the heme oxygenase-1 expressed in these UC-MSCs induced autophagy in the ovarian cells by activating the JNK/Bcl-2 signaling pathway. Overall, these studies show that modulation of autophagic activity in MSCs promotes self-renewal, enhances multipotent differentiation, delays aging, and improves immunomodulation.
Physical preconditioning factors include hypoxia, mechanical stimulation, and electromagnetic radiation. Hypoxic preconditioning can mimic the in vivo microenvironment and upregulate HIF-1α, which in turn promotes angiogenesis and metabolism in the MSCs by transcriptionally activating the downstream genes[58]. Mechanical stimulation, such as low intensity focused pulsed ultrasound, can effectively stimulate human UC-MSCs in vitro, reduce thyroid cell apoptosis, improve thyroid function, and reduce excessive accumulation of autoimmune antibodies in vivo[59]. Electromagnetic radiation generally includes low-intensity laser exposure and magnetic field stimulation. Magnetic field stimulation can affect cell membrane potential and ion channel activity in MSCs, which in turn regulates metabolism and cellular function[60].
Chemical pre-treatment includes drugs, active oxygen, etc. Some drugs can modulate intracellular signaling pathways and gene expression in the MSCs. For example, rapamycin pretreatment inhibits mTOR activity and upregulates autophagy in MSCs, thereby improving their survival under stressful conditions[39]. In addition, 5-azacytidine promotes the differentiation of MSCs to specific cell types[61]. Appropriate amounts of ROS activate intracellular stress responses and antioxidant defense mechanisms. ROS pretreatment can enhance the survival of MSCs and increase their resistance to apoptosis by modulating the antioxidant enzyme system and intracellular signaling pathways[62].
Cytokines play an important role in the proliferation, differentiation, and immunomodulation of MSCs. Inflammatory cytokines, such as TNF-α[63] and IL-1β[64], can activate the immunoregulatory and anti-inflammatory functions of MSCs. Simultaneous or sequential use of two or more pretreatment methods can synergistically enhance the therapeutic effects of MSCs. For example, drug preconditioning combined with mechanical stimulation protected MSCs against apoptosis and enhanced their function[65].
Pretreatment of MSCs maintains cellular homeostasis and survival, and regulates immune function through autophagy. Therefore, pretreated MSCs exhibit enhanced viability and differentiation abilities, which translate to improved therapeutic efficacy in tissue repair and regeneration. For example, Yang et al[66] established TNF α-licensed exosome-immobilized titanium surfaces to correct macrophage immune status and accelerate osseointegration in type 2 diabetic conditions by activating autophagy. Furthermore, pretreatment can also enhance the immunomodulatory and anti-inflammatory functions of MSCs. Pretreatment of MSCs by modulating the level of autophagy offers a novel treatment strategy for various diseases. For example, selenomethionine promoted the production of MSC-derived extracellular vesicles and increased the delivery of miR-125a-5p in MSC-derived extracellular vesicles, which enhanced the protective effects of MSC-derived extracellular vesicles on attenuating nucleus pulposus cellular senescence and mitigating disc degeneration[67].
Given their unique biological properties and wide range of applications, MSCs occupy a pivotal position in the field of medical research. Our study shows that autophagy plays a crucial role in MSCs, and interfering with the autophagic process, either through targeted drugs or other intervention strategies, can optimize their therapeutic effects. In the recent study “Pretreatment can alleviate programmed cell death in mesenchymal stem cells” published by Wan et al[9] in World Journal of Stem Cells, the researchers showed that reducing programmed death of MSCs by preconditioning can significantly enhance their engraftment, survival and differentiation potential, and modulate the immune microenvironment.
| 1. | Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ. 2015;22:367-376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 447] [Cited by in RCA: 575] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 2. | Pei F, Ma L, Jing J, Feng J, Yuan Y, Guo T, Han X, Ho TV, Lei J, He J, Zhang M, Chen JF, Chai Y. Sensory nerve niche regulates mesenchymal stem cell homeostasis via FGF/mTOR/autophagy axis. Nat Commun. 2023;14:344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 30] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 3. | Lin D, Chen H, Xiong J, Zhang J, Hu Z, Gao J, Gao B, Zhang S, Chen J, Cao H, Li Z, Lin B, Gao Z. Mesenchymal stem cells exosomal let-7a-5p improve autophagic flux and alleviate liver injury in acute-on-chronic liver failure by promoting nuclear expression of TFEB. Cell Death Dis. 2022;13:865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 4. | Vitale E, Perveen S, Rossin D, Lo Iacono M, Rastaldo R, Giachino C. Role of Chaperone-Mediated Autophagy in Ageing Biology and Rejuvenation of Stem Cells. Front Cell Dev Biol. 2022;10:912470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 5. | Kulkarni M, Hardwick JM. Programmed Cell Death in Unicellular Versus Multicellular Organisms. Annu Rev Genet. 2023;57:435-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 6. | Kashyap D, Garg VK, Goel N. Intrinsic and extrinsic pathways of apoptosis: Role in cancer development and prognosis. Adv Protein Chem Struct Biol. 2021;125:73-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 196] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 7. | Suraweera CD, Banjara S, Hinds MG, Kvansakul M. Metazoans and Intrinsic Apoptosis: An Evolutionary Analysis of the Bcl-2 Family. Int J Mol Sci. 2022;23:3691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 8. | Kesavardhana S, Malireddi RKS, Kanneganti TD. Caspases in Cell Death, Inflammation, and Pyroptosis. Annu Rev Immunol. 2020;38:567-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 621] [Article Influence: 124.2] [Reference Citation Analysis (0)] |
| 9. | Wan XX, Hu XM, Zhang Q, Xiong K. Pretreatment can alleviate programmed cell death in mesenchymal stem cells. World J Stem Cells. 2024;16:773-779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 10. | Jiang DT, Tuo L, Bai X, Bing WD, Qu QX, Zhao X, Song GM, Bi YW, Sun WY. Prostaglandin E1 reduces apoptosis and improves the homing of mesenchymal stem cells in pulmonary arterial hypertension by regulating hypoxia-inducible factor 1 alpha. Stem Cell Res Ther. 2022;13:316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 11. | Weinlich R, Oberst A, Beere HM, Green DR. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol. 2017;18:127-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 473] [Cited by in RCA: 739] [Article Influence: 82.1] [Reference Citation Analysis (0)] |
| 12. | Fang X, Wang Y, Zhang Y, Li Y, Kwak-Kim J, Wu L. NLRP3 Inflammasome and Its Critical Role in Gynecological Disorders and Obstetrical Complications. Front Immunol. 2020;11:555826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 13. | Du T, Gao J, Li P, Wang Y, Qi Q, Liu X, Li J, Wang C, Du L. Pyroptosis, metabolism, and tumor immune microenvironment. Clin Transl Med. 2021;11:e492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 199] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 14. | Yang F, Bettadapura SN, Smeltzer MS, Zhu H, Wang S. Pyroptosis and pyroptosis-inducing cancer drugs. Acta Pharmacol Sin. 2022;43:2462-2473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 86] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 15. | Jiang X, Stockwell BR, Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat Rev Mol Cell Biol. 2021;22:266-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 4251] [Article Influence: 1062.8] [Reference Citation Analysis (0)] |
| 16. | Liang D, Feng Y, Zandkarimi F, Wang H, Zhang Z, Kim J, Cai Y, Gu W, Stockwell BR, Jiang X. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell. 2023;186:2748-2764.e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 342] [Article Influence: 171.0] [Reference Citation Analysis (0)] |
| 17. | Chen L, Min J, Wang F. Copper homeostasis and cuproptosis in health and disease. Signal Transduct Target Ther. 2022;7:378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 675] [Article Influence: 225.0] [Reference Citation Analysis (0)] |
| 18. | Xue Q, Kang R, Klionsky DJ, Tang D, Liu J, Chen X. Copper metabolism in cell death and autophagy. Autophagy. 2023;19:2175-2195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 298] [Article Influence: 149.0] [Reference Citation Analysis (0)] |
| 19. | Xie J, Yang Y, Gao Y, He J. Cuproptosis: mechanisms and links with cancers. Mol Cancer. 2023;22:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 386] [Reference Citation Analysis (0)] |
| 20. | Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr, Doran KS, D'Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la Cruz V, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CMP, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4248] [Cited by in RCA: 4240] [Article Influence: 471.1] [Reference Citation Analysis (0)] |
| 21. | He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2982] [Cited by in RCA: 2880] [Article Influence: 180.0] [Reference Citation Analysis (0)] |
| 22. | Su M, Mei Y, Sinha S. Role of the Crosstalk between Autophagy and Apoptosis in Cancer. J Oncol. 2013;2013:102735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 159] [Cited by in RCA: 230] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 23. | Esclatine A, Chaumorcel M, Codogno P. Macroautophagy signaling and regulation. Curr Top Microbiol Immunol. 2009;335:33-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 24. | Wang L, Klionsky DJ, Shen HM. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol. 2023;24:186-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 250] [Article Influence: 125.0] [Reference Citation Analysis (0)] |
| 25. | Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 547] [Cited by in RCA: 944] [Article Influence: 157.3] [Reference Citation Analysis (0)] |
| 26. | Wang Y, Zhang H. Regulation of Autophagy by mTOR Signaling Pathway. Adv Exp Med Biol. 2019;1206:67-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 244] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 27. | Kaur S, Changotra H. The beclin 1 interactome: Modification and roles in the pathology of autophagy-related disorders. Biochimie. 2020;175:34-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 66] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 28. | Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 2063] [Article Influence: 343.8] [Reference Citation Analysis (0)] |
| 29. | Dong Y, Chen H, Gao J, Liu Y, Li J, Wang J. Molecular machinery and interplay of apoptosis and autophagy in coronary heart disease. J Mol Cell Cardiol. 2019;136:27-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 298] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 30. | Li Z, Hu X, Zhong JF. Mesenchymal Stem Cells: Characteristics, Function, and Application. Stem Cells Int. 2019;2019:8106818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 31. | Gong P, Zhang W, He Y, Wang J, Li S, Chen S, Ye Q, Li M. Classification and Characteristics of Mesenchymal Stem Cells and Its Potential Therapeutic Mechanisms and Applications against Ischemic Stroke. Stem Cells Int. 2021;2021:2602871. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Li N, Zhao L, Geng X, Liu J, Zhang X, Hu Y, Qi J, Chen H, Qiu J, Zhang X, Jin S. Stimulation by exosomes from hypoxia-preconditioned hair follicle mesenchymal stem cells facilitates mitophagy by inhibiting the PI3K/AKT/mTOR signaling pathway to alleviate ulcerative colitis. Theranostics. 2024;14:4278-4296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 23] [Reference Citation Analysis (0)] |
| 33. | Raza S, Siddiqui JA, Srivastava A, Chattopadhyay N, Sinha RA, Chakravarti B. Autophagy as a Therapeutic Target in Breast Tumors: The Cancer stem cell perspective. Autophagy Rep. 2024;3. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 34. | Li S, Ren W, Zheng J, Li S, Zhi K, Gao L. Role of O-linked N-acetylglucosamine protein modification in oxidative stress-induced autophagy: a novel target for bone remodeling. Cell Commun Signal. 2024;22:358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 35. | Chen Y, Yang F, Shi Y, Sheng J, Wang Y, Zhang L, Zhou J, Jin Y, Yan Y. RNF31 alleviates liver steatosis by promoting p53/BNIP3-related mitophagy in hepatocytes. Free Radic Biol Med. 2024;219:163-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 36. | Wang S, Li X, Wang T, Sun Z, Feng E, Jin Y. Overexpression of USP35 enhances the protective effect of hUC-MSCs and their extracellular vesicles in oxygen-glucose deprivation/reperfusion-induced SH-SY5Y cells via stabilizing FUNDC1. Commun Biol. 2024;7:1330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 37. | Shi Y, Wang S, Zhang W, Zhu Y, Fan Z, Huang Y, Li F, Yang R. Bone marrow mesenchymal stem cells facilitate diabetic wound healing through the restoration of epidermal cell autophagy via the HIF-1α/TGF-β1/SMAD pathway. Stem Cell Res Ther. 2022;13:314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 33] [Reference Citation Analysis (0)] |
| 38. | Mori T, Igarashi M, Onodera Y, Takehara T, Itokazu M, Teramura T. Fibrinogen supports self-renewal of mesenchymal stem cells under serum-reduced condition through autophagy activation. Biochem Biophys Res Commun. 2023;651:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 39. | Jia ZG, Li L, Zhao P, Fei G, Li SR, Song QQ, Liu GP, Liu JS. MicroRNA-451 from Human Umbilical Cord-Derived Mesenchymal Stem Cell Exosomes Inhibits Alveolar Macrophage Autophagy via Tuberous Sclerosis Complex 1/Mammalian Target of Rapamycin Pathway to Attenuate Burn-Induced Acute Lung Injury in Rats. Biomed Environ Sci. 2024;37:1030-1043. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 40. | Tong K, Wang P, Li Y, Tong Y, Li X, Yan S, Hu P. Resveratrol Inhibits Hepatocellular Carcinoma Progression through Regulating Exosome Secretion. Curr Med Chem. 2024;31:2107-2118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 41. | Deng J, Ouyang P, Li W, Zhong L, Gu C, Shen L, Cao S, Yin L, Ren Z, Zuo Z, Deng J, Yan Q, Yu S. Curcumin Alleviates the Senescence of Canine Bone Marrow Mesenchymal Stem Cells during In Vitro Expansion by Activating the Autophagy Pathway. Int J Mol Sci. 2021;22:11356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 42. | Ding DC, Chang YH, Shyu WC, Lin SZ. Human umbilical cord mesenchymal stem cells: a new era for stem cell therapy. Cell Transplant. 2015;24:339-347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 396] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 43. | Chu DT, Phuong TNT, Tien NLB, Tran DK, Thanh VV, Quang TL, Truong DT, Pham VH, Ngoc VTN, Chu-Dinh T, Kushekhar K. An Update on the Progress of Isolation, Culture, Storage, and Clinical Application of Human Bone Marrow Mesenchymal Stem/Stromal Cells. Int J Mol Sci. 2020;21:708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 44. | Pierrefite-Carle V, Santucci-Darmanin S, Breuil V, Camuzard O, Carle GF. Autophagy in bone: Self-eating to stay in balance. Ageing Res Rev. 2015;24:206-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 45. | Wang J, Zhang Y, Cao J, Wang Y, Anwar N, Zhang Z, Zhang D, Ma Y, Xiao Y, Xiao L, Wang X. The role of autophagy in bone metabolism and clinical significance. Autophagy. 2023;19:2409-2427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 143] [Reference Citation Analysis (0)] |
| 46. | Wu J, Niu J, Li X, Li Y, Wang X, Lin J, Zhang F. Hypoxia induces autophagy of bone marrow-derived mesenchymal stem cells via activation of ERK1/2. Cell Physiol Biochem. 2014;33:1467-1474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Zheng Y, Wu S, Ke H, Peng S, Hu C. Secretion of IL-6 and IL-8 in the senescence of bone marrow mesenchymal stem cells is regulated by autophagy via FoxO3a. Exp Gerontol. 2023;172:112062. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 48. | Li JR, Qu TT. Retraction: Into the eyes of bone marrow-derived mesenchymal stem cells therapy for myocardial infarction and other diseases. Stem Cell Investig. 2018;5:14. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 49. | Batsali AK, Kastrinaki MC, Papadaki HA, Pontikoglou C. Mesenchymal stem cells derived from Wharton's Jelly of the umbilical cord: biological properties and emerging clinical applications. Curr Stem Cell Res Ther. 2013;8:144-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 171] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 50. | Kacham S, Bhure TS, Eswaramoorthy SD, Naik G, Rath SN, Parcha SR, Basu S, Sangwan VS, Shukla S. Human Umbilical Cord-Derived Mesenchymal Stem Cells Promote Corneal Epithelial Repair In Vitro. Cells. 2021;10:1254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 51. | Tian G, Liu C, Wang H, Yu Z, Huang J, Gong Q, Zhang D, Cong H. Human umbilical cord mesenchymal stem cells prevent glucocorticoid-induced osteonecrosis of the femoral head by promoting angiogenesis. J Plast Surg Hand Surg. 2023;57:71-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 52. | He Q, Wang L, Zhao R, Yan F, Sha S, Cui C, Song J, Hu H, Guo X, Yang M, Cui Y, Sun Y, Sun Z, Liu F, Dong M, Hou X, Chen L. Retraction Note: Mesenchymal stem cell-derived exosomes exert ameliorative effects in type 2 diabetes by improving hepatic glucose and lipid metabolism via enhancing autophagy. Stem Cell Res Ther. 2022;13:505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 53. | Ma Z, Xie W, Luo T, Hu Z, Hua J, Zhou J, Yang T, Wang W, Song Z, Yu X, Xu J, Shi S. Exosomes from TNF-α preconditioned human umbilical cord mesenchymal stromal cells inhibit the autophagy of acinar cells of severe acute pancreatitis via shuttling bioactive metabolites. Cell Mol Life Sci. 2023;80:257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Reference Citation Analysis (0)] |
| 54. | Han Y, Sun T, Tao R, Han Y, Liu J. Clinical application prospect of umbilical cord-derived mesenchymal stem cells on clearance of advanced glycation end products through autophagy on diabetic wound. Eur J Med Res. 2017;22:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 55. | Wang W, Li X, Cui C, Yin G, Ren W, Wang X. Autophagy of umbilical cord mesenchymal stem cells induced by rapamycin conduces to pro-angiogenic function of the conditioned medium. Biochem Biophys Rep. 2023;36:101583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 56. | Ma S, Yin J, Hao L, Liu X, Shi Q, Diao Y, Yu G, Liu L, Chen J, Zhong J. Exosomes From Human Umbilical Cord Mesenchymal Stem Cells Treat Corneal Injury via Autophagy Activation. Front Bioeng Biotechnol. 2022;10:879192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 57. | Yin N, Wu C, Qiu J, Zhang Y, Bo L, Xu Y, Shi M, Zhu S, Yang G, Mao C. Protective properties of heme oxygenase-1 expressed in umbilical cord mesenchymal stem cells help restore the ovarian function of premature ovarian failure mice through activating the JNK/Bcl-2 signal pathway-regulated autophagy and upregulating the circulating of CD8(+)CD28(-) T cells. Stem Cell Res Ther. 2020;11:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 58. | Liu J, Wang Z, Lin A, Zhang N. Exosomes from Hypoxic Pretreatment ADSCs Ameliorate Cardiac Damage Post-MI via Activated circ-Stt3b/miR-15a-5p/GPX4 Signaling and Decreased Ferroptosis. Cardiovasc Toxicol. 2024;24:1215-1225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 59. | Ren Z, Fang R, Deng W, Long J, Liu D. Effect of Umbilical Cord Mesenchymal Stem Cell Transplantation Under LIFPUS Pretreatment on Thyroid Function in EAT Rats. Curr Stem Cell Res Ther. 2023;18:260-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 60. | Xie W, Xu R, Fan C, Yang C, Chen H, Cao Y. 900 MHz Radiofrequency Field Induces Mitochondrial Unfolded Protein Response in Mouse Bone Marrow Stem Cells. Front Public Health. 2021;9:724239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 61. | Akbar N, Anum H, Razzaq SS, Salim A, Usman S, Haneef K. Ascorbic acid and salvianolic acid B enhance the valproic acid and 5-azacytidinemediated cardiac differentiation of mesenchymal stem cells. Mol Biol Rep. 2023;50:7371-7380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 62. | Suwanmanee G, Tantrawatpan C, Kheolamai P, Paraoan L, Manochantr S. Fucoxanthin diminishes oxidative stress damage in human placenta-derived mesenchymal stem cells through the PI3K/Akt/Nrf-2 pathway. Sci Rep. 2023;13:22974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 63. | Wu J, Wu J, Xiang W, Gong Y, Feng D, Fang S, Wu Y, Liu Z, Li Y, Chen R, Zhang X, Li B, Chen L, Jin R, Li S, Zhang B, Zhang T, Yin L, Zhou Y, Huang S, Liu N, Xu H, Lian J, Wang Y, Zhou S, Ni Z. Engineering exosomes derived from TNF-α preconditioned IPFP-MSCs enhance both yield and therapeutic efficacy for osteoarthritis. J Nanobiotechnology. 2024;22:555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 64. | Liang W, Li Y, Ji Y, Kang R, Zhang K, Su X, Li J, Ji M, Wu T, Cao X, Chen J, Huo J. Exosomes derived from bone marrow mesenchymal stem cells induce the proliferation and osteogenic differentiation and regulate the inflammatory state in osteomyelitis in vitro model. Naunyn Schmiedebergs Arch Pharmacol. 2024. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 65. | Fu Q, Gao Q, Jiao S, Da F, Guo J, Liu Y, Liu J. Adipose-derived stem cells ameliorate radiation-induced lung injury by activating the DDAH1/ADMA/eNOS signaling pathway. Regen Ther. 2024;27:398-407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 66. | Yang Y, Wang J, Lin X, Zhang Z, Zhang M, Tang C, Kou X, Deng F. TNF-α-licensed exosome-integrated titaniumaccelerated T2D osseointegration by promoting autophagy-regulated M2 macrophage polarization. Biochem Biophys Res Commun. 2024;727:150316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 67. | Ma S, Xue R, Zhu H, Han Y, Ji X, Zhang C, Wei N, Xu J, Li F. Selenomethionine preconditioned mesenchymal stem cells derived extracellular vesicles exert enhanced therapeutic efficacy in intervertebral disc degeneration. Int Immunopharmacol. 2024;132:112028. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |