Published online Jul 26, 2023. doi: 10.4252/wjsc.v15.i7.734
Peer-review started: March 21, 2023
First decision: May 22, 2023
Revised: June 1, 2023
Accepted: June 21, 2023
Article in press: June 21, 2023
Published online: July 26, 2023
Processing time: 125 Days and 15.7 Hours
Haploid embryonic stem cells (haESCs) have been established in many species. Differentiated haploid cell line types in mammals are lacking due to spontaneous diploidization during differentiation that compromises lineage-specific screens.
To derive human haploid neural stem cells (haNSCs) to carry out lineage-specific screens.
Human haNSCs were differentiated from human extended haESCs with the help of Y27632 (ROCK signaling pathway inhibitor) and a series of cytokines to reduce diploidization. Neuronal differentiation of haNSCs was performed to examine their neural differentiation potency. Global gene expression analysis was con-ducted to compare haNSCs with diploid NSCs and haESCs. Fluorescence activated cell sorting was performed to assess the diploidization rate of extended haESCs and haNSCs. Genetic manipulation and screening were utilized to evaluate the significance of human haNSCs as genetic screening tools.
Human haESCs in extended pluripotent culture medium showed more compact and smaller colonies, a higher efficiency in neural differentiation, a higher cell survival ratio and higher stability in haploidy maintenance. These characteristics effectively facilitated the derivation of human haNSCs. These human haNSCs can be generated by differentiation and maintain haploidy and multipotency to neurons and glia in the long term in vitro. After PiggyBac transfection, there were multiple insertion sites in the human haNSCs’ genome, and the insertion sites were evenly spread across all chromosomes. In addition, after the cells were treated with manganese, we were able to generate a list of manganese-induced toxicity genes, demonstrating their utility as genetic screening tools.
This is the first report of a generated human haploid somatic cell line with a complete genome, proliferative ability and neural differentiation potential that provides cell resources for recessive inheritance and drug targeted screening.
Core Tip: Human embryonic stem cells are widely used in preclinical and genetic screening studies. Haploid cells are ideal tools to perform genetic screening. To date, no human haploid somatic cell lines have been successfully created. We converted human haploid embryonic stem cells to an extended pluripotency state by optimizing the culture medium. The derived haploid neural stem cells can proliferate as a haploid genome and maintain multipotency to generate functional neurons and glia. The haploid neural stem cells can also be easily used for gene editing to generate numerous homozygous mutations for lineage-specific screens.
- Citation: Wang HS, Ma XR, Niu WB, Shi H, Liu YD, Ma NZ, Zhang N, Jiang ZW, Sun YP. Generation of a human haploid neural stem cell line for genome-wide genetic screening. World J Stem Cells 2023; 15(7): 734-750
- URL: https://www.wjgnet.com/1948-0210/full/v15/i7/734.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i7.734
Haploid cells have one set of chromosomes that can be genetically analyzed. Compared to diploid cells, haploid cells are more prone to homozygous mutation. This characteristic is important for gene screening[1-5], especially for recessive gene screening, and the study of cellular and chromosomal fusion[6-8]. Although haploid genetics has been mostly restricted to unicellular organisms, recent reports of haploid embryonic stem cells (ESCs) have extended the research into animal species, including mice[9-13], rats[14] and monkeys[15]. More excitingly, haploid ESCs (haESCs) have been successfully derived from human[16-18] blastocysts. The success in producing human haESCs raises a particularly challenging question of whether these cells can be further differentiated while maintaining haploidy. Medaka (fish) haESCs can maintain haploidy without the need for cell sorting[19], whereas mammalian haESCs tend to diploidize in cell culture, which limits their potential for use in genetic screening. Almost all differentiated cells derived from mam-malian haESCs are diploid cells.
Neural stem cells (NSCs) are somatic stem cells that can differentiate into both neuronal and glial progeny. Although we can obtain haploid neural precursors and neurons by differentiating mouse and monkey haESCs[20,21], the pathogenesis of many diseases differs between humans and animals. For instance, the α7-nicotinic acetylcholine receptor is widely studied as a target for treating the cognitive symptoms of Alzheimer’s disease, attention deficit–hyperactivity disorder and schizophrenia[22]. However, the distribution of this receptor differs across species[23]. In general, there are many differences in gene expression among species[24].
This issue raises the question of how long human haESCs can maintain pluripotency in the stable haploid state. It also remains undetermined whether neural differentiation can be stably induced in haploid human cells, which would be useful to help clarify the specific pathogenesis of human neurological diseases. Previous studies have shown that haploid cells have poor differentiation potency. Yang et al[25] and Liu et al[26] indicated that by introducing a variety of small molecule compounds into the stem cell culture medium, traditional stem cells can be transformed into extended pluripotent stem (EPS) cells with higher pluripotency. Since their establishment, EPS cells have been widely used in different fields of research[27-32]. Recently, Zhang et al[33] demonstrated that inhibition of apoptosis reduced the diploidization of mouse haESCs during differentiation.
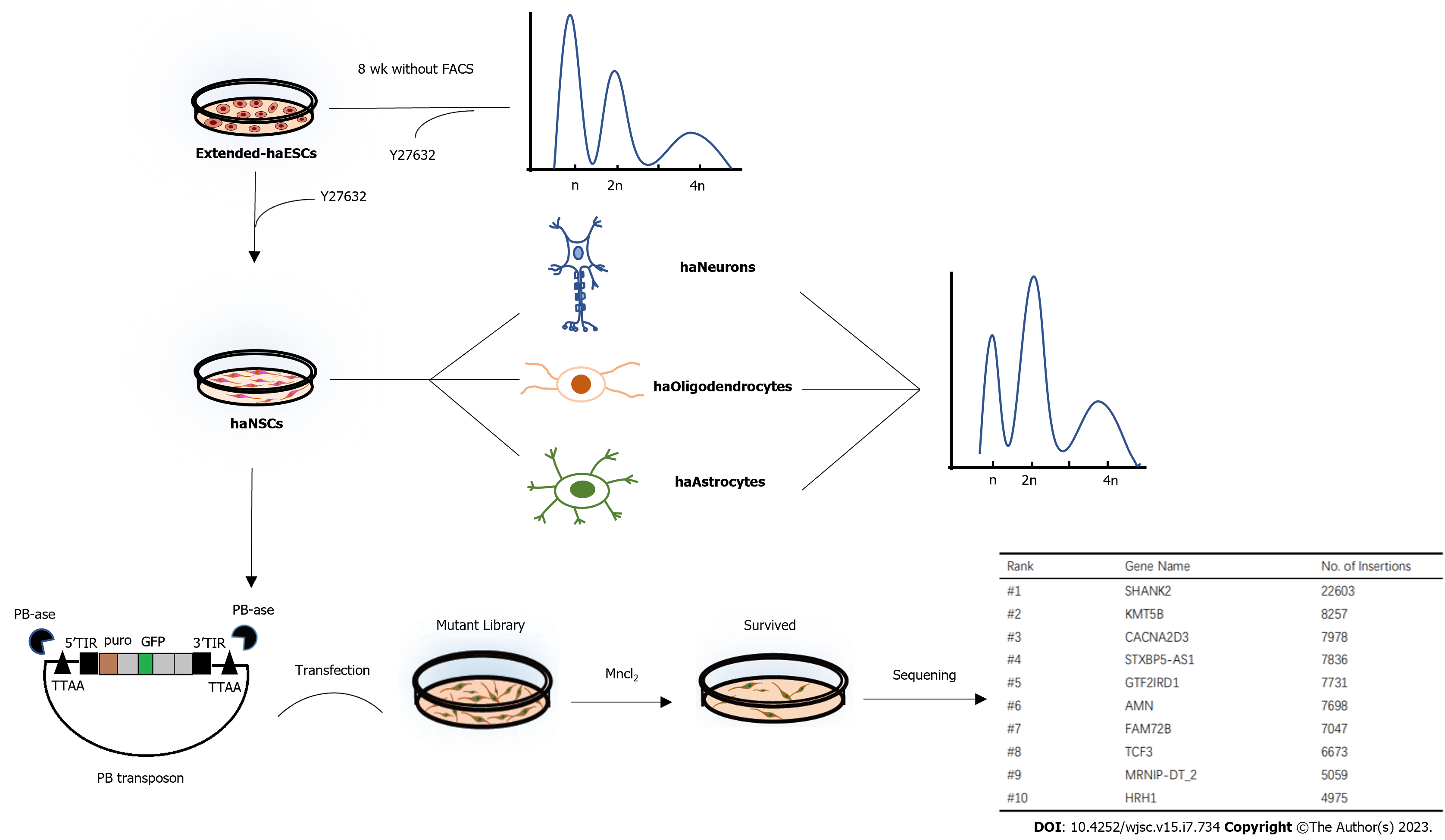
In this study, we introduced a small-molecule inhibitor during haESC culture and differentiation to restrain diploidization and enhance differential multipotency. Protocols were established for the derivation of human haploid neural stem cells (haNSCs) from human haESCs. These cells displayed neural differentiation potential in vitro. In addition, haNSCs can be maintained as predominantly haploid cells for more than 90 d without fluorescence activated cell sorting (FACS) enrichment. More importantly, we provided evidence that human haNSCs can be easily manipulated. These haNSCs are functional haploid adult stem cells and can be used for high-throughput genetic screening, providing a valuable resource for neural recessive gene studies.
The reagents and media used in the experiments were purchased from Life Technologies (Carlsbad, CA, United States) or Stem Cell Technologies, Inc. (Vancouver, Canada), unless otherwise indicated. Mouse embryonic fibroblasts were purchased from the Institute of Biochemistry and Cell Biology Shanghai Institutes for Biological Sciences Chinese Academy of Sciences.
Human haESCs were a kind gift from Dr. Jinsong Li of the State Key Laboratory of Cell Biology, Shanghai Key Laboratory of Molecular Andrology, Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences. The haESCs were cultured as previously reported[15,25,34]. Mouse embryonic fibroblasts were treated with 10 μg/mL mitomycin C for 3 h to generate a feeder layer and cultured in 10% fetal bovine serum (FBS) (BI) plus 1% penicillin-streptomycin Dulbecco’s modified Eagle medium (DMEM). The ESC medium before optimization contained DMEM/F12 + 20% KnockOut Serum Replacement (Gibco, Billings, MT, United States) + 0.1 mmol/L non-essential amino acids, 100 U/mL penicillin-0.1 mg/mL streptomycin, 0.1 mmol/L 2-mercaptoethanol, 2 mmol/L L-glutamine and 10 ng/mL basic fibroblast growth factor (bFGF). Human EPS cells were cultured in N2B27-LCDM (consisting of hLIF, CHIR99021, (S)-(+)-dimethindene maleate, minocycline hydrochloride) medium under 20% O2 and 5% CO2 at 37 °C. Then, 50 mL of N2B27 medium was prepared and included the following: 24 mL of DMEM/F12 (Gibco); 24 mL of Neurobasal (Gibco); 0.25 mL of N2 supplement (Gibco); 0.50 mL of B27 supplement (Gibco); 1% GlutaMAX (Sigma-Aldrich, St Louis, MO, United States); 1% non-essential amino acids (Gibco); 0.1 mmol/L β-mercaptoethanol (Sigma-Aldrich), penicillin-streptomycin (Gibco); and 5% KnockOut Serum Replacement (Gibco). The optimized medium was named N2B27-LCDM. Y-27632 (2 mmol/L; Peprotech, Rocky Hill, NJ, United States) was included in some cases. ESCs were passaged routinely using 1 mg/mL type IV collagenase (Sigma-Aldrich) (traditional culture ESCs) and 0.05 trypsin (optimized culture ESCs) every 3-4 d. The haNSCs were passaged every 4 d with 0.05% trypsin. The haNSC medium contained D/F12 + 1% N2 supplement + 20 ng/mL bFGF (Peprotech) + 20 ng/mL epidermal growth factor (EGF) (Peprotech). All cells were cultured in a 5% CO2 incubator at 37 °C.
Well-conditioned EhPGES1 were selected for differentiation, as the state of haESCs plays a decisive role in neural differentiation. EhPGES1 were digested to single cells with 1 mg/mL 0.05% trypsin. Then, these single cells were placed into gelatin-coated dishes for 30 min to allow the feeder cells to attach to the bottom of the dish. Single cells were suspended in embryoid body (EB) medium in a suspension petri dish for 4 d to form EBs. During the 1st d of differentiation, we added 10 μM ROCKi (Peprotech) to the EB medium. On day 4, the EB medium was replaced by neural induction medium (NIM) for another 2 d. On day 6, we transferred the EB from the suspension petri dishes to laminin-coated dishes and incubated overnight. The next day, EB paste was added to the bottom of the dish. We changed the NIM every other day.
The growth of flat cells and an increasing number of slender small cells were observed in the center of the implanted EB. After approximately 10 d in NIM, the central, small, lanky cells generated rosettes. Clusters were picked and expanded as described by Zhang et al[37] with slight optimizations[35-37]. In short, rosette clusters were attached to laminin-coated petri dishes in N2 medium containing human bFGF and EGF. Then, haNSCs were derived from haploid rosette clusters with continuous culture on polyornithine and laminin-coated dishes in N2 medium with 20 ng/mL bFGF and 20 ng/mL EGF for 3 mo. The differentiation efficiency from haploid rosette clusters to haNSCs under this condition was nearly 100% according to cell morphology evaluation.
haNSCs were examined for their neural differentiation potency. For differentiation into neurons, D/F12 medium was added as basal medium and supplemented with N2, B27 (Gibco), BDNF (20 ng/mL), glial-derived neurotrophic factor (20 ng/mL), dibutyryl-cAMP (1 mmol/L) and ascorbic acid (200 nM). Then, the haNSCs were replated on laminin-coated petri dishes at a density of 1 × 104 cells/cm2. The addition of 1% FBS (BI) to the primary medium was used to differentiate haNSCs into astrocytes and oligodendrocytes[36].
Human ESCs and NSCs were dissociated into single cells by treatment with 0.05% trypsin/EDTA at 37 °C for approximately 3 min. Cells were fixed in 75% ethyl alcohol at 4 °C for at least 12 h and stained with propidium iodide plus RNase A. Flow cytometry data were recorded by a BD Aria II system (BD Biosciences, Franklin Lakes, NJ, United States).
For sorting of haploid cells, haESCs and haNSCs were disaggregated by 0.05% trypsin/EDTA, washed with Dulbecco’s phosphate-buffered saline (PBS) and incubated with 4 μg/mL Hoechst 33342 in a 37 °C water bath for approximately 15 min. Subsequently, the haploid cells were collected using a BD FACSAria III (BD Biosciences) for further culture. ROCKi (10 μM Y27632) was added before and after sorting.
Total RNA from all cells was extracted with TRIzol reagent (Invitrogen, Waltham, MA, United States) and treated with DNase I (Promega, Madison, WI, United States). cDNA was obtained by first-strand synthesis using 2 μg of RNA and the ReverTra Ace system for reverse transcriptase-PCR (Toyobo, Osaka, Japan). Real-time PCR was performed as described in our previous report[21]. All reactions were repeated three times. Data are presented as the mean ± SD.
Immunostaining and alkaline phosphatase staining of ESCs and NSCs were performed as described previously[38]. In short, cells were fixed in formaldehyde, rinsed with PBS and permeabilized with Triton X-100 (Sigma-Aldrich). Then, the cells were incubated with blocking buffer, rinsed with PBS and incubated with primary and secondary antibodies (Abcam, Cambridge, and United Kingdom). After being rinsed with PBS, the cells were examined under a fluorescence microscope. The data represent three independent experiments.
Cells were incubated with 0.2 µg/mL colchicine for 1 h (ESCs) or 2 h (NSCs). Then, the cells were disaggregated with 0.05% trypsin/EDTA and resuspended in 5 mL of culture medium. The cells were then harvested from the 5 mL suspension in an automatic cell harvesting apparatus (Chromprep II; Sinochrome/Cytogenetics, Shanghai, China) according to the set procedure. Briefly, 4.5 mL of preheated potassium chloride (0.075 mmol/L) was added at 37 °C for 30 min. Hypotonic solution-treated cells were fixed in methanol:acetic acid (3:1 volume:volume) for 20 min at 37 °C, and the fixation step was repeated three times. Then, they were dropped onto precleaned charged slides by an automatic production machine (Chromprep AS; Sinochrome/Cytogenetics). After air drying, the cells were stained with Metaphase Auto-Stainer (Chromprep G; Sinochrome/Cytogenetics). More than 20 metaphase spread G-bands were analyzed by Ikaros karyotyping software with the MetaClient dashboard (MetaSystems, Altlussheim, Germany).
The Cell Counting Kit (CCK)-8 assays (Beyotime, Jiangsu Province, China) were performed as previously reported[15]. In short, cells were plated onto 96-well plates at a density of 1000 cells per well. Ten microliters of CCK-8 medium were added to each well on days 1, 2, 3, 4, and 5. The cells were incubated in a 37 °C 5% CO2 incubator for 1-4 h. Optical density values of each well at 450 nm wavelength were detected by an enzyme labeling instrument.
The RNA-seq data were sequenced by Novogene (Beijing, China). Total RNA was extracted using TRIzol Reagent (Invitrogen). Following the manufacturer’s instructions, raw data were processed with the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html) to remove noise and adaptors. The R heatmap function was used to perform hierarchical clustering.
PiggyBac [PB Dual promoter PB513B-1; Applied Biosystems Inc (ABI), Waltham, MA, United States] and PBase (Super PiggyBac Transposase PB200PA-1; ABI) were cotransfected into cells with Lipofectamine 3000 (Invitrogen). Puromycin (250 μg/mL) was used to culture and screen the transfected cells. To characterize the sites of the PB element insertion, genomic DNA was digested with BstYI (Thermo Fisher Scientific, Waltham, MA, United States) at 37 °C for 16 h, and the products were self-ligated. Then, with the linked DNA as a template, PCR was performed with the primers PB-RF and PB-RR to detect the previously reported insertion sites. PCR was performed as previously reported[39]. The first round was performed as follows: 98 °C for 75 s; 2 cycles of 98 °C for 20 s and 64 °C for 20 s; 30 cycles of 98 °C for 20 s, 58 °C for 15 s and 72 °C for 2 min; 72 °C for 7 min; and a final hold at 4 °C. The second round was conducted as follows: 98 °C for 75 s; 30 cycles of 98 °C for 20 s, 58 °C for 15 s and 72 °C for 90 s; 72 °C for 7 min; and a final hold at 4 °C. Splinkerette PCR and deep sequencing were also performed to identify the integration sites. All primers used are listed in Supple
After sequencing of the PB mutation library, adapters and PB tags were removed from the read data before mapping to the genome using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/commandline.html). HaSAPPy (https://github.com/gdiminin/HaSAPPy) was utilized to align the trimmed reads to the genome assembly from UCSC (mm10) (http://genome.ucsc.edu/cgi-bin/hgGateway? db = mm10)[40]. We analyzed the insertions for each gene with GENCODE[41] using the default parameters.
Student’s t-test was used to evaluate the differences in gene expression levels among groups. All statistical analyses were performed using SPSS software 13.0 (SPSS Inc., Chicago, IL, United States) and Image-Pro Plus (Media Cybernetics, Rockville, MD, United States).
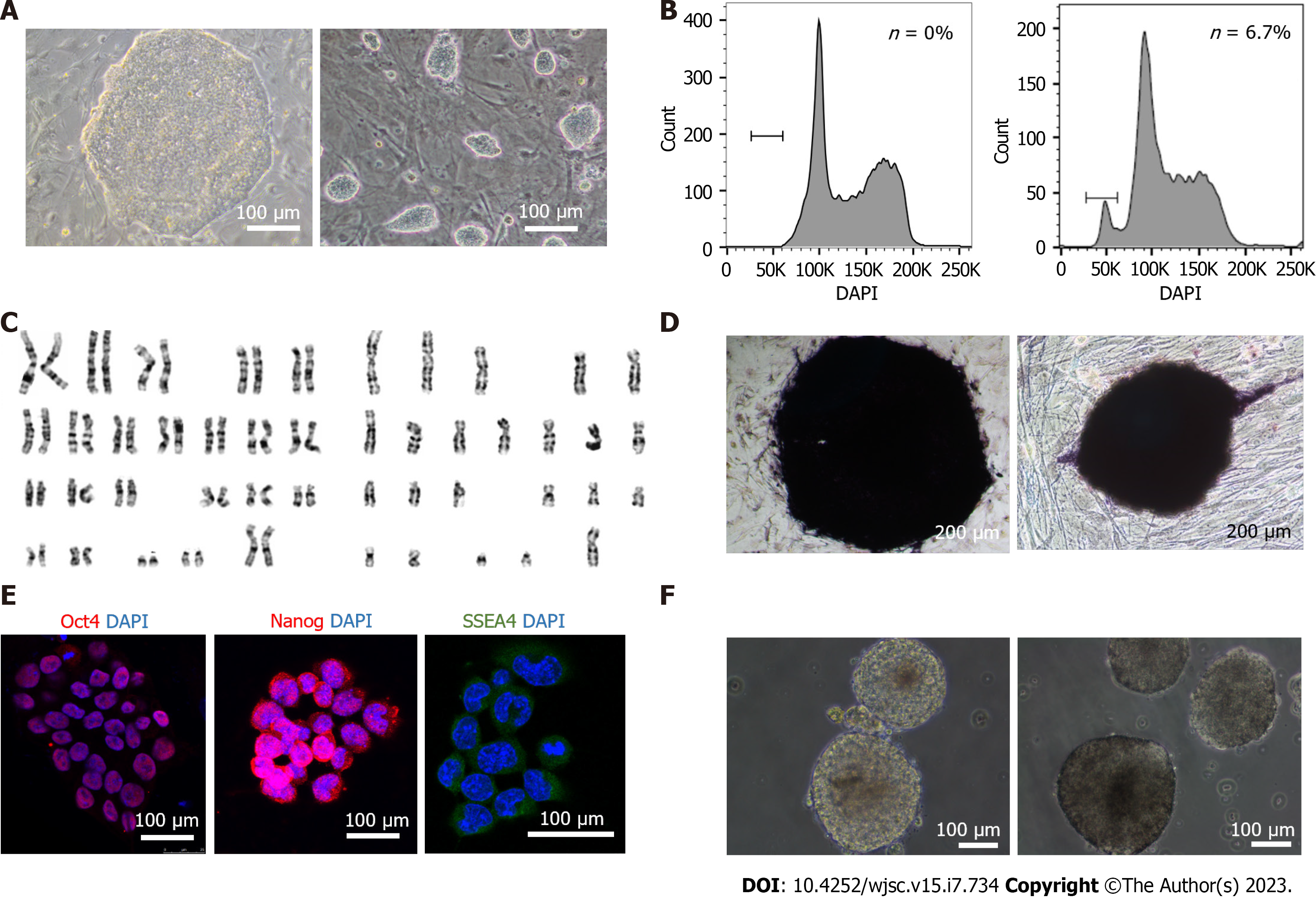
To determine whether traditional and extended pluripotent human haESCs exhibited pluripotency and intact haploid genomes, we compared them (cell line: hPGES1) with diploid human ESCs from zygotes (H9) in different respects. Traditional hPGES1 cells appeared as flat, compact colonies, a standard morphology for primed pluripotent cells (Supplementary Figure 1A) resembling that of diploid ESC H9 (Figure 1A), whereas extended pluripotent hPGES1 cells appeared as domed compact and small colonies, a standard morphology for naïve pluripotent cells resembling that of mouse ESCs (Figure 1A). After FACS of haploid cells, traditional and extended pluripotent hPGES1 cells maintained an acceptable percentage of haploidy for more than 6 wk without further sorting according to DNA content analysis (Figure 1B, Supplementary Figure 1B). We further assessed the chromosome numbers and morphology in traditional and extended pluripotent hPGES1 cells by G-band analysis and found that most cells were haploid (23 chromosomes) cells. A few cells were found to be haploid and aneuploid (Figure 1C, Supplementary Figure 1C and D). Next, we assessed the pluripotency of traditional and extended pluripotent hPGES1 cells from different aspects in vitro. We observed (by immunostaining) strong alkaline phosphatase activity (Figure 1D, Supplementary Figure 1D) and strong expression of pluripotent markers, including Oct4, Nanog, Sox2 and SSEA-4 (Figure 1E, Supplementary Figure 1E). After withdrawal of bFGF and feeder cells, traditional and extended pluripotent hPGES1 cells formed typical EBs, guaranteeing their utility for subsequent differentiation experiments (Figure 1F, Supplementary Figure 1F).
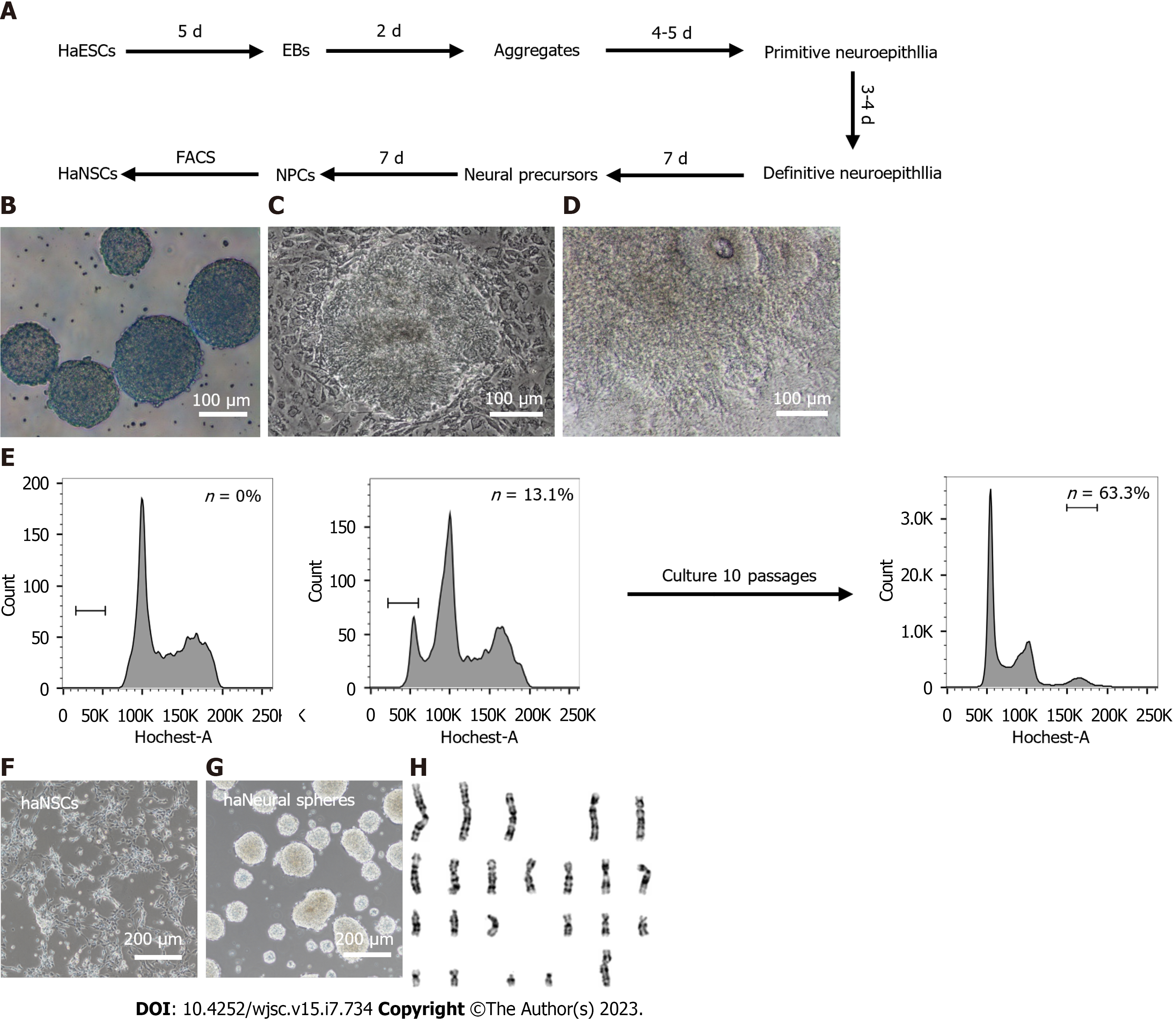
Suspension aggregation and posterior adherent culture are the most common methods for neural differentiation of human ESCs[37,42]. We thus developed a scheme to derive haNSCs based on a previously reported protocol with slight modifications (Figure 2A). During the first 5 d, well-formed human haESCs (Supplementary Figure 2A) were suspended and aggregated in EBs. EBs with better morphology (Supplementary Figure 2B) were cultured in suspension in NIM for 2 d. Neuroectodermal structures were observed in the middle part of the EB on day 7 (Figure 2B). All aggregates were seeded in laminin-coated dishes for adherence-induced culture. Aggregates adhering to laminin began to form primitive neuroepithelium after 4-5 d (Figure 2C). After approximately 15 d of differentiation, a large number of neuroepithelial cells had accumulated, and rosette structures were formed in the centers of the adherent cells (Figure 2D). The rosettes were manually selected and expanded in NSC medium for 14 d (Supplementary Figure 2C) before FACS. Haploid haNSCs in G0/G1 phase were predominant at the first round of sorting (Figure 2E). Interestingly, purified haNSCs could be amplified in NSC medium for more than 10 generations without sorting, and the proportion of haploid cells was still very high. haNSCs retained the standard NSC morphology (microscopic and bipolar) before and after sorting (Supplementary Figure 2D, Figure 2F) and were able to aggregate into typical neurosphere structures in suspension in petri dishes (Figure 2G). We also examined the chromosomes of haNSCs by G-banding analysis and found that haNSCs maintained intact haploid genomes without any aneuploid mutations (Figure 2H, Supplementary Figure 2E). We compared the cell growth rates of haNSCs and diploid NSCs using the CCK-8 method. The results showed no significant difference between the two cell types (Supplementary Figure 2F).
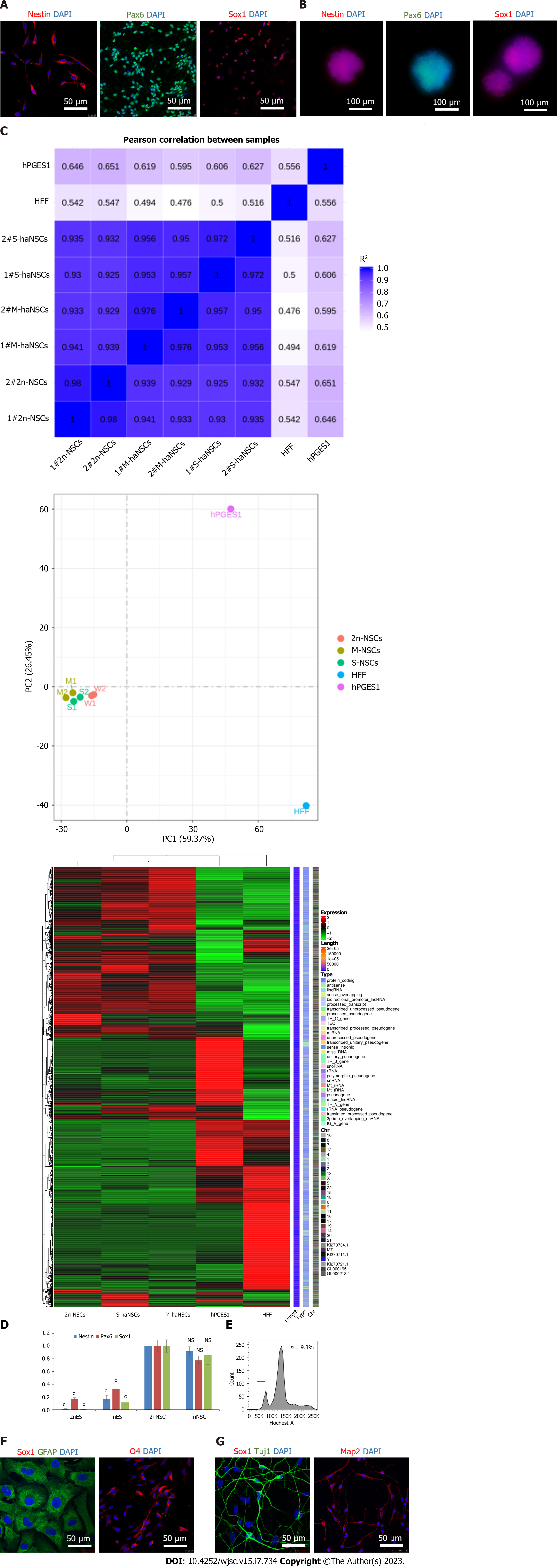
NSCs are proliferative adult stem cells that have the potential to differentiate into functional neurons or neuroglial cells, making them useful resources for cell replacement therapy and screening research[43]. To determine whether the haNSCs that we had generated possessed neural lineage properties, we assessed NSC-specific markers by immunostaining. The results showed that haNSCs expressed NSC markers (including Nestin, Sox1, and Pax6) in both monolayer cells (Figure 3A) and neural spheres (Figure 3B), similar to that of the diploid NSCs used as a control (Supple
Next, to evaluate the differentiation potential of haNSCs, we induced them to differentiate into astrocytes, oligodendrocytes and neurons by applying specific growth factors for approximately 3 wk and then sorted out the haploid cells by FACS (Figure 3E). Immunostaining results demonstrated that astrocytes (GFAP) and oligodendrocytes (O4) could be obtained from haNSCs and maintained as haploid cells (Figure 3F). In a parallel experiment, diverse neurons (Tuj1- and Map2-positive) were also generated from haNSCs (Figure 3G). To evaluate the purity of the haNSCs, we randomly selected haNSC spheres for differentiation induction (Supplementary Table 2). The results showed that each haNSC sphere could differentiate into neurons, astrocytes and oligodendrocytes. Therefore, the haNSCs were deemed to have good homogeneity.
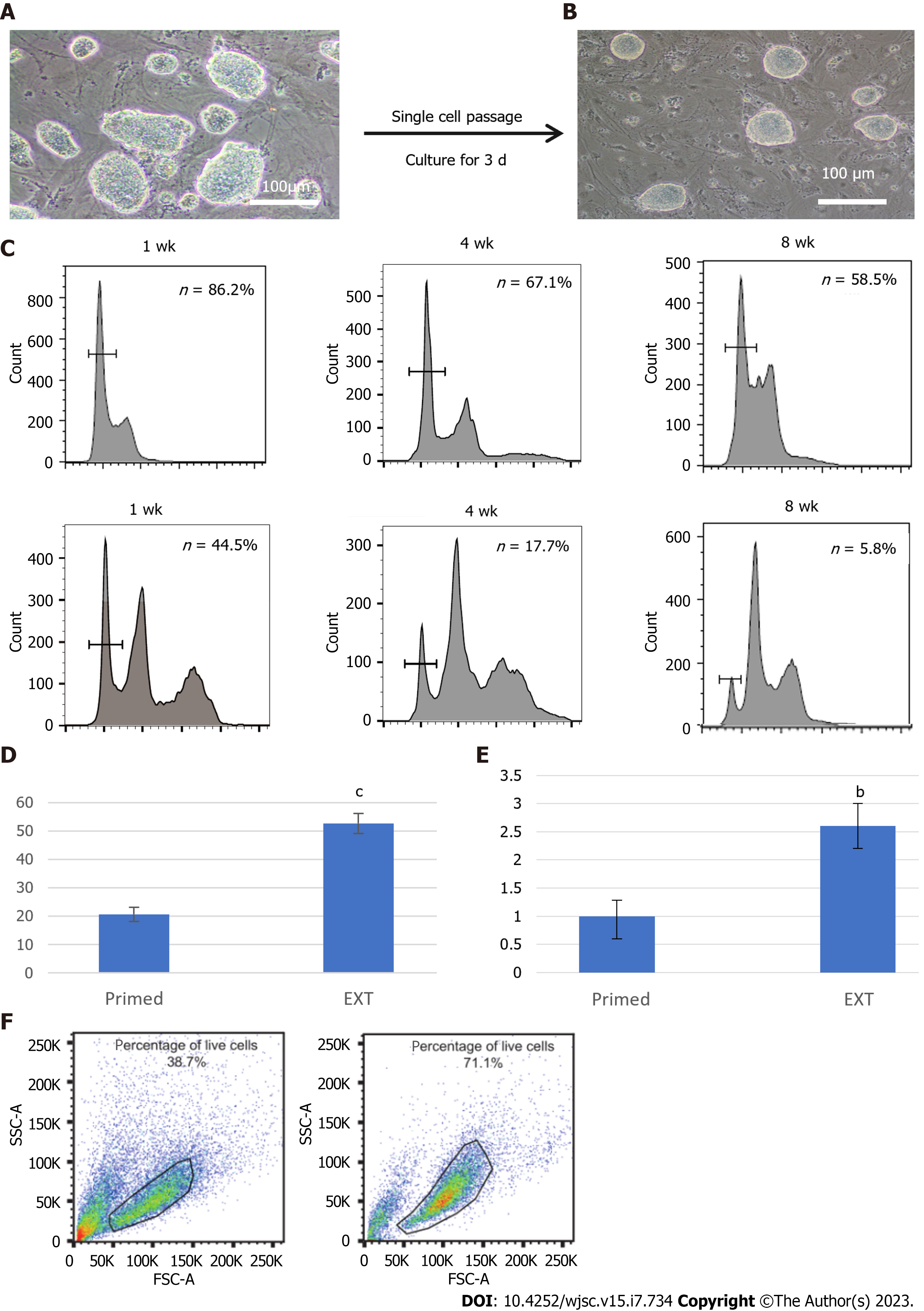
Recently, primate ESCs, including human ESCs, were converted to a better state by introducing small molecule compounds[25,44] or regulating transcription[45], with morphology and molecular levels similar to those of mouse ESCs. Whether extended haESCs in primate species show better pluripotency than traditional primed state ESCs has not been clarified. Herein, we optimized the culture system of human haESCs on the basis of previous research[25] with slight improvements (Supplementary Figure 4A). After culture in the optimized extended medium for approximately three passages, dome-shaped human pluripotent stem cell colonies formed, which is a morphological feature characteristic of mouse ESCs (Figure 4A). Optimized hPGES1 could be digested into single cells by 0.05% trypsin-EDTA in the absence of Y27632 (Figure 4B). The purified haESCs were divided into two populations and cultured in traditional and optimized media. FACS analysis showed that the proportion of haploid cells was higher in the optimized medium, indicating that the optimized medium could inhibit diploidy to some extent (Figure 4C, Supplementary Table 3).
To compare the EB formation ability between the optimized extended haESCs and traditional primed haESCs, we suspended approximately 106 cells from each group and allowed them to aggregate in untreated dishes. The number of EBs generated from optimized haESCs was more than two-fold that generated from the traditional group (Figure 4D). Moreover, the aggregate-generating neural rosettes from the optimized group were larger and more abundant than those from the traditional group (Figure 4E). When first sorting the haNSCs from optimized and traditional haESCs separately, the number of surviving cells from the optimized group was greater than that from the traditional group, according to the analysis of the side scatter and forward scatter plots (Figure 4F), indicating that haESCs in optimized medium could produce more NSCs with higher survival rates. A similar trend was found in harvested cells; we found that sorted cells from the traditional group had lower survival rates (Supplementary Figure 4B). Therefore, sorted haNSCs from the traditional group were limited in number and survival rate and readily became diploid NSCs. Overall, we generated two haNSC lines from four parallel differentiation lines established from the optimized groups. No haNSC lines were obtained from six parallel differentiation lines established from the traditional groups (Supplementary Figure 4C).
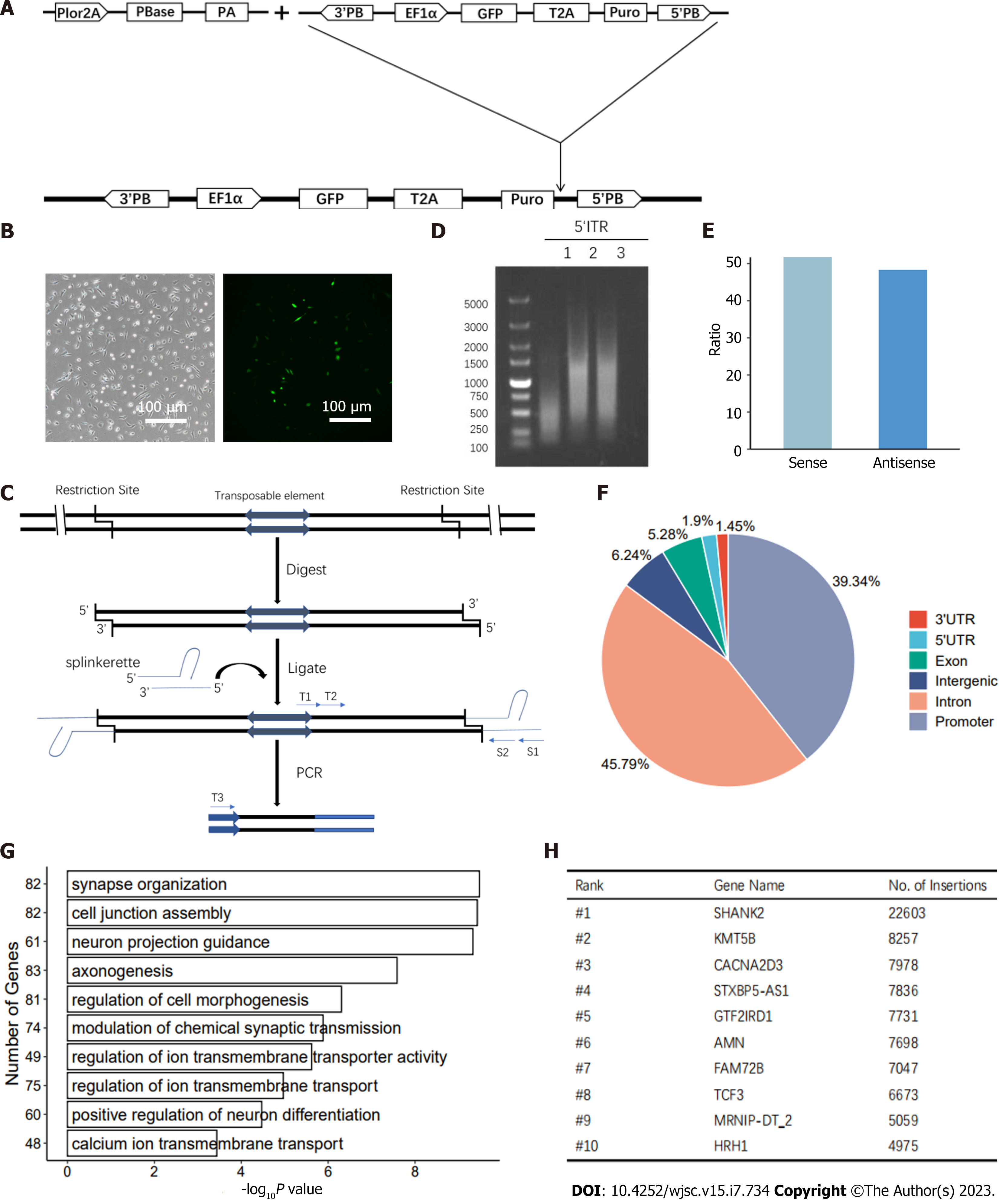
Next, we studied the feasibility of gene manipulation in human haNSCs. We initially applied transposable element-based strategies to establish transgenic haNSCs (Figure 5A). First, we employed the transposon and splinkerette PCR system, which has been applied for a variety of genetic manipulations in cells from mice[46,47], nonhuman primates[21] and humans[48]. We performed gene trapping in haNSCs to generate mutation libraries, which might be useful for neural lineage genetic screening.
Specifically, haNSCs were transfected with PB transposon-based gene-trap vectors[46] carrying the enhanced green fluorescent protein (eGFP) gene. After transfection, haNSCs exhibited a PB insertion and expressed eGFP (Figure 5B). We then added the nerve agent manganese chloride to haNSCs carrying the mutant library. Five days later, all cells in the control group had died. We harvested the surviving haNSCs and amplified the integration sites by performing splinkerette PCR (Figure 5C). The PCR products were analyzed by agarose gel electrophoresis, and the 5’ inverted terminal repeat PCR results showed that the bands were relatively diffuse, indicating that there were multiple insertion sites in the haNSC genome (Figure 5D).
Deep sequencing analysis identified approximately 4 million independent insertions across more than 20000 genes, of which 51.8% were in the sense orientation (Figure 5E). We analyzed the hits across the whole genome, and the results showed that the insertion sites were evenly spread across all chromosomes, indicating genome-wide coverage of mutations (Supplementary Figure 5A). In addition, 93.00% of the insertions were located in intragenic regions (exon + intron + promoter + 5’/3’ untranslated region), whereas 6.24% of insertions occurred in intergenic regions (Figure 5F).
Gene Ontology enrichment analysis showed that insertions occurred preferentially in genes with specific functions in neural activity-related pathways, such as neuron projection guidance, neuron projection guidance and regulation of transmembrane ion transporters (Figure 5G). Ten genes with especially frequent insertions were identified by PB screening (Figure 5H). To comprehensively assess the inserted genes, we analyzed them for roles in previously reported neurotoxicity-related pathways (MAPK, Notch, Wnt and other pathways) (Supplementary Figure 5B). In summary, the derived haNSCs were found to be beneficial for genome-wide genetic screening.
Collectively, these results indicated that the optimized conditions supported the generation of human haNSCs, which can act as an ideal tool for screening key genes during neurogenesis.
Researchers have obtained haESCs from a variety of species, including mammals[9-13], rhesus monkeys[15] and even humans[21]. Because there are obvious differences in the pathogenesis of many diseases between humans and other species[23,24], we urgently need to increase the number of available human cell models to more directly and efficiently explain the pathogenesis of human diseases. In this study, we demonstrated that tissue-specific adult cells (haNSCs) can be generated in vitro by the conversion of haESCs without chromosome doubling. Although severe diploidization was observed during ESC-NSC conversion, it was possible to optimize the differentiation system without gene modification through the addition of a number of small molecule inhibitors to the culture medium. With these modifications, we were able to establish haNSC lines relatively easily.
As the derived haNSCs were generated from haESCs in vitro, their purity and NSC properties needed to be confirmed. Nestin, SOX1 and PAX6 are NSC-specific markers that have been widely used in research related to neural cell fate[37]. Our results showed that these haNSCs express NSC markers and are similar to brain-derived NSCs in morphology. After functional verification, we found that these cells can differentiate into neurons and glial cells in vitro. In this regard, haNSCs have differentiation potential resembling that of brain-derived NSCs.
HaESCs are an important tool for mammalian genetic research. Human haESCs can differentiate into haploid somatic cells of the ectoderm, mesoderm and endoderm[16]. This result suggests that primate species may have a potential superiority for a haploidy maintenance mechanism. Our experiments with human haNSCs confirmed this finding, as this cell group maintained a stable haploid state during the culture process. These advantages guaranteed their utility in a subsequent gene trapping procedure.
The PB transposon can insert a large number of random mutations in the genome[21,48]. Therefore, we combined the PB transposon and haNSCs to conduct genetic screening. Previous work demonstrated that neural cells are more susceptible to manganese-induced toxicity[49]. We performed genetic screening to uncover the target genes of manganese in haNSCs. The results of the gene trapping using the PB transposon system showed that random insertions could be generated efficiently in these haploid somatic cells. This method could be used in combination with high-throughput sequencing analysis to easily identify the gene insertion sites and insertion frequencies in all of the 23 chromosomes.
As described previously, human haESCs could differentiate into Tuj1-positive cells with a haploid genome. However, the cells lacked stability and proliferation capacity to act as an ideal tool for screening key genes.
There were limitations of this study. The diploidization of the haploid cells was not completely terminated; it was only slowed down. The underlying mechanism remains to be further studied and explored.
In conclusion, we optimized a culture system for human haESCs and achieved the derivation of human haNSCs (Figure 6). This is the first human haploid somatic stem cell line reported with good proliferative ability and pluripotency toward neural subtypes. Moreover, through chromosome G-band karyotype analysis, we confirmed for the first time that the differentiated haNSCs maintained 23 chromosomes while not exhibiting fragmentation, deletion, duplication, translocation, inversion or other balanced variations. Meanwhile, we identified manganese-induced toxicity genes with the help of this haploid cell line. This cell line will be valuable for genetic screening relevant to human nervous system function and drug targeting research.
Recently, haploid embryonic stem cells (haESCs) have been established in many species and are widely used in forward and reverse genetic screening. Differentiated haploid cell line types in mammals are lacking due to spontaneous diploidization during differentiation that compromises lineage-specific screens.
No human haploid somatic cell line has been generated, although there is an urgent need for lineage-specific screens.
This study aimed to derive haploid neural stem cells (haNSCs) from human extended haESCs by optimizing differentiation methods and verifying human haNSCs to easily carry out lineage-specific screens.
Human haploid NSCs were differentiated from human extended haESCs with the help of Y27632 to reduce the diploidization. Neuronal differentiation of haNSCs was performed to examine their neural differentiation potential. Global gene expression analysis was performed to compare haNSCs with diploid NSCs and haESCs. Fluorescence activated cell sorting was conducted to assess the diploidization rate of extended haESCs and haNSCs. Genetic manipulation and screening were utilized to evaluate the significance of human haNSCs as genetic screening tools.
haNSCs can be generated by differentiation in vitro, and they maintain long-term haploidy with a complete genome and multipotency towards neurons and glia. Human haESCs cultured in extended pluripotency medium formed more compact colonies and exhibited more efficient neural differentiation and more stable maintenance of haploidy, which effectively facilitated the derivation of haNSCs. After PiggyBac transfection, there were multiple insertion sites in the haNSC genome and the insertion sites were evenly spread across all chromosomes.
In this work, we demonstrated that human haNSCs can be generated by differentiation from extended haESCs. This is the first human haploid somatic cell line with a complete genome, acceptable proliferative ability and neural differentiation potential, which provides a cell resource for studies of recessive inheritance and drug targeted screening.
This cell line will be valuable for genetic screening relevant to the function of the human nervous system and drug targeting research. Future research will be related to identifying key neurogenesis genes with the help of this cell line. Another research focus is to identify the underlying mechanisms of diploidization.
We thank Dr. Ling Shuai (State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China) for the PiggyBac-GFP vector. Dr. Jinsong Li (GTP Center and Stem Cell Bank, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences) is acknowledged for assistance with human parthenogenetic ESCs.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Durán Alonso MB, Spain; Tabarzad M, Iran S-Editor: Chen YL L-Editor: A P-Editor: Chen YX
| 1. | Xu M, Zhao Y, Zhang W, Geng M, Liu Q, Gao Q, Shuai L. Genome-scale screening in a rat haploid system identifies Thop1 as a modulator of pluripotency exit. Cell Prolif. 2022;55:e13209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 2. | Bar S, Vershkov D, Keshet G, Lezmi E, Meller N, Yilmaz A, Yanuka O, Nissim-Rafinia M, Meshorer E, Eldar-Geva T, Benvenisty N. Identifying regulators of parental imprinting by CRISPR/Cas9 screening in haploid human embryonic stem cells. Nat Commun. 2021;12:6718. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 3. | Zhang J, Zhao Y, Tian Y, Geng M, Liu Y, Zhang W, Shuai L. Genome-wide screening in the haploid system reveals Slc25a43 as a target gene of oxidative toxicity. Cell Death Dis. 2022;13:284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 4. | Cui T, Li Z, Zhou Q, Li W. Current advances in haploid stem cells. Protein Cell. 2020;11:23-33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 5. | Gao Q, Zhang W, Zhao Y, Tian Y, Wang Y, Zhang J, Geng M, Xu M, Yao C, Wang H, Li L, Liu Y, Shuai L. High-throughput screening in postimplantation haploid epiblast stem cells reveals Hs3st3b1 as a modulator for reprogramming. Stem Cells Transl Med. 2021;10:743-755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Wang LB, Li ZK, Wang LY, Xu K, Ji TT, Mao YH, Ma SN, Liu T, Tu CF, Zhao Q, Fan XN, Liu C, Shu YJ, Yang N, Zhou Q, Li W. A sustainable mouse karyotype created by programmed chromosome fusion. Science. 2022;377:967-975. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 7. | Gu Z, Guo J, Wang H, Wen Y, Gu Q. Bioengineered microenvironment to culture early embryos. Cell Prolif. 2020;53:e12754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 8. | Li X, Cui XL, Wang JQ, Wang YK, Li YF, Wang LY, Wan HF, Li TD, Feng GH, Shuai L, Li ZK, Gu Q, Hao J, Wang L, Zhao XY, Liu ZH, Wang XJ, Li W, Zhou Q. Generation and Application of Mouse-Rat Allodiploid Embryonic Stem Cells. Cell. 2016;164:279-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 9. | Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 204] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 10. | Li W, Shuai L, Wan H, Dong M, Wang M, Sang L, Feng C, Luo GZ, Li T, Li X, Wang L, Zheng QY, Sheng C, Wu HJ, Liu Z, Liu L, Wang XJ, Zhao XY, Zhou Q. Androgenetic haploid embryonic stem cells produce live transgenic mice. Nature. 2012;490:407-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 11. | Yang H, Shi L, Wang BA, Liang D, Zhong C, Liu W, Nie Y, Liu J, Zhao J, Gao X, Li D, Xu GL, Li J. Generation of genetically modified mice by oocyte injection of androgenetic haploid embryonic stem cells. Cell. 2012;149:605-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 12. | Wutz A. Haploid mouse embryonic stem cells: rapid genetic screening and germline transmission. Annu Rev Cell Dev Biol. 2014;30:705-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 13. | Elling U, Taubenschmid J, Wirnsberger G, O'Malley R, Demers SP, Vanhaelen Q, Shukalyuk AI, Schmauss G, Schramek D, Schnuetgen F, von Melchner H, Ecker JR, Stanford WL, Zuber J, Stark A, Penninger JM. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 197] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 14. | Li W, Li X, Li T, Jiang MG, Wan H, Luo GZ, Feng C, Cui X, Teng F, Yuan Y, Zhou Q, Gu Q, Shuai L, Sha J, Xiao Y, Wang L, Liu Z, Wang XJ, Zhao XY. Genetic modification and screening in rat using haploid embryonic stem cells. Cell Stem Cell. 2014;14:404-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 84] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 15. | Yang H, Liu Z, Ma Y, Zhong C, Yin Q, Zhou C, Shi L, Cai Y, Zhao H, Wang H, Tang F, Wang Y, Zhang C, Liu XY, Lai D, Jin Y, Sun Q, Li J. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 2013;23:1187-1200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 16. | Sagi I, Chia G, Golan-Lev T, Peretz M, Weissbein U, Sui L, Sauer MV, Yanuka O, Egli D, Benvenisty N. Derivation and differentiation of haploid human embryonic stem cells. Nature. 2016;532:107-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 122] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 17. | Zhang XM, Wu K, Zheng Y, Zhao H, Gao J, Hou Z, Zhang M, Liao J, Zhang J, Gao Y, Li Y, Li L, Tang F, Chen ZJ, Li J. In vitro expansion of human sperm through nuclear transfer. Cell Res. 2020;30:356-359. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Zhong C, Zhang M, Yin Q, Zhao H, Wang Y, Huang S, Tao W, Wu K, Chen ZJ, Li J. Generation of human haploid embryonic stem cells from parthenogenetic embryos obtained by microsurgical removal of male pronucleus. Cell Res. 2016;26:743-746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 19. | Yi M, Hong N, Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 145] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 20. | He ZQ, Xia BL, Wang YK, Li J, Feng GH, Zhang LL, Li YH, Wan HF, Li TD, Xu K, Yuan XW, Li YF, Zhang XX, Zhang Y, Wang L, Li W, Zhou Q. Generation of Mouse Haploid Somatic Cells by Small Molecules for Genome-wide Genetic Screening. Cell Rep. 2017;20:2227-2237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 21. | Wang H, Zhang W, Yu J, Wu C, Gao Q, Li X, Li Y, Zhang J, Tian Y, Tan T, Ji W, Li L, Yu Y, Shuai L. Genetic screening and multipotency in rhesus monkey haploid neural progenitor cells. Development. 2018;145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Levin ED. α7-Nicotinic receptors and cognition. Curr Drug Targets. 2012;13:602-606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Quik M, Polonskaya Y, Gillespie A, Jakowec M, Lloyd GK, Langston JW. Localization of nicotinic receptor subunit mRNAs in monkey brain by in situ hybridization. J Comp Neurol. 2000;425:58-69. [PubMed] [DOI] [Full Text] |
| 24. | Mashiko H, Yoshida AC, Kikuchi SS, Niimi K, Takahashi E, Aruga J, Okano H, Shimogori T. Comparative anatomy of marmoset and mouse cortex from genomic expression. J Neurosci. 2012;32:5039-5053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 25. | Yang Y, Liu B, Xu J, Wang J, Wu J, Shi C, Xu Y, Dong J, Wang C, Lai W, Zhu J, Xiong L, Zhu D, Li X, Yang W, Yamauchi T, Sugawara A, Li Z, Sun F, Li C, He A, Du Y, Wang T, Zhao C, Li H, Chi X, Zhang H, Liu Y, Duo S, Yin M, Shen H, Belmonte JCI, Deng H. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell. 2017;169:243-257.e25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 356] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 26. | Liu B, Chen S, Xu Y, Lyu Y, Wang J, Du Y, Sun Y, Liu H, Zhou H, Lai W, Xue A, Yin M, Li C, Bai Y, Xu J, Deng H. Chemically defined and xeno-free culture condition for human extended pluripotent stem cells. Nat Commun. 2021;12:3017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 27. | Li H, Zhao C, Xu J, Xu Y, Cheng C, Liu Y, Wang T, Du Y, Xie L, Zhao J, Han Y, Wang X, Bai Y, Deng H. Rapid generation of gene-targeted EPS-derived mouse models through tetraploid complementation. Protein Cell. 2019;10:20-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Wang Q, Sun D, Liang Z, Wang J, Zhong X, Lyu Y, Cao J, Lin Z, Du Y, Miao Z, Lu S, Li C, Xu J, Shi Y, Deng H. Generation of human hepatocytes from extended pluripotent stem cells. Cell Res. 2020;30:810-813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Tan T, Wu J, Si C, Dai S, Zhang Y, Sun N, Zhang E, Shao H, Si W, Yang P, Wang H, Chen Z, Zhu R, Kang Y, Hernandez-Benitez R, Martinez Martinez L, Nuñez Delicado E, Berggren WT, Schwarz M, Ai Z, Li T, Deng H, Esteban CR, Ji W, Niu Y, Izpisua Belmonte JC. Chimeric contribution of human extended pluripotent stem cells to monkey embryos ex vivo. Cell. 2021;184:3589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Li R, Zhong C, Yu Y, Liu H, Sakurai M, Yu L, Min Z, Shi L, Wei Y, Takahashi Y, Liao HK, Qiao J, Deng H, Nuñez-Delicado E, Rodriguez Esteban C, Wu J, Izpisua Belmonte JC. Generation of Blastocyst-like Structures from Mouse Embryonic and Adult Cell Cultures. Cell. 2019;179:687-702.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 186] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 31. | Sozen B, Cox AL, De Jonghe J, Bao M, Hollfelder F, Glover DM, Zernicka-Goetz M. Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid. Dev Cell. 2019;51:698-712.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 32. | Zhang Y, An C, Yu Y, Lin J, Jin L, Li C, Tan T, Fan Y. Epidermal growth factor induces a trophectoderm lineage transcriptome resembling that of human embryos during reconstruction of blastoids from extended pluripotent stem cells. Cell Prolif. 2022;55:e13317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 33. | Zhang W, Tian Y, Gao Q, Li X, Li Y, Zhang J, Yao C, Wang Y, Wang H, Zhao Y, Zhang Q, Li L, Yu Y, Fan Y, Shuai L. Inhibition of Apoptosis Reduces Diploidization of Haploid Mouse Embryonic Stem Cells during Differentiation. Stem Cell Reports. 2020;15:185-197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 34. | Fang R, Liu K, Zhao Y, Li H, Zhu D, Du Y, Xiang C, Li X, Liu H, Miao Z, Zhang X, Shi Y, Yang W, Xu J, Deng H. Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell. 2014;15:488-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 35. | Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 514] [Cited by in RCA: 528] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 36. | Zhao Y, Ji S, Wang J, Huang J, Zheng P. mRNA-Seq and microRNA-Seq whole-transcriptome analyses of rhesus monkey embryonic stem cell neural differentiation revealed the potential regulators of rosette neural stem cells. DNA Res. 2014;21:541-554. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1451] [Cited by in RCA: 1347] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 38. | Marin Navarro A, Pronk RJ, van der Geest AT, Oliynyk G, Nordgren A, Arsenian-Henriksson M, Falk A, Wilhelm M. p53 controls genomic stability and temporal differentiation of human neural stem cells and affects neural organization in human brain organoids. Cell Death Dis. 2020;11:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 39. | Uren AG, Mikkers H, Kool J, van der Weyden L, Lund AH, Wilson CH, Rance R, Jonkers J, van Lohuizen M, Berns A, Adams DJ. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat Protoc. 2009;4:789-798. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 125] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 40. | Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, Tharp GK, Marçais G, Roberts M, Ferguson B, Fox HS, Treangen T, Salzberg SL, Yorke JA, Norgren RB Jr. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct. 2014;9:20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 143] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 41. | Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C, Hubbard T, Antonarakis SE, Guigo R. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7 Suppl 1:S4.1-S4.9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 464] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 42. | Jensen MB, Yan H, Krishnaney-Davison R, Al Sawaf A, Zhang SC. Survival and differentiation of transplanted neural stem cells derived from human induced pluripotent stem cells in a rat stroke model. J Stroke Cerebrovasc Dis. 2013;22:304-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 43. | Zhang SC. Neural subtype specification from embryonic stem cells. Brain Pathol. 2006;16:132-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 118] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 44. | Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, Rais Y, Shipony Z, Mukamel Z, Krupalnik V, Zerbib M, Geula S, Caspi I, Schneir D, Shwartz T, Gilad S, Amann-Zalcenstein D, Benjamin S, Amit I, Tanay A, Massarwa R, Novershtern N, Hanna JH. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 764] [Cited by in RCA: 834] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 45. | Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, Reik W, Bertone P, Smith A. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254-1269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 770] [Cited by in RCA: 721] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
| 46. | Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 761] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 47. | Li X, Zhu L, Yang A, Lin J, Tang F, Jin S, Wei Z, Li J, Jin Y. Calcineurin-NFAT signaling critically regulates early lineage specification in mouse embryonic stem cells and embryos. Cell Stem Cell. 2011;8:46-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 48. | Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1423] [Cited by in RCA: 1263] [Article Influence: 78.9] [Reference Citation Analysis (0)] |
| 49. | Chen P, Chakraborty S, Peres TV, Bowman AB, Aschner M. Manganese-induced Neurotoxicity: From C. elegans to Humans. Toxicol Res (Camb). 2015;4:191-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |