Published online Jun 26, 2023. doi: 10.4252/wjsc.v15.i6.530
Peer-review started: February 16, 2023
First decision: February 28, 2023
Revised: March 14, 2023
Accepted: April 27, 2023
Article in press: April 24, 2023
Published online: June 26, 2023
Processing time: 130 Days and 11.7 Hours
Brain diseases affect 1 in 6 people worldwide. These diseases range from acute neurological conditions such as stroke to chronic neurodegenerative disorders such as Alzheimer’s disease. Recent advancements in tissue-engineered brain disease models have overcome many of the different shortcomings associated with the various animal models, tissue culture models, and epidemiologic patient data that are commonly used to study brain disease. One innovative method by which to model human neurological disease is via the directed differentiation of human pluripotent stem cells (hPSCs) to neural lineages including neurons, astrocytes, and oligodendrocytes. Three-dimensional models such as brain organoids have also been derived from hPSCs, offering more physiological relevance due to their incorporation of various cell types. As such, brain orga
Core Tip: This review discusses recent advances in the field of disease modeling using human-induced pluripotent stem cell-derived neural cell types as well as organoids. It also discusses challenges that exist with current approaches, in addition to considerations for possible improvements that will further advance the field of disease modeling.
- Citation: Yan YW, Qian ES, Woodard LE, Bejoy J. Neural lineage differentiation of human pluripotent stem cells: Advances in disease modeling. World J Stem Cells 2023; 15(6): 530-547
- URL: https://www.wjgnet.com/1948-0210/full/v15/i6/530.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i6.530
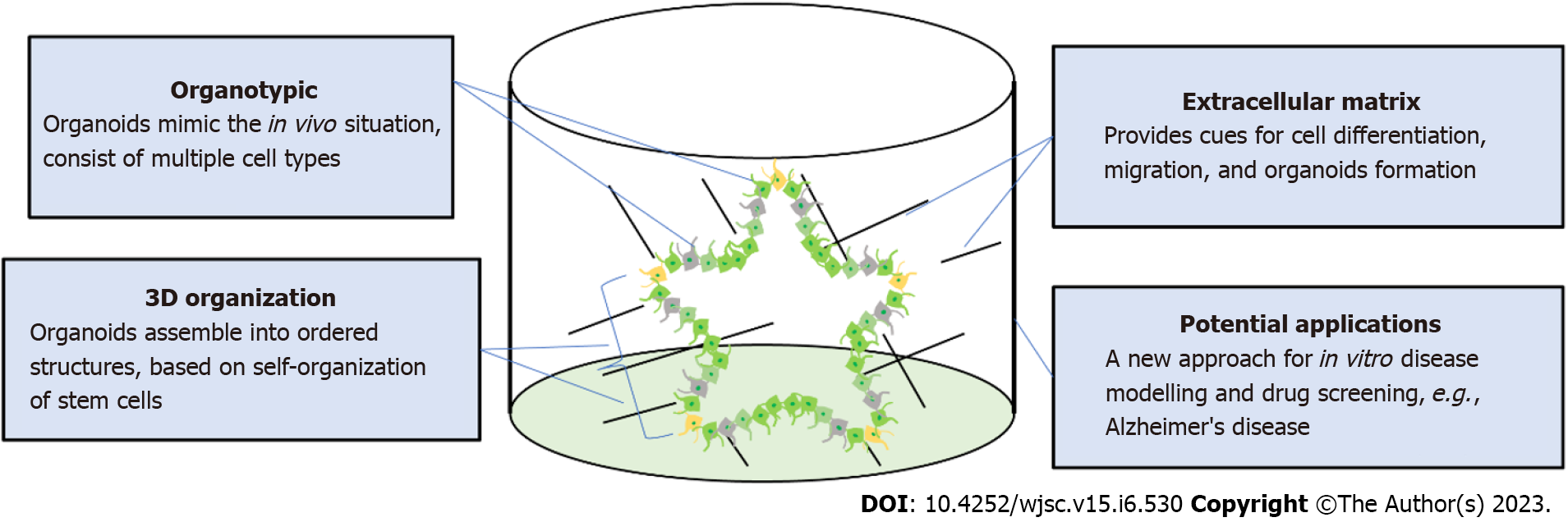
Human pluripotent stem cells (hPSCs) include both human embryonic stem cells (hESCs) and human-induced PSCs (hiPSCs). Current in vitro disease models that use hiPSCs begin with skin or blood cells that have been reprogramed with the four transcriptional elements octamer-binding transcription factor 4, SRY-box 2, Krüppel-like factor 4, and MYC[1]. Through differentiation, these hPSCs are the starting material to create models for different organs including the brain[2], kidney[3], liver[4,5], lung[6], and pancreas[7]. These models are able to simulate a “disease-in-a-dish”, mimicking different disease phenotypes in vitro[8,9]. Both genetically modified hiPSCs and patient-derived hiPSCs can generate disease models[10-12]. These models are advantageous because of their accessibility, quick processing, and species-specific human attributes. Patient-derived hiPSCs can also be used to test personalized medicine approaches to effectively model gene mutations and chromosomal abnormalities. To study neurological diseases, scientists have generated multiple neural cell types from hPSCs including neurons[13], astrocytes[14], and oligodendrocytes (OLs)[15]. In the past decade, advancements in disease modeling and tissue engineering have also led to the “brain organoid” model[16]. Brain organoids are self-assembled structures that resemble the fetal human brain and are composed of progenitor, neuronal, and glial cells. A related system is the spheroid, a circular aggregate of cells that may reflect biological properties of an organ system but ultimately lacks structural complexity. Perhaps the most cutting-edge form of modeling technology in stem-cell research is the assembloid, which are three-dimensional (3D) structures made by fusing and integrating two or more cell types or organoids from different organ culture protocols[17]. These assembloids can model the organ crosstalk interactions that occur across physiological systems in the human body.
In this review, we discuss recent advancements in the field of disease modeling using hiPSC-derived neural cell types as well as organoids. We also discuss challenges that exist with current approaches, in addition to considerations for possible improvements that will further advance the field of disease modeling.
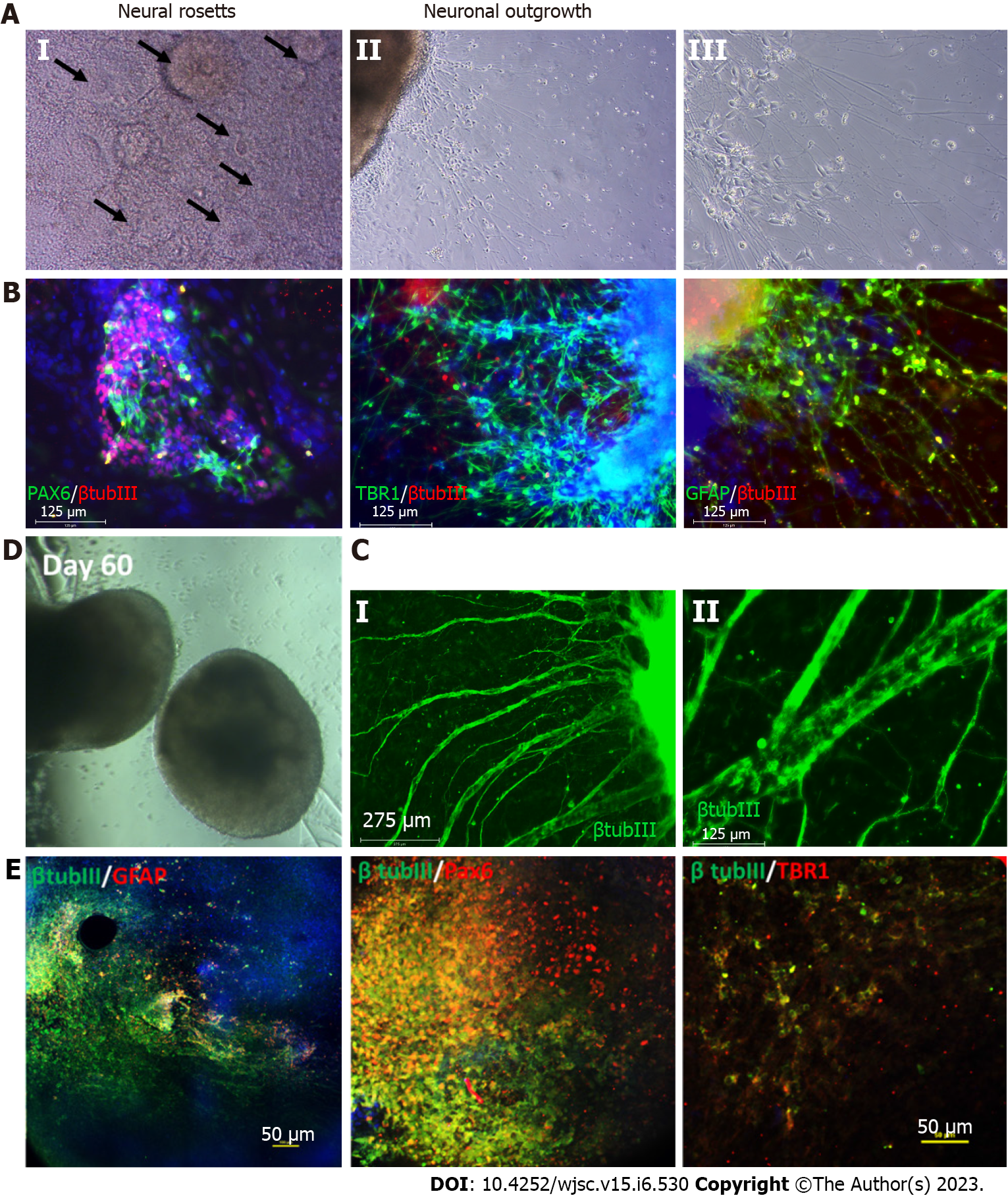
The development of the mammalian brain initially occurs at the gastrula stage when the ectoderm differentiates to form the neural tube. This process is called neural induction, wherein neural tube cells become neural progenitors[18,19]. These progenitors subsequently give rise to specific neuronal subtypes along the rostral-caudal axis and dorsal-ventral axis[20]. Protocols for neural progenitor differentiation from hPSCs have been developed that reflect this neural induction principle (Table 1). In 2001, the first protocol of neural progenitor differentiation from hPSCs was developed using the embryoid body (EB) method, which was a combined two-dimensional (2D) monolayer and 3D suspension culture[21]. Without extrinsic factors, the EB method mainly derived dorsal forebrain cortical neurons. In 2008, advances in the EB method eventually gave rise to a complete 3D culture system called the serum-free floating culture of EB-like aggregates with quick aggregation (SFEBq)[22]. The SFEBq method generated neural tissues with self-organized structure using hPSCs, paving the way for the development of more complex systems such as brain spheroids and organoids. Following this advance, in 2009 the dual SMAD inhibition method was developed, which successfully directed over 80% of hESCs to induce neural differentiation[23]. The dual SMAD inhibition method was initially intended for 2D monolayer culture by inhibiting the bone morphogenetic protein (BMP) and transforming growth factor beta (TGF-β) signaling pathways, but it has been widely applied to 3D culture for neural progenitor differentiation from hPSCs. It is worth noting that both the SFEBq and dual SMAD inhibition methods can enable the generation of cortical spheroids and organoids[24-26]. In 2017, Studer’s group modified the dual SMAD inhibition protocol to also block mitogen-activated protein kinase (MAPK), fibroblast growth factor (FGF), and Notch signaling, thereby accelerating forebrain cortical neuron derivation[27]. Although these protocols yield primarily deep-layer cortical neurons, deriving upper layer cortical neurons such as L2/3 and L4 cells is still a challenge.
| Method | Neural induction outcomes | Significance | Ref. |
| Embryoid bodies; selected neural rosettes; 2D and 3D culture | Neural tube-like rosettes stained with Nestin, Musashi-1 and NCAM; positive neuronal markers MAP2 and TUJ1 expression | First study of neural progenitor differentiation from hPSCs | Zhang et al[22], 2001 |
| SFEBq aggregate; sorting cells; 3D culture | Self-organized structure with four distinct zones: ventricular, early and late cortical-plate, and Cajal-Retzius cell zones | Pure 3D culture, provides the basis for the brain organoid method | Eiraku et al[23], 2008 |
| Dual SMAD inhibition; 2D monolayer culture | Complete neural conversion of > 80% of hESCs | Mostly wild used method; also enables neural induction in 3D culture | Chambers et al[24], 2009 |
| Dual SMAD inhibition combined with retinoid signaling; 2D monolayer culture | More than 95% of hPSCs were PAX6 and OTX1/2 cortical progenitor cells in 15 d | Improved the dual SMAD inhibition protocol and higher neural induction efficiency | Shi et al[62], 2012 |
| Cortical organoid/spheroid; 3D culture | Form layered structure tissues partially mimicking human cerebral cortex | Mostly brain-like tissue with some functions | Lancaster et al[17], 2013; Paşca et al[26], 2015; Qian et al[27], 2016 |
| Dual SMAD inhibition combined with Wnt, FGF and Notch inhibition | Generate functional cortical neuron in 16 d | Improved the dual SMAD inhibition protocol and accelerated neural induction | Qi et al[28] |
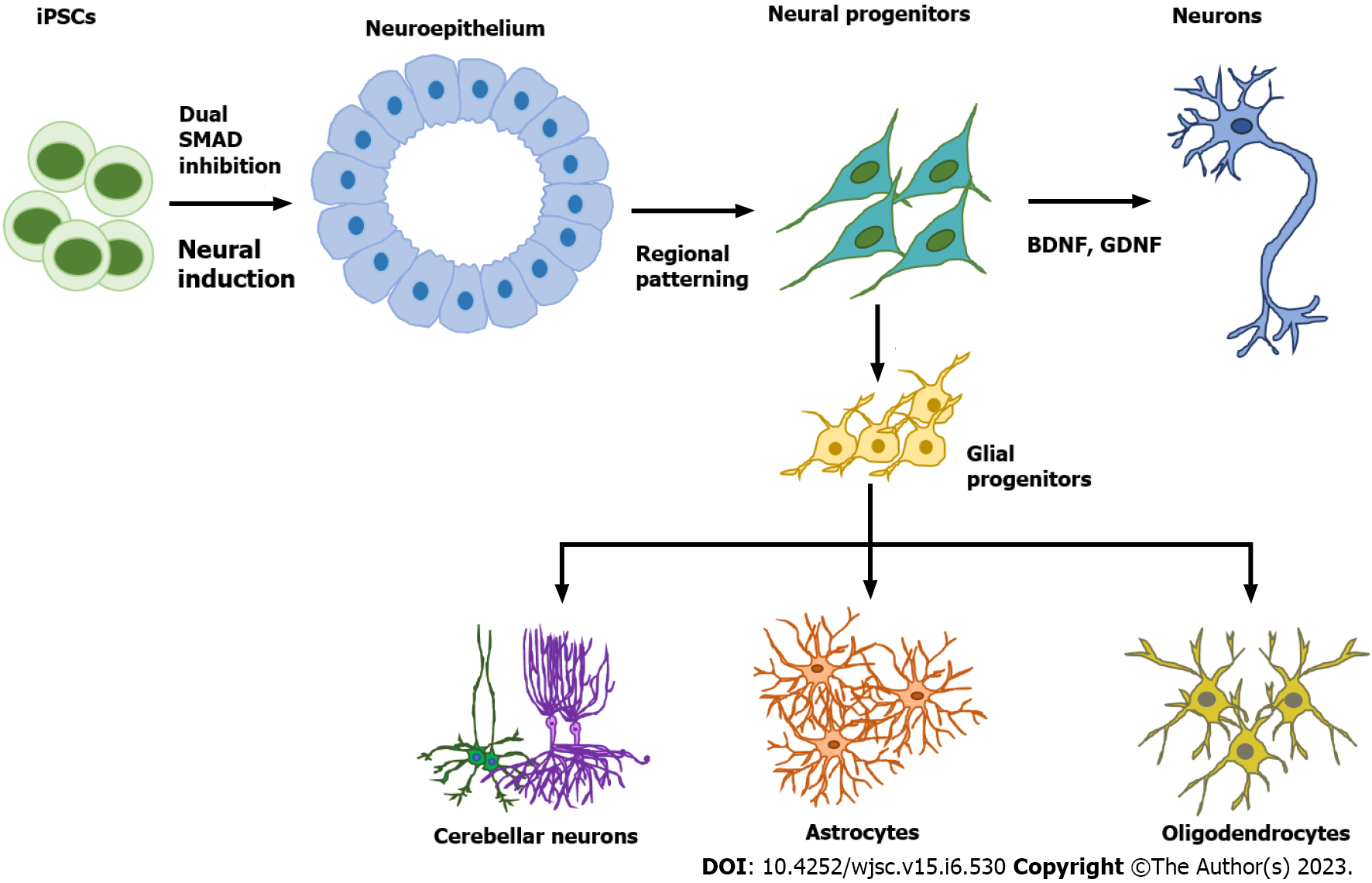
Astrocytes are star-shaped populations of glial cells that help maintain homeostatic balance and support neuron growth within the central nervous system. There are two distinct groups of astrocytes: The highly branching protoplasmic astrocytes of the grey matter and the fibrous astrocytes found in the white matter that interact with OLs and axons[28]. Activated astrocytes can release neuroinflammatory cytokines and chemokines that mediate intercellular communication with microglia and invoke various neuroinflammatory responses. Similar to neurons, there are many subtypes of astrocytes depending on their location, morphology, molecular signature, and physiological function. The differentiation of glia cells from hPSCs usually takes more time and is more complicated than differentiating a neuron (Figure 1).
During brain development, astrocytes differentiate from radial glia or neural progenitors at the subventricular zone. It is currently unknown what signaling regulates the regional identity of astrocytes. The differentiation of astrocytes is usually initiated by inhibition of dual SMAD signaling using small molecules or by the EB method to generate neuroepithelial cells. Glial progenitors expressing nuclear factor IA (NFIA), S100β, and cluster of differentiation 44 (CD44) are derived from these neuroepithelial cells[29]. Ultimately, mature astrocytes are generated from radial glia by activating the signal transducer and activator of transcription 2 signaling pathway using ciliary neurotrophic factor. The most common marker for astrocytes is glial fibrillary acidic protein (GFAP)[30]. Mature astrocytes express aldehyde dehydrogenase family 1 member L1, aldolase C, glutamate transporter-1, and aquaporin 4[31]. In 2011, the first reported protocol for hPSC-derived astrocytes in a chemically defined system required long-term culture of up to 6 mo[29,32]. This protocol used the EB method and supported differentiation through the addition of the factors FGF2 and epidermal growth factor (EGF). To attenuate the culture time, shorter 4-6 wk long accelerated protocols for generating functional astrocytes through overexpression of the transcription factors SOX9 and NFIB were developed[33,34]. In 2017, the Pasca lab found a method to derive functional astrocytes using 3D cortical organoids. However, this protocol required up to 590 d, limiting its application[30]. The majority of recent studies use commercially available astrocyte differentiation medium to differentiate astrocytes from neural progenitor cells[35,36].
Astrocytopathies including Alexander disease[37,38], Aicardi-Goutières syndrome (AGS)[39], and vanishing white matter disease[40] can be effectively modeled with hiPSC-derived astrocytes[41]. Neurodegenerative diseases including Alzheimer’s disease (AD)[36], Parkinson’s disease (PD)[42], and Huntington’s disease have also been modeled using similar methods. The familial presenilin-1 (PS1) mutation along with PD familial leucine-rich repeat kinase 2 (LRRK2) G2019S mutations were both modeled using hiPSC-derived astrocytes. The results showed the crucial role of astrocytes in the disease pathogenesis of AD and PD, respectively[36,42]. When co-cultured with neurons, astrocytes generated from Huntington’s disease patient-derived hiPSCs displayed decreased electrophysiological activity and diminished neuroprotection consistent with Huntington’s disease[43]. Regarding in vitro stroke modeling, ischemia-like conditions can be simulated by replacing normal oxygen (O2)/carbon dioxide (CO2) conditions with nitrogen (N2)/CO2 and subjecting cells to glucose deprivation[44,45]. However, cultures in 2D cannot effectively model stroke due logistical difficulties in restricting oxygenation as well as maintaining nutrition deprivation. However, Wevers et al[46] used neurovascular unit on-a-chip, which included a triculture of brain vascular cells, hiPSC-astrocytes, and hiPSC-neurons to model ischemic stroke. The study used antimycin-A, an inhibitor of complex III of the electron transport chain, to induce hypoxic conditions[45]. Modeling the motor neuron pathology linked to amyotrophic lateral sclerosis (ALS) was achieved using hiPSC-derived astrocytes from a patient who had the C9ORF72 mutation[46,47]. Recent studies also reported generation of ventral spinal cord-like astrocytes, which better reflect ALS pathophysiology[48]. Zika virus targeting of astrocytes has also been studied using hiPSC-derived astrocytes, which corroborated the reactive oxygen species imbalance, mitochondrial abnormalities, and DNA damage observed after Zika virus infection[49]. Astrocytes derived from hiPSCs are also beneficial in modeling neurodevelopment disorders including Down’s syndrome[50-53], Rett syndrome[54-57], and schizophrenia[58,59]. Rare genetic diseases such as the lysosomal storage disorder Gaucher disease can be modeled using patient hiPSC-derived astrocytes[60,61] (Figure 1, Table 1).
Protocols to differentiate hiPSCs into pre-OL progenitors were first established in 2012[62]. Retinoic acid (RA) and purmorphamine, a small-molecule agonist of Sonic Hedgehog (Shh) signaling, were used to make pre-OL progenitors that express the markers oligodendrocyte transcription factor 2 (OLIG2) and NK2 homeobox 2. Pre-OL progenitors were then further differentiated into bipotential OL progenitor cells (OPCs) that expressed markers SOX10 and platelet-derived growth factor receptor alpha (PDGFRA) using PDGF-AA, triiodothyronine, and neurotrophin-3. The OPCs at this stage were further developed into either O4+ and myelin basic protein-positive (MBP+) human-induced OLs or GFAP+ astrocytes. OPCs ameliorate neurological deterioration and support survival of shiverer mice after engraftment[62]. However, this protocol requires a lengthy 120-d culture period. Efficient and robust generation of hiPSC-derived OPCs in 95 d has been achieved more recently[63,64]. Improved differentiation of myelinating OLs was obtained using brain extracellular matrix from decellularized human brain tissue[65]. Fast and efficient OL generation has additionally been achieved with SOX10 overexpression, either by introducing lentiviral vectors at the neuroepithelial stage or by direct transfection of hiPSCs prior to differentiation[66,67].
Shaker et al[68] published a 42-d protocol to derive organoids containing myelinating human OLs and astrocytes. Differentiated OLs that were produced using hiPSCs from primary progressive multiple sclerosis patients were found to be functional and supported in vivo myelination in shiverer mice[63]. Death of OLs is a hallmark of Pelizaeus-Merzbacher disease, an X-linked leukodystrophy caused by mutations in proteolipid protein 1 (PLP1)[63]. Human-induced OLs from individuals with PLP1 mutations have helped to identify important subgroups based on cell-intrinsic phenotypes and to elucidate the pathogenesis of various PLP1 mutations[15,68]. Involvement of OLs in neurodegenerative diseases, including AD and PD as well as multiple system atrophy[69], have also been studied using human-induced OLs.
Although murine models have been the main tool for studying the genetics and function of microglia, there are important distinctions between murine microglia and human microglia when it comes to aging and associated diseases[70,71]. Historically, viable microglia cells have been obtained by extracting them from brain tumors or epileptic foci removed from surgery, but this procedure is logistically very challenging. These hurdles were reduced when multiple methods to differentiate microglia from hPSCs were developed[70-75]. Muffat et al[73] published the first protocol by producing microglia-like cells from regular and patient hESCs and hiPSCs. This method used serum-free neuroglial differentiation media, which contained various components with concentrations adjusted to biologically match human cerebrospinal fluid. Abud et al[76] described a two-step method to successfully derive microglia-like cells (iMGLs) from 10 different hiPSC lines in 5 wk. The transcriptome profile of the derived iMGLs was strikingly similar to that of both adult human and fetal microglia[75]. Most microglial directed differentiation protocols involve hematopoiesis[73,75,76]. Some reported studies use chemically-defined protocols to generate human microglia through the formation of myeloid progenitors in 30 d[77]. Ionized calcium binding adapter molecule 1, a protein that belongs to the calcium-binding protein family, is one of the main markers of microglia[78]. It is primarily involved in rearranging cytoskeleton and has been used as a marker for the 3D reconstruction of microglial cells[79,80]. Other general markers used for microglial identification are CD45 and CX3C motif chemokine receptor 1. In a recent study, Dräger et al[81] described an effective 8-d protocol for generating induced transcription factor microglia-like cells (iTF-Microglia) based on the inducible expression of six transcription factors (human MAF BZIP transcription factor B, CCAAT enhancer-binding protein, interferon regulatory factor (IRF8) PU.1, and IRF5).
The risk of developing late-onset AD is linked to several genes, including triggering receptor expressed on myeloid cells 2 (TREM2) and CD33 expressed by microglia. Microglia accumulate around amyloid plaques during AD and exacerbate pathophysiology by secreting cytokines and chemokines that induce inflammation. Microglia that have been generated using hiPSCs can be effectively used to model neurological diseases in vitro[80,81]. Alternatively, microglia derived from patient hiPSCs have also been used for modeling neurodegenerative diseases. Recently, patient hiPSCs expressing the AD-linked R47Hhet TREM2 variant was used to elucidate the signal transduction deficit observed during AD progression[82]. Another study using microglia derived from AD patient hiPSCs reported that dysregulated peroxisome proliferator-activated receptor gamma (PPARγ)/p38 MAPK signaling causes the phenotypic deficits observed in TREM2 variants. The results of this study concluded that the activation of PPARγ/p38MAPK signaling can ameliorate metabolic deficits within these cells and consequently rescue critical microglial cellular functions such as β-amyloid phagocytosis[83].
Protocols for producing brain organoids were derived from the EB and SFEBq methods for neural induction. The formation of brain organoids is based on the self-organization and self-renewal of stem cells to generate a mixed cell population in 3D suspension culture (Figure 2). In 2013, the first study on brain organoids was reported to generate whole brain tissues with regional specific structures using an EB-based culture involving Matrigel support in a spinning bioreactor[24]. The organoids were used to model microcephaly, a neurodevelopmental disease whose pathologic features are difficult to recapitulate using animal models. This was the first work done to generate brain-like tissue in vitro and apply them to study human pathological disorders. In the following years, a simpler method was developed to generate cortical spheroids in 3D static culture without Matrigel and agitation[25]. This method first derived neural progenitors using dual SMAD inhibition and then induced regional specific patterning by supplementing culture with the growth factors FGF2 and EGF. The last step of this protocol extended cultivation of brain aggregates and replaced the growth factors with the neurotrophic factors brain-derived neurotrophic factor (BDNF) and glial cell line-derived neurotrophic factor (GDNF) for up to 3 mo. The generated cortical spheroids exhibited a cortical layer-like structure and were developmentally comparable with the human fetal cortex. Brain spheroids and organoids have been widely used to model human brain development and neurological diseases in vitro and have provided a promising platform for drug screening[84] (Table 2, Figure 2).
| Organoid type or brain region modeled | Method brief description | Model application | Ref. |
| EB-like aggregates; cerebral cortex | SFEBq, static suspension culture with cell sorting | Form self-organized structure mimicking the early cortiogenesis | Eiraku et al[23], 2008 |
| Cerebral organoid; whole brain | Spinning bioreactor with Matrigel supporting | Form pyramidal identities with spatial separation mimicking the developing human brain at early stage; modeling microcephaly | Lancaster et al[17], 2013 |
| Cortical neuroepithelium; cerebral cortex | Improved SFEBq, in 40% oxygen in Lumox plates | Inside-out layer pattern for human cortex | Kadoshima et al[86], 2013 |
| Cortical spheroid; cerebral cortex | Static suspension culture with FGF-2 and EGF | Generated laminated cerebral cortex-like structure with some functions | Paşca et al[26], 2015 |
| Cerebellar-plate-like neuroepithelium; cerebellum | Static suspension culture with FGF-19 and SDF-1 | Mimicking the early development of human cerebellum | Muguruma et al[129], 2015 |
| Telencephalic organoids; forebrain | Static suspension culture after neural rosettes isolation manually | Modeling autism spectrum disorder | Mariani et al[130], 2015 |
| Dorsomedial telencephalic-like tissue; hippocampus | Improved SFEBq, in 40% oxygen | Modeling the development of human hippocampus | Sakaguchi et al[107], 2015 |
| Forebrain organoids; cerebral cortex | Miniaturized spinning bioreactor | Zika virus exposure | Qian et al[27], 2016 |
| Midbrain organoids; midbrain | Miniaturized spinning bioreactor | Midbrain organoids contained TH+ cells | Qian et al[27], 2016 |
| Hypothalamic organoids; hypothalamus | Miniaturized spinning bioreactor | Modeling early hypothalamus development | Qian et al[27], 2016 |
| Midbrain organoids; midbrain | Static suspension culture on orbital shaker | Midbrain produced neuromelanin and dopamine | Jo et al[131], 2016 |
| Pituitary organoid; anterior pituitary | Improved SFEBq | Formed pituitary placode with pituitary hormone-producing cells | Ozone et al[132], 2016 |
| Cerebral organoid; cerebral cortex | Microfilament-engineered organoids under agitation | Formed polarized cortical plate and radial units | Lancaster et al[133], 2017 |
| Cerebral organoid; whole brain | Spinning bioreactor with Matrigel supporting | Brain organoids formed spontaneously active neuronal networks | Quadrato et al[134], 2017 |
| Brain assembloids; assembly dorsal and ventral forebrain organoids | Static suspension culture | Modelling migration of human interneurons and their functional integration into microcircuits using healthy and timothy syndrome cell line | Birey et al[99], 2017 |
| Fused cerebral organoids; assembly dorsal and ventral forebrain organoids | Static suspension culture with Matrigel supporting on orbital shaker | Modelling migration of human interneurons in cerebral cortex | Birey et al[99], 2017 |
| Fused cortical organoids and MGE organoids | Static suspension culture on orbital shaker | Modelling migration of human interneurons | Xiang et al[101], 2017 |
| Neoplastic cerebral organoid | Static suspension culture with Matrigel supporting on orbital shaker | Modelling brain tumorigenesis | Bian et al[135], 2018 |
| Granted brain organoids in mouse | Spinning bioreactor | Formed functional networks and blood vessels in the grafts | Mansour et al[136], 2018 |
| Cortical spheroid | Static suspension culture | Modelling Alzheimer’s disease | Yan et al[87], 2018 |
| Cerebral organoids | Static suspension culture with Geltrex supporting on orbital shaker | Modelling Alzheimer’s disease | Gonzalez et al[93], 2018 |
| Neuromuscular organoid | Static suspension culture supporting on orbital shaker | Formed functional neuromuscular junctions and modelling myasthenia gravis | Faustion Martins et al[137], 2020 |
| Section spherical organoid | Manually slicing forebrain organoids | Sliced organoids exhibited separated upper and deep cortical layer | Qian et al[90], 2020 |
| Cortico-motor assembloids; assembly cortical spheroids, spinal spheroids, and skeletal muscle spheroids | Static suspension culture | Modeling cortical-motor circuits | Andersen et al[18], 2020 |
| Cortico-striatal assembloids; assembly cortical spheroids and striatal spheroids | Static suspension culture | Modeling cortical-striatal circuits and 22q13.3 deletion syndrome | Miura et al[102], 2020 |
| Air-liquid interface cerebral organoids | Slicing mature organoids and cultured in air-liquid interface not completely submerged in liquid | Formed network with functional output | Giandomenico et al[138], 2019 |
The areas of the brain originating from the telencephalon and diencephalon are referred to as the forebrain. The telencephalic region consists of the cerebral cortex and cerebellum, whereas the diencephalon includes the thalamus, hypothalamus, and pituitary glands. Self-organized cortical organoids or dorsal forebrain organoids were first reported using the SFEBq method. Multiple cortical layer tissues were generated through inhibition of TGF-β and Wnt signaling, resulting in dorsal-ventral patterning[22,85]. Lancaster et al[17] reported the first 3D culture system for deriving cortical organoids from hPSCs. Later, the Pasca lab generated more complex cortical spheroids and organoids from hPSCs, containing both neurons and astrocytes[25] (Figure 3). In following years, several groups attempted to develop protocols to derive cortical organoids from hPSCs. However, a common problem faced by many of these approaches was the presence of multiple ventricular subtypes within each organoid[26,86-91]. Cortical organoids derived from familial AD patient hiPSCs show increased levels of phosphorylated Tau and cytoplasmic neurofibrillary tangle-like deposits[92,93] (Table 1). Recent studies have shown that culture variations have an impact on the AD phenotypes seen in cerebral organoids and should be considered when using these models[94]. Cortical organoids were also used in stroke modeling to study the effects of oxygen-glucose deprivation (OGD), neuronal death that followed, and damaged neural networks[95]. These models use 2-8 h of OGD with O2 (0.1%), CO2 (5.0%), and N2 (95.0%) gas levels and deoxygenated glucose-free medium to induce ischemia[95]. It has also been discovered that hypoxic conditions can reduce the number of progenitors and impair the differentiation of immature neurons during the development stage of brain organoids[96,97]. Ventral forebrain tissue-like medial ganglionic eminence (MGE) organoids are usually patterned by high Shh and low Wnt signals[85,98]. These MGE organoids contain diverse GABAergic interneurons subtypes including somatostatin, parvalbumin, calretinin, and calbindin. These MGE organoids were assembled to model the migration of human interneurons towards the cerebral cortex[98-100]. In 2020, the Pasca lab reported a method to generate striatal organoids expressing medium spiny neuron markers such as DARPP32 using activin A, IWP-2, and SR11237[101]. Brain organoid technology has also been utilized to generate organoids that can model other regions of forebrain tissue including thalamic organoids[102], hypothalamic organoids[103-105], and hippocampal organoids[106,107].
The hippocampus plays a significant role in learning, memory, and emotion. Hippocampal atrophy or hyperexcitability can cause neurological disorders such as schizophrenia and neurodegenerative diseases like AD. Hippocampal spheroids can be derived from hiPSCs using dual SMAD inhibition, Shh, and Wnt pathway inhibition followed with Wnt activation[107,108]. Commonly reported hippocampal markers include zinc finger and BTB domain-containing 20 and prospero homeobox 1. Hippocampal spheroids can be used to model AD pathology either by the exogenous addition of amyloid beta 42 oligomer[107,108] or by using amyloid precursor protein/PS1 variant hiPSCs. Current hippocampal organoids reflect the early stages of embryonic hippocampus development and successfully can create dentate gyrus granule and carbonic anhydrase 3 (CA3) pyramidal-like neurons, but are unable to produce CA1 pyramidal-like neurons.
The protocol to differentiate human midbrain-like organoids (hMLOs) employs several molecules to mediate the differentiation of neuroepithelial cells. These factors include hBDNF, hGDNF, dibutyryl cyclic adenosine monophosphate, ascorbic acid, TGF-β3, and 1 purmorphamine[109,110]. The presence of dopamine transporter tyrosine hydroxylase as well as the expression of G-protein-regulated inward-rectifier potassium channel 2 are both characteristics of midbrain dopaminergic (mDA) neurons in hMLOs. Common midbrain genes including engrailed, nuclear receptor 4A2, LIM homeobox transcription factor 1 beta (LMX1B), LMX1A, monoamine oxidase B, calbindin 1, tyrosine hydroxylase, catechol-O-methyltransferase, and dopa-decarboxylase have also been detected in these organoids. Additionally, neurons in hMLOs have been found to exhibit action potentials with large sag currents, indicating the existence of mDA neurons[111] (Figure 3).
According to single cell sequencing studies, hMLOs replicate early embryonic neurodevelopment and recapitulate disease characteristics[112,113]. However, the methods to generate midbrain organoids can take a significant amount of time, and can vary from batch to batch. To scale up the generation of midbrain organoids, Mohamed et al[114] recently published microfabricated disk technology using eNUVIO EB-Disks. Another study found that the use of recombinant spider-silk microfibers functionalized with full-length human laminin produced similar ventral midbrain organoids with lower inter-organoid variability[114,115]. Alternatively, an automated approach, termed automated midbrain organoids, was published by Renner et al[116] that produced high-throughput 3D midbrain organoids. The high-throughput production of hMLOs from hPSCs in spinner flasks was also reported using TH-TdTomato reporter hPSC lines as well[116].
As the second most prevalent neurodegenerative disease worldwide[117], PD is frequently studied using hPSC-derived hMLOs[87,111,112,115,118-120]. The disease is characterized by the loss of dopaminergic neurons in the substantia nigra and is mainly caused by mutations in glucocerebrosidase and LRRK2 genes in addition to α-synuclein (α-syn; SNCA) gene triplications[111,121,122]. The hMLOs generated from patients with these mutations display PD traits such as oligomeric and fibrillar α-syn aggregates, loss of mDA neurons, and Lewy body-like inclusions[111,118,119]. Since hPSCs can be edited using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 technology, SNCA gene genome correction has been demonstrated to revert PD patient hPSCs back to wild-type phenotypes[119,123]. These hMLOs have also been successfully generated from hiPSCs carrying the LRRK2-G2019S mutation[124]. Biallelic pathogenic variations in the phosphatase and tensin homolog-induced kinase 1 gene that controls mitochondrial function is also connected to the etiology of PD[125]. Human Parkinsonism can also have a more direct cause such as the toxicity of some drugs including 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine, which has also been modeled using hMLOs[126-128] (Table 2)[129-138].
The medulla, pons, and cerebellum make up the hindbrain region, which is developed from the metencephalon and myelencephalon. Methods to generate hindbrain organoids commonly involve purmorphamine-mediated Shh signaling activation to convert neuroepithelial cells into ventral identity neurons. RA is used as a potent caudalizing agent to promote the fate of hindbrain cells instead of Wnt signaling activation, which is required for midbrain patterning[138]. Markers of hindbrain neurons include serotonergic neuron marker serotonin, human fifth Ewing variant, gastrulation brain homeobox 2, choline acetyltransferase (ChAT), and HB9. Due to their location, cerebellar neurons cannot be easily studied at the cellular or molecular level. Thus, using hiPSC-derived technologies is advantageous in this situation. The cerebellum can be divided into inhibitory GABAergic neurons known as Purkinje cells, which are derived from pancreas-specific transcription factor 1α progenitors, and excitatory glutamatergic neurons known as granule cells, which are descended from atonal homolog 1 (also known as MATH1) progenitors[139].
The first published granule cell differentiation protocol using hiPSCs involved several factors including FGF8B, Wnt proteins, BMPs, and RA. This method recapitulates anteroposterior and dorsoventral patterning and thereby induces MATH1-expressing mitotic neural progenitors, which can later be differentiated into cerebellar granule cells. Purkinje cells derived from hiPSCs initially had an immature phenotype, and thus needed to be co-cultured with mouse cerebellar granule cell precursors to allow for maturation[129,140]. However, cells made using this approach had substantial functional variability. Silva et al[141] recently published a protocol that generates mature cerebellar neurons without the need of such a co-culture system. This method involves stimulating the development of cerebellar precursors with FGF19, followed by self-organization and differentiation using stromal cell-derived factor 1 and BDNF/GDNF. Cerebellar neurons derived from hiPSCs are also helpful for modeling diseases, particularly cerebellar ataxia, a neurodegenerative disease that affects cerebellar neurons and eventually leads to motor incoordination. Cerebellar neurons derived from hiPSCs of either healthy human participants or ataxia patients were used in several recent studies to create an in vitro disease model. Spinocerebellar ataxia type 6 patient hiPSC-derived Purkinje cells have been used to model both thyroid hormone depletion-dependent degeneration and downregulation of the transcriptional targets TATA-Box Binding Protein Associated Factor 1 and BTG anti-proliferation factor 1, indicating their potential as a pathogenesis tool[141].
Recently, protocols for generating brain stem organoids have also been published, offering a new tool for evaluating the pathophysiology of disorders that impact the brainstem. Human brain stem organoids express the medullary marker ChAT, the pons marker hydroxylase, and the mature and functioning excitatory and inhibitory neuron markers vesicular glutamate transporter 1 and glutamic acid decarboxylase 67 in addition to various other relevant markers[142]. Both OLIG2+ and MBP+ OLs, as well as S100+ astrocytes, are expressed in brain stem organoids[142].
Assembloids are systems that combine one type of spheroid or organoid with another type of spheroid or organoid. For example, assembloids can be produced by combining the dorsal and ventral forebrain, the cerebral cortex with the thalamus, or the cerebral cortex with any other non-neural cell type such as microglia, immunological cells, pericytes, and endothelial cells. The Pasca lab published the first assembloid study in 2017, in which human cortical spheroids were mixed with human subpallium spheroids[98]. The assembloids were created using human subpallium spheroids and cortical spheroids differentiated from hiPSCs from timothy syndrome (TS) patients with mutations in the α1c subunit of the L-type calcium channel (CACNA1C) gene. The two types of spheroids were combined in simple conical tubes and left undisturbed for 3 d to produce assembloids. The interneurons within the assembloids migrated, suggesting high potential for the study of certain aspects of migratory disorders such as TS. These patient-derived assembloids showed less effective interneuron movement, which was reversed by the administration of L-type calcium channel blockers.
Since then, many labs have sought to use assembloids to elucidate the interactions that occur between different physiological systems. The Knoblich lab reported the use of fused cerebral organoids that combined dorsal and ventral forebrain tissue cultures. They showed migration of C-X-C chemokine receptor type 4-dependent GABAergic interneurons from the ventral forebrain to the dorsal forebrain, which had a more MGE identity[143]. Assembloids of cortical organoids with integrated pericyte-like cells which express angiotensin-converting enzyme 2 have also been shown to enhance severe acute respiratory syndrome coronavirus 2 infection, suggesting the involvement of multiple cell types[100]. Another study employed cortico-striatal assembloids to recapitulate neurodevelopmental disorders that impair the cortico-striatal pathway, including schizophrenia, obsessive-compulsive disorder, and autism spectrum disorder[144-146]. These cortico-striatal assembloids were developed from patients with Phelan-McDermid Syndrome, a severe developmental disorder also known as 22q13.3DS. It is important to note that these patient-derived assembloids had a higher number of calcium spike events than striatal organoids, offering a better representation of altered neural activity. Interneuron migration has also been reported in a separate assembloids study that fused human MGE organoids with human cortical organoids[147].
The extended culture times required by current methods to produce neural cell types as well as organoids restrict their application. Another consideration is the cell-line-to-cell-line and batch-to-batch variabilities of hiPSC differentiation. Therefore, accelerated protocols with less variable outcomes should be developed. For hiPSC-derived astrocytes, the major drawback is their lack of regional identity. Most protocols derive astrocytes with cortical identity, which may not be useful for modeling disease pathophysiology affecting the ventral part of the brain. Therefore, it is essential to employ experimental approaches that can produce astrocyte subtypes with the appropriate rostro-caudal and dorso-ventral identities. In the case of hiPSC-derived OLs, the lack of advanced OL disease models created using genetically-modified hiPSCs also limits their application. Finally, the lack of vascularization in current organoid and assembloid systems prevents the important study of cell-type crosstalk. Therefore, incorporating vasculature as well as reducing culture time would benefit multiple methods of neural lineage disease modeling.
Research in hPSCs has proven to be extremely helpful in creating disease models that can corroborate results gleaned from animal models and overcome their associated limitations. Distinct brain cell types can be produced using hPSCs including neurons, astrocytes, OLs, microglia, in addition to more advanced heterogeneous systems such as brain organoids. These systems have contributed to the development of models for neurological diseases such as AD, PD, and many others. Current models that employ hPSCs have certain shortcomings related to the absence of vasculature as well as microglia. However, developing research in the field of tissue engineering that use cocultures, organ-on-chip and assembloids may be able to get around these limitations in the years to come.
The VA Tennessee Valley Healthcare System provided resources and facilities for the research that was reported in this article. Images were obtained with the resources of the Vanderbilt Cell Imaging Shared Resource.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Neurosciences
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bagheri-Mohammadi S, Iran; Liu QS, China S-Editor: Chen YL L-Editor: Filipodia P-Editor: Liu JH
| 1. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14312] [Article Influence: 841.9] [Reference Citation Analysis (0)] |
| 2. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18186] [Article Influence: 957.2] [Reference Citation Analysis (0)] |
| 3. | Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329-2340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 1127] [Article Influence: 102.5] [Reference Citation Analysis (0)] |
| 4. | Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, Little MH. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 904] [Cited by in RCA: 1076] [Article Influence: 107.6] [Reference Citation Analysis (0)] |
| 5. | Wang Y, Wang H, Deng P, Chen W, Guo Y, Tao T, Qin J. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip. 2018;18:3606-3616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 138] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 6. | Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1763] [Cited by in RCA: 1515] [Article Influence: 126.3] [Reference Citation Analysis (0)] |
| 7. | Dye BR, Hill DR, Ferguson MA, Tsai YH, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, Deutsch GH, Spence JR. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 456] [Cited by in RCA: 565] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 8. | Hohwieler M, Illing A, Hermann PC, Mayer T, Stockmann M, Perkhofer L, Eiseler T, Antony JS, Müller M, Renz S, Kuo CC, Lin Q, Sendler M, Breunig M, Kleiderman SM, Lechel A, Zenker M, Leichsenring M, Rosendahl J, Zenke M, Sainz B Jr, Mayerle J, Costa IG, Seufferlein T, Kormann M, Wagner M, Liebau S, Kleger A. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017;66:473-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 163] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 9. | Gerakis Y, Hetz C. Brain organoids: a next step for humanized Alzheimer’s disease models? Mol Psychiatry. 2019;24:474-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 10. | Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1502] [Cited by in RCA: 1913] [Article Influence: 173.9] [Reference Citation Analysis (0)] |
| 11. | Shimizu T, Mae SI, Araoka T, Okita K, Hotta A, Yamagata K, Osafune K. A novel ADPKD model using kidney organoids derived from disease-specific human iPSCs. Biochem Biophys Res Commun. 2020;529:1186-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 12. | Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH. Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS One. 2016;11:e0161969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 382] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 13. | Li R, Sun L, Fang A, Li P, Wu Q, Wang X. Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell. 2017;8:823-833. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 14. | Dolmetsch R, Geschwind DH. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 247] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 15. | Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, Ming GL, Song H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol Brain. 2012;5:17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 183] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 16. | Douvaras P, Wang J, Zimmer M, Hanchuk S, O’Bara MA, Sadiq S, Sim FJ, Goldman J, Fossati V. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports. 2014;3:250-259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 237] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 17. | Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373-379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3083] [Cited by in RCA: 3520] [Article Influence: 293.3] [Reference Citation Analysis (0)] |
| 18. | Andersen J, Revah O, Miura Y, Thom N, Amin ND, Kelley KW, Singh M, Chen X, Thete MV, Walczak EM, Vogel H, Fan HC, Paşca SP. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell. 2020;183:1913-1929.e26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 327] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 19. | Muñoz-Sanjuán I, Brivanlou AH. Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci. 2002;3:271-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 446] [Cited by in RCA: 439] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 20. | Stern CD. Neural induction: old problem, new findings, yet more questions. Development. 2005;132:2007-2021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 292] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 21. | Suzuki IK, Vanderhaeghen P. Is this a brain which I see before me? Development. 2015;142:3138-3150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 22. | Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1451] [Cited by in RCA: 1347] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 23. | Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 951] [Cited by in RCA: 1083] [Article Influence: 67.7] [Reference Citation Analysis (1)] |
| 24. | Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3036] [Cited by in RCA: 2677] [Article Influence: 167.3] [Reference Citation Analysis (0)] |
| 25. | Hattori N. Cerebral organoids model human brain development and microcephaly. Mov Disord. 2014;29:185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Paşca SP. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671-678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1174] [Cited by in RCA: 1095] [Article Influence: 109.5] [Reference Citation Analysis (0)] |
| 27. | Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1610] [Cited by in RCA: 1545] [Article Influence: 171.7] [Reference Citation Analysis (0)] |
| 28. | Qi Y, Zhang XJ, Renier N, Wu Z, Atkin T, Sun Z, Ozair MZ, Tchieu J, Zimmer B, Fattahi F, Ganat Y, Azevedo R, Zeltner N, Brivanlou AH, Karayiorgou M, Gogos J, Tomishima M, Tessier-Lavigne M, Shi SH, Studer L. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol. 2017;35:154-163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 29. | Tabata H. Diverse subtypes of astrocytes and their development during corticogenesis. Front Neurosci. 2015;9:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 30. | Krencik R, Weick JP, Liu Y, Zhang ZJ, Zhang SC. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat Biotechnol. 2011;29:528-534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 358] [Cited by in RCA: 315] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 31. | Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, Paşca SP. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron. 2017;95:779-790.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 408] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 32. | Jurga AM, Paleczna M, Kadluczka J, Kuter KZ. Beyond the GFAP-Astrocyte Protein Markers in the Brain. Biomolecules. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 160] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 33. | Krencik R, Zhang SC. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat Protoc. 2011;6:1710-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 203] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 34. | Li X, Tao Y, Bradley R, Du Z, Kong L, Dong Y, Jones J, Yan Y, Harder CRK, Friedman LM, Bilal M, Hoffmann B, Zhang SC. Fast Generation of Functional Subtype Astrocytes from Human Pluripotent Stem Cells. Stem Cell Reports. 2018;11:998-1008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 35. | Canals I, Ginisty A, Quist E, Timmerman R, Fritze J, Miskinyte G, Monni E, Hansen MG, Hidalgo I, Bryder D, Bengzon J, Ahlenius H. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods. 2018;15:693-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 149] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 36. | Soubannier V, Maussion G, Chaineau M, Sigutova V, Rouleau G, Durcan TM, Stifani S. Characterization of human iPSC-derived astrocytes with potential for disease modeling and drug discovery. Neurosci Lett. 2020;731:135028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 37. | Brezovakova V, Sykova E, Jadhav S. Astrocytes Derived from Familial and Sporadic Alzheimer’s Disease iPSCs Show Altered Calcium Signaling and Respond Differently to Misfolded Protein Tau. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 38. | Kondo T, Funayama M, Miyake M, Tsukita K, Era T, Osaka H, Ayaki T, Takahashi R, Inoue H. Modeling Alexander disease with patient iPSCs reveals cellular and molecular pathology of astrocytes. Acta Neuropathol Commun. 2016;4:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 39. | Li L, Tian E, Chen X, Chao J, Klein J, Qu Q, Sun G, Huang Y, Warden CD, Ye P, Feng L, Li X, Cui Q, Sultan A, Douvaras P, Fossati V, Sanjana NE, Riggs AD, Shi Y. GFAP Mutations in Astrocytes Impair Oligodendrocyte Progenitor Proliferation and Myelination in an hiPSC Model of Alexander Disease. Cell Stem Cell. 2018;23:239-251.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 40. | Thomas CA, Tejwani L, Trujillo CA, Negraes PD, Herai RH, Mesci P, Macia A, Crow YJ, Muotri AR. Modeling of TREX1-Dependent Autoimmune Disease using Human Stem Cells Highlights L1 Accumulation as a Source of Neuroinflammation. Cell Stem Cell. 2017;21:319-331.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 239] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 41. | Zhou L, Li P, Chen N, Dai LF, Gao K, Liu YN, Shen L, Wang JM, Jiang YW, Wu Y. Modeling vanishing white matter disease with patient-derived induced pluripotent stem cells reveals astrocytic dysfunction. CNS Neurosci Ther. 2019;25:759-771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 42. | Lanciotti A, Brignone MS, Macioce P, Visentin S, Ambrosini E. Human iPSC-Derived Astrocytes: A Powerful Tool to Study Primary Astrocyte Dysfunction in the Pathogenesis of Rare Leukodystrophies. Int J Mol Sci. 2021;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | di Domenico A, Carola G, Calatayud C, Pons-Espinal M, Muñoz JP, Richaud-Patin Y, Fernandez-Carasa I, Gut M, Faella A, Parameswaran J, Soriano J, Ferrer I, Tolosa E, Zorzano A, Cuervo AM, Raya A, Consiglio A. Patient-Specific iPSC-Derived Astrocytes Contribute to Non-Cell-Autonomous Neurodegeneration in Parkinson’s Disease. Stem Cell Reports. 2019;12:213-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 182] [Cited by in RCA: 259] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 44. | Garcia VJ, Rushton DJ, Tom CM, Allen ND, Kemp PJ, Svendsen CN, Mattis VB. Huntington’s Disease Patient-Derived Astrocytes Display Electrophysiological Impairments and Reduced Neuronal Support. Front Neurosci. 2019;13:669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 45. | Holloway PM, Gavins FN. Modeling Ischemic Stroke In Vitro: Status Quo and Future Perspectives. Stroke. 2016;47:561-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 92] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 46. | Wevers NR, Nair AL, Fowke TM, Pontier M, Kasi DG, Spijkers XM, Hallard C, Rabussier G, van Vught R, Vulto P, de Vries HE, Lanz HL. Modeling ischemic stroke in a triculture neurovascular unit on-a-chip. Fluids Barriers CNS. 2021;18:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 47. | Zhao C, Devlin AC, Chouhan AK, Selvaraj BT, Stavrou M, Burr K, Brivio V, He X, Mehta AR, Story D, Shaw CE, Dando O, Hardingham GE, Miles GB, Chandran S. Mutant C9orf72 human iPSC-derived astrocytes cause non-cell autonomous motor neuron pathophysiology. Glia. 2020;68:1046-1064. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 48. | Birger A, Ben-Dor I, Ottolenghi M, Turetsky T, Gil Y, Sweetat S, Perez L, Belzer V, Casden N, Steiner D, Izrael M, Galun E, Feldman E, Behar O, Reubinoff B. Human iPSC-derived astrocytes from ALS patients with mutated C9ORF72 show increased oxidative stress and neurotoxicity. EBioMedicine. 2019;50:274-289. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 128] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 49. | Soubannier V, Chaineau M, Gursu L, Haghi G, Franco Flores AK, Rouleau G, Durcan TM, Stifani S. Rapid Generation of Ventral Spinal Cord-like Astrocytes from Human iPSCs for Modeling Non-Cell Autonomous Mechanisms of Lower Motor Neuron Disease. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 50. | Ledur PF, Karmirian K, Pedrosa CDSG, Souza LRQ, Assis-de-Lemos G, Martins TM, Ferreira JCCG, de Azevedo Reis GF, Silva ES, Silva D, Salerno JA, Ornelas IM, Devalle S, Madeiro da Costa RF, Goto-Silva L, Higa LM, Melo A, Tanuri A, Chimelli L, Murata MM, Garcez PP, Filippi-Chiela EC, Galina A, Borges HL, Rehen SK. Zika virus infection leads to mitochondrial failure, oxidative stress and DNA damage in human iPSC-derived astrocytes. Sci Rep. 2020;10:1218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 99] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 51. | Araujo BHS, Kaid C, De Souza JS, Gomes da Silva S, Goulart E, Caires LCJ, Musso CM, Torres LB, Ferrasa A, Herai R, Zatz M, Okamoto OK, Cavalheiro EA. Down Syndrome iPSC-Derived Astrocytes Impair Neuronal Synaptogenesis and the mTOR Pathway In Vitro. Mol Neurobiol. 2018;55:5962-5975. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 52. | Chen C, Jiang P, Xue H, Peterson SE, Tran HT, McCann AE, Parast MM, Li S, Pleasure DE, Laurent LC, Loring JF, Liu Y, Deng W. Role of astroglia in Down’s syndrome revealed by patient-derived human-induced pluripotent stem cells. Nat Commun. 2014;5:4430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 175] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 53. | Mizuno GO, Wang Y, Shi G, Sun J, Papadopoulos S, Broussard GJ, Unger EK, Deng W, Weick J, Bhattacharyya A, Chen CY, Yu G, Looger LL, Tian L. Aberrant Calcium Signaling in Astrocytes Inhibits Neuronal Excitability in a Human Down Syndrome Stem Cell Model. Cell Rep. 2018;24:355-365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 54. | Ponroy Bally B, Farmer WT, Jones EV, Jessa S, Kacerovsky JB, Mayran A, Peng H, Lefebvre JL, Drouin J, Hayer A, Ernst C, Murai KK. Human iPSC-derived Down syndrome astrocytes display genome-wide perturbations in gene expression, an altered adhesion profile, and increased cellular dynamics. Hum Mol Genet. 2020;29:785-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 55. | Williams EC, Zhong X, Mohamed A, Li R, Liu Y, Dong Q, Ananiev GE, Mok JC, Lin BR, Lu J, Chiao C, Cherney R, Li H, Zhang SC, Chang Q. Mutant astrocytes differentiated from Rett syndrome patients-specific iPSCs have adverse effects on wild-type neurons. Hum Mol Genet. 2014;23:2968-2980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 152] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 56. | Yasui T, Uezono N, Nakashima H, Noguchi H, Matsuda T, Noda-Andoh T, Okano H, Nakashima K. Hypoxia Epigenetically Confers Astrocytic Differentiation Potential on Human Pluripotent Cell-Derived Neural Precursor Cells. Stem Cell Reports. 2017;8:1743-1756. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 57. | Andoh-Noda T, Akamatsu W, Miyake K, Matsumoto T, Yamaguchi R, Sanosaka T, Okada Y, Kobayashi T, Ohyama M, Nakashima K, Kurosawa H, Kubota T, Okano H. Differentiation of multipotent neural stem cells derived from Rett syndrome patients is biased toward the astrocytic lineage. Mol Brain. 2015;8:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 58. | Kim JJ, Savas JN, Miller MT, Hu X, Carromeu C, Lavallée-Adam M, Freitas BCG, Muotri AR, Yates JR 3rd, Ghosh A. Proteomic analyses reveal misregulation of LIN28 expression and delayed timing of glial differentiation in human iPS cells with MECP2 Loss-of-function. PLoS One. 2019;14:e0212553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 59. | Liu Z, Osipovitch M, Benraiss A, Huynh NPT, Foti R, Bates J, Chandler-Militello D, Findling RL, Tesar PJ, Nedergaard M, Windrem MS, Goldman SA. Dysregulated Glial Differentiation in Schizophrenia May Be Relieved by Suppression of SMAD4- and REST-Dependent Signaling. Cell Rep. 2019;27:3832-3843.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 60. | Akkouh IA, Ueland T, Hansson L, Inderhaug E, Hughes T, Steen NE, Aukrust P, Andreassen OA, Szabo A, Djurovic S. Decreased IL-1β-induced CCL20 response in human iPSC-astrocytes in schizophrenia: Potential attenuating effects on recruitment of regulatory T cells. Brain Behav Immun. 2020;87:634-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 61. | Aflaki E, Stubblefield BK, McGlinchey RP, McMahon B, Ory DS, Sidransky E. A characterization of Gaucher iPS-derived astrocytes: Potential implications for Parkinson’s disease. Neurobiol Dis. 2020;134:104647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 62. | Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477-486, S1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 659] [Cited by in RCA: 608] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 63. | Wang S, Bates J, Li X, Schanz S, Chandler-Militello D, Levine C, Maherali N, Studer L, Hochedlinger K, Windrem M, Goldman SA. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 452] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 64. | Nobuta H, Yang N, Ng YH, Marro SG, Sabeur K, Chavali M, Stockley JH, Killilea DW, Walter PB, Zhao C, Huie P Jr, Goldman SA, Kriegstein AR, Franklin RJM, Rowitch DH, Wernig M. Oligodendrocyte Death in Pelizaeus-Merzbacher Disease Is Rescued by Iron Chelation. Cell Stem Cell. 2019;25:531-541.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 65. | Douvaras P, Fossati V. Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat Protoc. 2015;10:1143-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 180] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 66. | Cho AN, Jin Y, Kim S, Kumar S, Shin H, Kang HC, Cho SW. Aligned Brain Extracellular Matrix Promotes Differentiation and Myelination of Human-Induced Pluripotent Stem Cell-Derived Oligodendrocytes. ACS Appl Mater Interfaces. 2019;11:15344-15353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 67. | Neyrinck K, García-León JA. Single Transcription Factor-Based Differentiation Allowing Fast and Efficient Oligodendrocyte Generation via SOX10 Overexpression. Methods Mol Biol. 2021;2352:149-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 68. | Shaker MR, Pietrogrande G, Martin S, Lee JH, Sun W, Wolvetang EJ. Rapid and Efficient Generation of Myelinating Human Oligodendrocytes in Organoids. Front Cell Neurosci. 2021;15:631548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 69. | Numasawa-Kuroiwa Y, Okada Y, Shibata S, Kishi N, Akamatsu W, Shoji M, Nakanishi A, Oyama M, Osaka H, Inoue K, Takahashi K, Yamanaka S, Kosaki K, Takahashi T, Okano H. Involvement of ER stress in dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes. Stem Cell Reports. 2014;2:648-661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 94] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 70. | Azevedo C, Teku G, Pomeshchik Y, Reyes JF, Chumarina M, Russ K, Savchenko E, Hammarberg A, Lamas NJ, Collin A, Gouras GK, Klementieva O, Hallbeck M, Taipa R, Vihinen M, Roybon L. Parkinson’s disease and multiple system atrophy patient iPSC-derived oligodendrocytes exhibit alpha-synuclein-induced changes in maturation and immune reactive properties. Proc Natl Acad Sci U S A. 2022;119:e2111405119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 26] [Reference Citation Analysis (0)] |
| 71. | Friedman BA, Srinivasan K, Ayalon G, Meilandt WJ, Lin H, Huntley MA, Cao Y, Lee SH, Haddick PCG, Ngu H, Modrusan Z, Larson JL, Kaminker JS, van der Brug MP, Hansen DV. Diverse Brain Myeloid Expression Profiles Reveal Distinct Microglial Activation States and Aspects of Alzheimer’s Disease Not Evident in Mouse Models. Cell Rep. 2018;22:832-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 461] [Article Influence: 76.8] [Reference Citation Analysis (0)] |
| 72. | Ueda Y, Gullipalli D, Song WC. Modeling complement-driven diseases in transgenic mice: Values and limitations. Immunobiology. 2016;221:1080-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 73. | Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai LH, Aubourg P, Ransohoff RM, Jaenisch R. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med. 2016;22:1358-1367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 471] [Cited by in RCA: 488] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 74. | Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, Chintawar S, Schnell C, Antel JP, Allen ND, Cader MZ, Wade-Martins R, James WS, Cowley SA. A Highly Efficient Human Pluripotent Stem Cell Microglia Model Displays a Neuronal-Co-culture-Specific Expression Profile and Inflammatory Response. Stem Cell Reports. 2017;8:1727-1742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 357] [Cited by in RCA: 366] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 75. | Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, Chakarov S, See P, Low D, Low G, Garcia-Miralles M, Zeng R, Zhang J, Goh CC, Gul A, Hubert S, Lee B, Chen J, Low I, Shadan NB, Lum J, Wei TS, Mok E, Kawanishi S, Kitamura Y, Larbi A, Poidinger M, Renia L, Ng LG, Wolf Y, Jung S, Önder T, Newell E, Huber T, Ashihara E, Garel S, Pouladi MA, Ginhoux F. Induced-Pluripotent-Stem-Cell-Derived Primitive Macrophages Provide a Platform for Modeling Tissue-Resident Macrophage Differentiation and Function. Immunity. 2017;47:183-198.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 255] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 76. | Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, Caraway CA, Fote GM, Madany AM, Agrawal A, Kayed R, Gylys KH, Cahalan MD, Cummings BJ, Antel JP, Mortazavi A, Carson MJ, Poon WW, Blurton-Jones M. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017;94:278-293.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 769] [Article Influence: 96.1] [Reference Citation Analysis (0)] |
| 77. | Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, Kim G, Brown MA, Elkahloun AG, Maric D, Sweeney CL, Gossa S, Malech HL, McGavern DB, Park JK. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci. 2017;20:753-759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 289] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 78. | Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, Terrenoire C, Zhang B, Gandy S, Schadt E, Freytes DO, Noggle S, Fossati V. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Reports. 2017;8:1516-1524. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 263] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 79. | McQuade A, Blurton-Jones M. Human Induced Pluripotent Stem Cell-Derived Microglia (hiPSC-Microglia). Methods Mol Biol. 2022;2454:473-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 80. | Bejoy J, Yuan X, Song L, Hua T, Jeske R, Sart S, Sang QA, Li Y. Genomics Analysis of Metabolic Pathways of Human Stem Cell-Derived Microglia-Like Cells and the Integrated Cortical Spheroids. Stem Cells Int. 2019;2019:2382534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 81. | Dräger NM, Sattler SM, Huang CT, Teter OM, Leng K, Hashemi SH, Hong J, Aviles G, Clelland CD, Zhan L, Udeochu JC, Kodama L, Singleton AB, Nalls MA, Ichida J, Ward ME, Faghri F, Gan L, Kampmann M. A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat Neurosci. 2022;25:1149-1162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 134] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 82. | Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, Kohsaka S. Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res Mol Brain Res. 1998;57:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 1195] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 83. | Andreone BJ, Przybyla L, Llapashtica C, Rana A, Davis SS, van Lengerich B, Lin K, Shi J, Mei Y, Astarita G, Di Paolo G, Sandmann T, Monroe KM, Lewcock JW. Alzheimer’s-associated PLCγ2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. Nat Neurosci. 2020;23:927-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 158] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 84. | McQuade A, Blurton-Jones M. Microglia in Alzheimer’s Disease: Exploring How Genetics and Phenotype Influence Risk. J Mol Biol. 2019;431:1805-1817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 155] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 85. | Tan HY, Cho H, Lee LP. Human mini-brain models. Nat Biomed Eng. 2021;5:11-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 86. | Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci USA. 2013;110:20284-20289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 715] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 87. | Yan Y, Song L, Bejoy J, Zhao J, Kanekiyo T, Bu G, Zhou Y, Li Y. Modeling Neurodegenerative Microenvironment Using Cortical Organoids Derived from Human Stem Cells. Tissue Eng Part A. 2018;24:1125-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 59] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 88. | Boreström C, Jonebring A, Guo J, Palmgren H, Cederblad L, Forslöw A, Svensson A, Söderberg M, Reznichenko A, Nyström J, Patrakka J, Hicks R, Maresca M, Valastro B, Collén A. A CRISP(e)R view on kidney organoids allows generation of an induced pluripotent stem cell-derived kidney model for drug discovery. Kidney Int. 2018;94:1099-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 89. | Yan Y, Song L, Tsai AC, Ma T, Li Y. Generation of Neural Progenitor Spheres from Human Pluripotent Stem Cells in a Suspension Bioreactor. Methods Mol Biol. 2016;1502:119-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 90. | Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, Su K, Li S, Lu L, Jacob F, Nguyen PTT, Huh S, Hoke A, Swinford-Jackson SE, Wen Z, Gu X, Pierce RC, Wu H, Briand LA, Chen HI, Wolf JA, Song H, Ming GL. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell. 2020;26:766-781.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 304] [Article Influence: 60.8] [Reference Citation Analysis (0)] |
| 91. | Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, Domissy A, Vandenberghe M, Devor A, Yeo GW, Voytek B, Muotri AR. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell. 2019;25:558-569.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 531] [Cited by in RCA: 514] [Article Influence: 85.7] [Reference Citation Analysis (0)] |
| 92. | Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J, Valencia AM, Horvath S, Xiao X, Huguenard JR, Pașca SP, Geschwind DH. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat Neurosci. 2021;24:331-342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 249] [Cited by in RCA: 229] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 93. | Gonzalez C, Armijo E, Bravo-Alegria J, Becerra-Calixto A, Mays CE, Soto C. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol Psychiatry. 2018;23:2363-2374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 261] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 94. | Alić I, Goh PA, Murray A, Portelius E, Gkanatsiou E, Gough G, Mok KY, Koschut D, Brunmeir R, Yeap YJ, O’Brien NL, Groet J, Shao X, Havlicek S, Dunn NR, Kvartsberg H, Brinkmalm G, Hithersay R, Startin C, Hamburg S, Phillips M, Pervushin K, Turmaine M, Wallon D, Rovelet-Lecrux A, Soininen H, Volpi E, Martin JE, Foo JN, Becker DL, Rostagno A, Ghiso J, Krsnik Ž, Šimić G, Kostović I, Mitrečić D; LonDownS Consortium, Francis PT, Blennow K, Strydom A, Hardy J, Zetterberg H, Nižetić D. Patient-specific Alzheimer-like pathology in trisomy 21 cerebral organoids reveals BACE2 as a gene dose-sensitive AD suppressor in human brain. Mol Psychiatry. 2021;26:5766-5788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 95. | Hernández D, Rooney LA, Daniszewski M, Gulluyan L, Liang HH, Cook AL, Hewitt AW, Pébay A. Culture Variabilities of Human iPSC-Derived Cerebral Organoids Are a Major Issue for the Modelling of Phenotypes Observed in Alzheimer’s Disease. Stem Cell Rev Rep. 2022;18:718-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 96. | De Paola M, Pischiutta F, Comolli D, Mariani A, Kelk J, Lisi I, Cerovic M, Fumagalli S, Forloni G, Zanier ER. Neural cortical organoids from self-assembling human iPSC as a model to investigate neurotoxicity in brain ischemia. J Cereb Blood Flow Metab. 2023;43:680-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 97. | Boisvert EM, Means RE, Michaud M, Madri JA, Katz SG. Minocycline mitigates the effect of neonatal hypoxic insult on human brain organoids. Cell Death Dis. 2019;10:325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 98. | Pașca AM, Park JY, Shin HW, Qi Q, Revah O, Krasnoff R, O’Hara R, Willsey AJ, Palmer TD, Pașca SP. Human 3D cellular model of hypoxic brain injury of prematurity. Nat Med. 2019;25:784-791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 99. | Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O’Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Paşca SP. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 684] [Cited by in RCA: 907] [Article Influence: 113.4] [Reference Citation Analysis (0)] |
| 100. | Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA. Fused cerebral organoids model interactions between brain regions. Nat Methods. 2017;14:743-751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 538] [Article Influence: 67.3] [Reference Citation Analysis (0)] |
| 101. | Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, Zhong M, Stanley EG, Elefanty AG, Naegele JR, Lee SH, Weissman SM, Park IH. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell. 2017;21:383-398.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 477] [Article Influence: 59.6] [Reference Citation Analysis (0)] |
| 102. | Miura Y, Li MY, Birey F, Ikeda K, Revah O, Thete MV, Park JY, Puno A, Lee SH, Porteus MH, Pașca SP. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat Biotechnol. 2020;38:1421-1430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 245] [Cited by in RCA: 251] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 103. | Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, Lee SH, Weissman SM, Park IH. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell. 2019;24:487-497.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 326] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 104. | Wataya T, Ando S, Muguruma K, Ikeda H, Watanabe K, Eiraku M, Kawada M, Takahashi J, Hashimoto N, Sasai Y. Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc Natl Acad Sci U S A. 2008;105:11796-11801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 204] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 105. | Merkle FT, Maroof A, Wataya T, Sasai Y, Studer L, Eggan K, Schier AF. Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development. 2015;142:633-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 103] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 106. | Huang WK, Wong SZH, Pather SR, Nguyen PTT, Zhang F, Zhang DY, Zhang Z, Lu L, Fang W, Chen L, Fernandes A, Su Y, Song H, Ming GL. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell. 2021;28:1657-1670.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 108] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 107. | Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, Takahashi J, Eiraku M, Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 2015;6:8896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 292] [Cited by in RCA: 378] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 108. | Pomeshchik Y, Klementieva O, Gil J, Martinsson I, Hansen MG, de Vries T, Sancho-Balsells A, Russ K, Savchenko E, Collin A, Vaz AR, Bagnoli S, Nacmias B, Rampon C, Sorbi S, Brites D, Marko-Varga G, Kokaia Z, Rezeli M, Gouras GK, Roybon L. Human iPSC-Derived Hippocampal Spheroids: An Innovative Tool for Stratifying Alzheimer Disease Patient-Specific Cellular Phenotypes and Developing Therapies. Stem Cell Reports. 2020;15:256-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 109. | Bejoy J, Song L, Wang Z, Sang QX, Zhou Y, Li Y. Neuroprotective Activities of Heparin, Heparinase III, and Hyaluronic Acid on the Aβ42-Treated Forebrain Spheroids Derived from Human Stem Cells. ACS Biomater Sci Eng. 2018;4:2922-2933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 110. | Sozzi E, Nilsson F, Kajtez J, Parmar M, Fiorenzano A. Generation of Human Ventral Midbrain Organoids Derived from Pluripotent Stem Cells. Curr Protoc. 2022;2:e555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 111. | Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, Jarazo J, Walter J, Brüggemann I, Boussaad I, Berger E, Fleming RMT, Bolognin S, Schwamborn JC. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports. 2017;8:1144-1154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 300] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 112. | Jo J, Yang L, Tran HD, Yu W, Sun AX, Chang YY, Jung BC, Lee SJ, Saw TY, Xiao B, Khoo ATT, Yaw LP, Xie JJ, Lokman H, Ong WY, Lim GGY, Lim KL, Tan EK, Ng HH, Je HS. Lewy Body-like Inclusions in Human Midbrain Organoids Carrying Glucocerebrosidase and α-Synuclein Mutations. Ann Neurol. 2021;90:490-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 113. | Zagare A, Barmpa K, Smajic S, Smits LM, Grzyb K, Grünewald A, Skupin A, Nickels SL, Schwamborn JC. Midbrain organoids mimic early embryonic neurodevelopment and recapitulate LRRK2-p.Gly2019Ser-associated gene expression. Am J Hum Genet. 2022;109:311-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 114. | Mohamed NV, Lépine P, Lacalle-Aurioles M, Sirois J, Mathur M, Reintsch W, Beitel LK, Fon EA, Durcan TM. Microfabricated disk technology: Rapid scale up in midbrain organoid generation. Methods. 2022;203:465-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 115. | Fiorenzano A, Sozzi E, Birtele M, Kajtez J, Giacomoni J, Nilsson F, Bruzelius A, Sharma Y, Zhang Y, Mattsson B, Emnéus J, Ottosson DR, Storm P, Parmar M. Single-cell transcriptomics captures features of human midbrain development and dopamine neuron diversity in brain organoids. Nat Commun. 2021;12:7302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 80] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 116. | Renner H, Grabos M, Becker KJ, Kagermeier TE, Wu J, Otto M, Peischard S, Zeuschner D, TsyTsyura Y, Disse P, Klingauf J, Leidel SA, Seebohm G, Schöler HR, Bruder JM. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 117. | Sarrafha L, Parfitt GM, Reyes R, Goldman C, Coccia E, Kareva T, Ahfeldt T. High-throughput generation of midbrain dopaminergic neuron organoids from reporter human pluripotent stem cells. STAR Protoc. 2021;2:100463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 118. | Maserejian N, Vinikoor-Imler L, Dilley A. Estimation of the 2020 Global Population of Parkinson’s Disease (PD) [abstract]. Mov Disord 2020. [cited 16 March 2023]. Available from: https://www.mdsabstracts.org/abstract/estimation-of-the-2020-global-population-of-parkinsons-disease-pd/. |
| 119. | Modamio Chamarro, Saraiva C, Gomez Giro G, Louise Nickels S, Jarazo J, M A Antony P, Barbuti P, Halder R, Jäger C, Krüger R, Glaab E. Synaptic decline precedes dopaminergic neuronal loss in human midbrain organoids harboring a triplication of the SNCA gene. July 15, 2021. [cited 16 March 2023]. Available from: https://www.researchgate.net/publication/353290998_Synaptic_decline_precedes_dopaminergic_neuronal_loss_in_human_midbrain_organoids_harboring_a_triplication_of_the_SNCA_gene. |
| 120. | Mohamed NV, Sirois J, Ramamurthy J, Mathur M, Lépine P, Deneault E, Maussion G, Nicouleau M, Chen CX, Abdian N, Soubannier V, Cai E, Nami H, Thomas RA, Wen D, Tabatabaei M, Beitel LK, Singh Dolt K, Karamchandani J, Stratton JA, Kunath T, Fon EA, Durcan TM. Midbrain organoids with an SNCA gene triplication model key features of synucleinopathy. Brain Commun. 2021;3:fcab223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 121. | Smits LM, Reinhardt L, Reinhardt P, Glatza M, Monzel AS, Stanslowsky N, Rosato-Siri MD, Zanon A, Antony PM, Bellmann J, Nicklas SM, Hemmer K, Qing X, Berger E, Kalmbach N, Ehrlich M, Bolognin S, Hicks AA, Wegner F, Sterneckert JL, Schwamborn JC. Modeling Parkinson’s disease in midbrain-like organoids. NPJ Parkinsons Dis. 2019;5:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 206] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 122. | Alexander GE. Biology of Parkinson’s disease: pathogenesis and pathophysiology of a multisystem neurodegenerative disorder. Dialogues Clin Neurosci. 2004;6:259-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 310] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 123. | Poewe W, Seppi K, Tanner CM, Halliday GM, Brundin P, Volkmann J, Schrag AE, Lang AE. Parkinson disease. Nat Rev Dis Primers. 2017;3:17013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2206] [Cited by in RCA: 3088] [Article Influence: 386.0] [Reference Citation Analysis (0)] |
| 124. | Jarazo J, Barmpa K, Modamio J, Saraiva C, Sabaté-Soler S, Rosety I, Griesbeck A, Skwirblies F, Zaffaroni G, Smits LM, Su J, Arias-Fuenzalida J, Walter J, Gomez-Giro G, Monzel AS, Qing X, Vitali A, Cruciani G, Boussaad I, Brunelli F, Jäger C, Rakovic A, Li W, Yuan L, Berger E, Arena G, Bolognin S, Schmidt R, Schröder C, Antony PMA, Klein C, Krüger R, Seibler P, Schwamborn JC. Parkinson’s Disease Phenotypes in Patient Neuronal Cultures and Brain Organoids Improved by 2-Hydroxypropyl-β-Cyclodextrin Treatment. Mov Disord. 2022;37:80-94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 125. | Zanetti C, Spitz S, Berger E, Bolognin S, Smits LM, Crepaz P, Rothbauer M, Rosser JM, Marchetti-Deschmann M, Schwamborn JC, Ertl P. Monitoring the neurotransmitter release of human midbrain organoids using a redox cycling microsensor as a novel tool for personalized Parkinson’s disease modelling and drug screening. Analyst. 2021;146:2358-2367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 126. | Larsen SB, Hanss Z, Krüger R. The genetic architecture of mitochondrial dysfunction in Parkinson’s disease. Cell Tissue Res. 2018;373:21-37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 127. | Goldman SM. Environmental toxins and Parkinson’s disease. Annu Rev Pharmacol Toxicol. 2014;54:141-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 351] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 128. | Langston JW, Ballard P, Tetrud JW, Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3580] [Cited by in RCA: 3323] [Article Influence: 79.1] [Reference Citation Analysis (0)] |
| 129. | Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 486] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 130. | Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko EL, Chawarska K, Pelphrey KA, Howe JR, Vaccarino FM. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162:375-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 719] [Cited by in RCA: 802] [Article Influence: 80.2] [Reference Citation Analysis (0)] |
| 131. | Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Göke J, Tan ZY, Saw TY, Tan CP, Lokman H, Lee Y, Kim D, Ko HS, Kim SO, Park JH, Cho NJ, Hyde TM, Kleinman JE, Shin JH, Weinberger DR, Tan EK, Je HS, Ng HH. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016;19:248-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 588] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 132. | Ozone C, Suga H, Eiraku M, Kadoshima T, Yonemura S, Takata N, Oiso Y, Tsuji T, Sasai Y. Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat Commun. 2016;7:10351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 141] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 133. | Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol. 2017;35:659-666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 549] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 134. | Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 959] [Cited by in RCA: 868] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 135. | Bian S, Repic M, Guo Z, Kavirayani A, Burkard T, Bagley JA, Krauditsch C, Knoblich JA. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods. 2018;15:631-639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 189] [Cited by in RCA: 292] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 136. | Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, Gage FH. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. 2018;36:432-441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 810] [Article Influence: 115.7] [Reference Citation Analysis (0)] |
| 137. | Faustino Martins JM, Fischer C, Urzi A, Vidal R, Kunz S, Ruffault PL, Kabuss L, Hube I, Gazzerro E, Birchmeier C, Spuler S, Sauer S, Gouti M. Self-Organizing 3D Human Trunk Neuromuscular Organoids. Cell Stem Cell. 2020;26:172-186.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 182] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 138. | Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, Paulsen O, Lakatos A, Lancaster MA. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22:669-679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 396] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 139. | Valiulahi P, Vidyawan V, Puspita L, Oh Y, Juwono VB, Sittipo P, Friedlander G, Yahalomi D, Sohn JW, Lee YK, Yoon JK, Shim JW. Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs. Stem Cell Reports. 2021;16:1938-1952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 140. | Bekman E, Silva TP, Cotovio JP, de Almeida RM. Concepts and Applications of Stem Cell Biology: A Guide for Students. Rodrigues G, Roelen BAG, editors. Cham: Springer International Publishing, 2020: 213-228. [DOI] [Full Text] |
| 141. | Silva TP, Bekman EP, Fernandes TG, Vaz SH, Rodrigues CAV, Diogo MM, Cabral JMS, Carmo-Fonseca M. Maturation of Human Pluripotent Stem Cell-Derived Cerebellar Neurons in the Absence of Co-culture. Front Bioeng Biotechnol. 2020;8:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 142. | Ishida Y, Kawakami H, Kitajima H, Nishiyama A, Sasai Y, Inoue H, Muguruma K. Vulnerability of Purkinje Cells Generated from Spinocerebellar Ataxia Type 6 Patient-Derived iPSCs. Cell Rep. 2016;17:1482-1490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 77] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 143. | Eura N, Matsui TK, Luginbühl J, Matsubayashi M, Nanaura H, Shiota T, Kinugawa K, Iguchi N, Kiriyama T, Zheng C, Kouno T, Lan YJ, Kongpracha P, Wiriyasermkul P, Sakaguchi YM, Nagata R, Komeda T, Morikawa N, Kitayoshi F, Jong M, Kobashigawa S, Nakanishi M, Hasegawa M, Saito Y, Shiromizu T, Nishimura Y, Kasai T, Takeda M, Kobayashi H, Inagaki Y, Tanaka Y, Makinodan M, Kishimoto T, Kuniyasu H, Nagamori S, Muotri AR, Shin JW, Sugie K, Mori E. Brainstem Organoids From Human Pluripotent Stem Cells. Front Neurosci. 2020;14:538. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 144. | Wang L, Sievert D, Clark AE, Lee S, Federman H, Gastfriend BD, Shusta EV, Palecek SP, Carlin AF, Gleeson JG. A human three-dimensional neural-perivascular ‘assembloid’ promotes astrocytic development and enables modeling of SARS-CoV-2 neuropathology. Nat Med. 2021;27:1600-1606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 145. | Shepherd GM. Corticostriatal connectivity and its role in disease. Nat Rev Neurosci. 2013;14:278-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 601] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 146. | Milad MR, Rauch SL. Obsessive-compulsive disorder: beyond segregated cortico-striatal pathways. Trends Cogn Sci. 2012;16:43-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 518] [Cited by in RCA: 593] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 147. | Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437-442. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1279] [Cited by in RCA: 1146] [Article Influence: 81.9] [Reference Citation Analysis (0)] |