Published online Mar 26, 2023. doi: 10.4252/wjsc.v15.i3.71
Peer-review started: December 16, 2022
First decision: February 3, 2023
Revised: February 10, 2023
Accepted: March 15, 2023
Article in press: March 15, 2023
Published online: March 26, 2023
Processing time: 97 Days and 1.1 Hours
Cardiomyopathy is a pathological condition characterized by cardiac pump failure due to myocardial dysfunction and the major cause of advanced heart failure requiring heart transplantation. Although optimized medical therapies have been developed for heart failure during the last few decades, some patients with cardiomyopathy exhibit advanced heart failure and are refractory to medical therapies. Desmosome, which is a dynamic cell-to-cell junctional component, maintains the structural integrity of heart tissues. Genetic mutations in desmo
Core Tip: Prevention of advanced heart failure caused by cardiomyopathy is an urgent unmet need in the field of cardiovascular medicine. Desmosome, a cell-to-cell junctional component, maintains the structural integrity of heart tissues. Genetic mutations in desmosomal genes cause desmosome-related cardiomyopathy, an intractable disease refractory to standard medical therapies. This review introduces the recent advances in disease modeling of desmosome-related cardiomyopathy caused by PKP2 mutations using induced pluripotent stem cell-derived cardiomyocytes.
- Citation: Higo S. Disease modeling of desmosome-related cardiomyopathy using induced pluripotent stem cell-derived cardiomyocytes. World J Stem Cells 2023; 15(3): 71-82
- URL: https://www.wjgnet.com/1948-0210/full/v15/i3/71.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i3.71
Heart failure is a clinical syndrome characterized by dyspnea, malaise, swelling, and/or decreased exercise capacity owing to impaired cardiac pumping function[1]. The established optimal medical therapies for heart failure have increased the survival rates of patients in the last few decades[2-4]. However, some patients are refractory to medical therapies and develop symptoms that are diagnosed as advanced heart failure. Currently, the therapeutic strategies available for these patients are heart transplantation and implantation of the ventricular assisting device[1,5]. Cardiomyopathy is a disease of cardiac pump failure due to myocardial dysfunction and is the major cause of advanced heart failure requiring heart transplantation[6-11]. Cardiomyopathies are differentially diagnosed mainly by using imaging modalities, including echocardiography, scintigraphy, computed tomography, magnetic resonance imaging, and cardiac catheterization. Based on the findings of these modalities, cardiomyopathies are classified into dilated cardiomyopathy (DCM), hypertrophic cardiomyopathy (HCM), restrictive cardiomyopathy (RCM), or other rare cardiomyopathies, such as arrhythmogenic right ventricular cardiomyopathy (ARVC)[12]. Among 36,883 heart transplantation recipients registered in the International Society for Heart and Lung Transplantation Thoracic Organ Transplant Registry between 2010 and 2018, the major primary diagnoses were non-ischemic DCM (50.8%), ischemic cardiomyopathy (ICM) (32.4%) with coronary artery disease, RCM (3.5%), and HCM (3.4%)[13]. In Japan, cardiomyopathies [DCM (64%), end-stage HCM with left ventricular systolic dysfunction (12%), and ICM (9%)] account for more than three-quarters of underlying diseases among heart transplant recipients[14]. ARVC, a rare inherited disease, is characterized by the risk of life-threatening arrhythmias, myocardial dysfunction, and fibrofatty replacement of myocardial tissue, predisposing the patients to sudden cardiac death and heart failure[9,11]. The prevalence of ARVC among the registrants for heart transplantation is rare (0.3% and 1%-2% in the United Network for Organ Sharing registry[15] and Japan Organ Transplant Network[14], respectively).
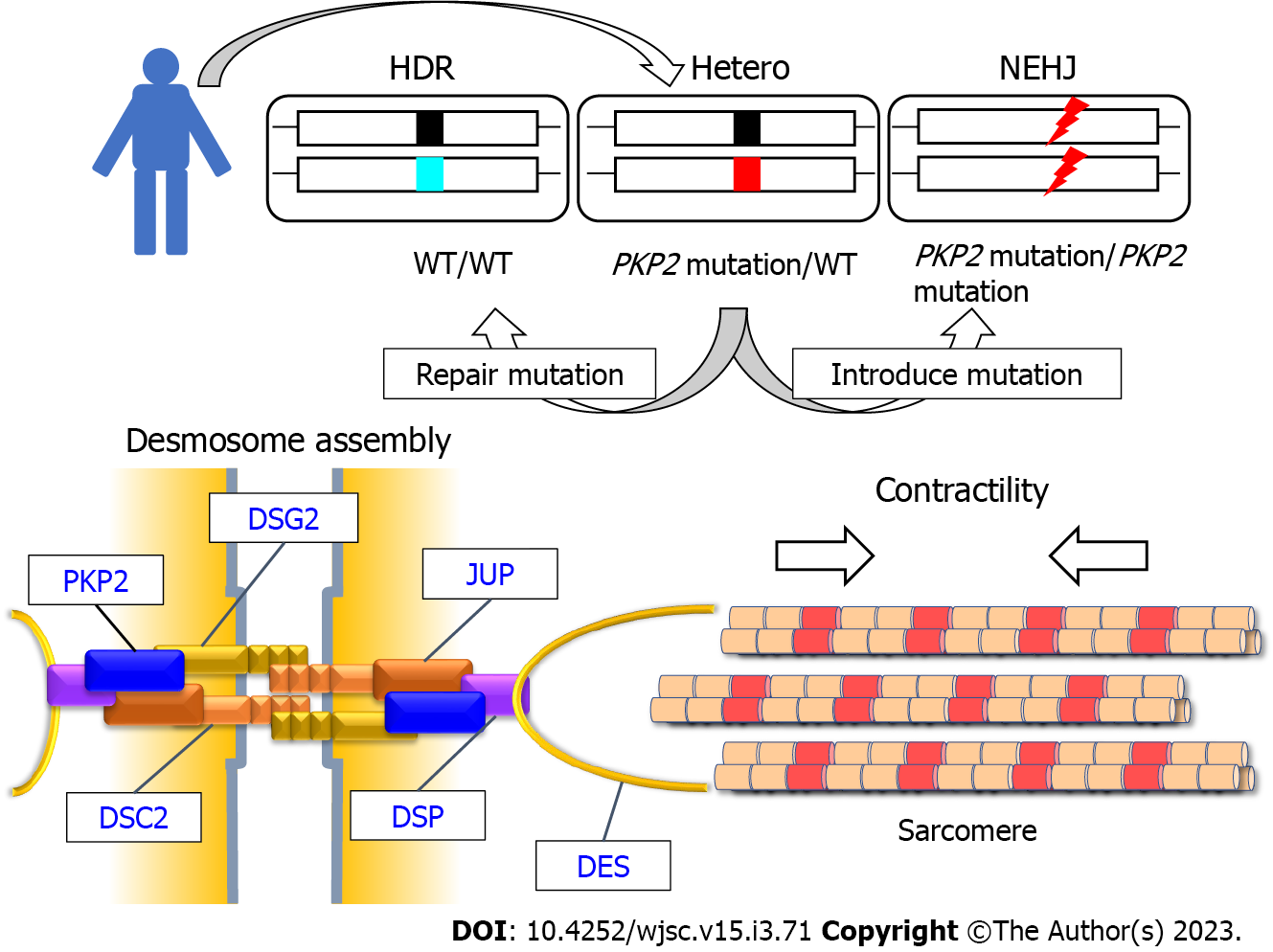
Recent clinical studies utilizing high-throughput sequencing technologies have elucidated the genetic basis of cardiomyopathies, identified various causative genetic variants, and revealed the correlation between genetic factors and clinical phenotypes or cardiac morphologies in patients with cardiomyopathies[16-20]. ARVC is an inherited disease caused by mutations in desmosomal genes (PKP2, JUP, DSC2, DSG2, and DSP) (Figure 1)[11,21,22]. These genes encode the structural components of the desmosome, a dynamic junction between cells that maintain the structural integrity of heart tissues[23,24]. The original disease phenotypes of ARVC are characterized by predominant right ventricular enlargement and contractile dysfunction. However, recent studies have reported left ventricular or biventricular involvement in patients with ARVC, resulting in the use of a broad phrase [arrhythmogenic cardiomyopathy (AC)][9,11]. Although the prevalence of AC in patients with advanced heart failure is rare, recent genetic analyses in large cohorts have demonstrated an increased incidence of desmosomal gene mutations in patients with DCM[18,25,26], which is the most frequent basal disease among heart transplantation registrants. Furthermore, homozygosity and compound or digenic heterozygosity of desmosomal genes are not rare, and patients with combined mutations exhibit a severe phenotype[27-30]. Recently, we identified DSG2-deficient cardiomyopathy caused by a rare homozygous stop-gain mutation in a patient initially diagnosed with idiopathic sporadic DCM[30]. Dsg2 deficiency is associated with embryonic lethality in mice. Additionally, Dsg2-depleted embryonic stem cells do not proliferate[31]. However, a human male patient with a complete lack of DSG2 expression did not exhibit pathological phenotypes at birth but developed advanced heart failure during the teenage years. Immunohistochemical and transmission electron microscopy analyses of left ventricular heart tissues revealed that the loss of DSG2 leads to aberrant deposition of desmosomal proteins and disruption of intercalated discs in cardiomyocytes. These findings suggest that desmosome-related cardiomyopathy is concealed in patients with advanced heart failure who are diagnosed with idiopathic DCM. As desmosome impairment is the most upstream molecular change in these patients, experimental studies must focus on elucidating the molecular mechanisms underlying the instability of cell-to-cell junctions to overcome advanced heart failure caused by desmosome-related cardiomyopathy. For disease modeling, patient-derived induced pluripotent stem cells (iPSCs) in combination with genome editing, which allows precise genomic modification of the targeted mutations, are powerful experimental tools to recapitulate pathological phenotypes based on the molecular factors of inherited cardiomyopathies[30,32-35].
PKP2, which is encoded by PKP2, is a desmosomal protein localized to the outer dense plaque and functions as a scaffold for the other desmosome proteins DSG2, DSC2, JUP, and DSP[23,36] (Figure 1). Among the desmosomal genes, mutations in PKP2 are most frequently identified in patients with AC[11,37-39], and have been extensively studied using patient-derived iPSC-CMs compared to other desmosomal genes (DSG2[30,40,41], DSP[42,43], and DSC2[44,45]). Various clinical phenotypes and pathological characteristics observed in patients with AC harboring PKP2 mutations, downregulated desmosomal protein expression, upregulated lipogenesis, and increased apoptosis in heart tissues have been recapitulated using genetically engineered mouse models[11] and human cardiomyocytes differentiated from iPSCs[46-54] (Table 1). Most known mutations of PKP2 are heterozygous and are missense, nonsense, and frameshift mutations. Studies on patient-derived iPSCs have identified that PKP2 variants are heterozygous missense[48], heterozygous frameshift[46,47,49,50,54], homozygous fram
| Genetic mutation | Origin of disease-specific iPSC | Experimental control | Desmosome proteins | Lipid accumulation | Apoptosis | Electrophysiology | Ultrastructure of desmosome | Contractility | Phenotypic rescue by gene replacement | Ref. |
| Heterozygous missense (c.1841T>C, p.L614P) | Dermal fibroblasts from a 30-yr-old male patient with AC | iPSCs from a 32-yr-old healthy male donor | Decreased JUP; No change in DSP, CDH2, and GJA1 (immunofluorescence staining at weeks 4-5) | Increased oil red O staining after exposure to adipogenic differentiation medium for 2 wk (oil red O staining) | NA | Ventricular-like action potential profile (single-cell patch-clamp recording (without control)) | Increased cell width (TEM at weeks 4-5) | NA | NA | Ma et al[48] |
| Heterozygous frameshift (c.971_972ins, pA324fs335X); Heterozygous frameshift (c.148_151delACAG, p.T50SfsX110) | Dermal fibroblasts from a 30-yr-old male patient with AC | iPSCs from a healthy control | Decreased JUP and GJA1 (immunofluorescence staining) | Lipid droplet accumulation (TEM on day 40) | Increased apoptosis after serum starvation (TUNEL) | Prolonged field potential rise time (multielectrode array) | Widened and distorted desmosomes (TEM on day 40) | NA | NA | Caspi et al[46] |
| Homozygous frameshift (c.2484C>T leading to cryptic splicing); Heterozygous frameshift (c.2013delC, p.Lys672ArgfsX12) | Fibroblasts from a female patient with AC; Fibroblasts from a patient with AC | H9 human embryonic stem cell; iPSCs from cardiac fibroblasts of aborted fetus without a family history of AC | Nuclear translocation of JUP (immunofluorescence staining) | Increased lipogenesis after adipogenic stimulation for 4-5 wk (Nile red staining) | Increased apoptosis after adipogenic stimulation for 4-5 wk (TUNEL) | Slow intracellular calcium relaxation; Prolonged relaxation time (calcium imaging using Fura-2 acetoxymethyl on day 60) | NA | NA | NA | Kim et al[47] |
| Heterozygous frameshift (c.1760delT, p.V587Afsx655) | Dermal keratinocytes from a male patient with AC | iPSCs from dermal keratinocytes of a healthy control | Interrupted expression of DSP (immunofluorescence staining) | Lipid droplet accumulation after adipogenic stimulation for 4 wk (oil red O staining at months 3-4) | Genes associated with apoptosis remained unchanged (quantitative real-time PCR) | NA | NA | NA | NA | Dorn et al[49] |
| Homozygous frameshift (c.2484C>T leading to cryptic splicing) | Fibroblasts from a female patient with AC | iPSCs from a healthy control | Reduced JUP (immunofluorescence staining) | NA | NA | NA (decreased co-localization of NaV1.5 with PKP2) | NA | NA (increased pro-fibrotic gene expression after stretch) | NA | Martewicz et al[51] |
| Heterozygous frameshift (c.971_972InsT, p.A324fs335X) | A patient with AC | H9 human embryonic stem cells | Decreased membrane-localized JUP (immunofluorescence staining on day 34) | Increased lipid content (Nile red staining on day 34) | NA | Short action potential and slow spontaneous beat rate in engineered heart slices [optical mapping (relative to monolayer cardiomyocytes)] | NA | NA | NA | Blazeski et al[50] |
| Compound heterozygous frameshift and missense (c.354delT, p.Y119MfsX23 and p.K859R) | Adipose tissue-derived mesenchymal multipotent stromal cells from a 14-yr-old female patient with AC | Gender-matched healthy donor | Increased cytoplasmic and nuclear JUP levels (immunofluorescence staining on days 24-30) | No presence of lipid droplets (oil red O staining on day 24) | Not increased (PI staining at day 24-30) | Reduced sodium current density; Decreased action potential upstroke velocity (whole-cell patch-clamp and microelectrodes on days 24-30) | NA | NA | Restored sodium current after lentiviral transduction of PKP2 | Khudiakov et al[52] |
| Heterozygous and homozygous frameshift mutation (p.D109AfsX10, introduced mutation via genome editing) | Wild-type iPSC lines from two different donors with introduced heterozygous and homozygous frameshift mutations | Isogenic wild-type iPSCs | Decreased junctional localization of DSP and GJA1 (immunofluorescence staining); Impaired stability of junctional CDH2 (fluorescence recovery after photobleaching) | NA | NA | Prolonged action potential duration (optical voltage recording on day 30) | NA | Decreased systolic force (three-dimensional cardiac microtissues on day 40) | NA | Zhang et al[53] |
| Heterozygous frameshift mutation (c.1228dupG, p.D410fsX425) | Peripheral blood mononuclear cells from a female patient with AC | Isogenic iPSCs with corrected mutation (wild-type) and introduced homozygous frameshift mutations | Decreased area of desmosomes (DSG2, DSC2, and DSP) (immunofluorescence staining on day 14) | Lipid droplet accumulation in iPSC-CMs with homozygous frameshift mutations (TEM on day 28) | Increased apoptosis in iPSC-CMs with homozygous frameshift mutations (cleaved CASP3 expression on day 28) | Decreased propagation speed in iPSC-CMs with homozygous frameshift mutations (motion vector analysis on day 28) | Increased desmosome gap width (TEM on day 28) | Decreased contractility (contraction velocity and deformation distance evaluated using motion vector analysis on days 14 and 28) | Recovered contractility and desmosome assembly via AAV-mediated PKP2 delivery | Inoue et al[54] |
We established iPSCs from a patient with AC harboring a heterozygous frameshift PKP2 mutation (c.1228dupG, p.D410fsX425) and generated an isogenic set of iPSC clones harboring three genotypes [heterozygous mutation (Hetero), homozygously corrected with homology-directed repair (HDR), and homozygously introduced frameshift mutations via non-homologous end joining (NHEJ)] using genome editing[54] (Figure 1). These isogenic sets of iPSCs comprise patient-derived Hetero-iPSCs, HDR-iPSCs with two-fold higher PKP2 expression relative to Hetero-iPSCs, and NHEJ-iPSCs, which do not express PKP2, recapitulating both haploinsufficiency and complete loss of PKP2. After cardiomyocyte differentiation using the monolayer protocol with chemically defined medium[58], NHEJ-iPSC-CMs lacking PKP2 expression exhibit lipid droplet accumulation, increased apoptosis, and decreased propagation rate (Table 1). However, patient-derived Hetero-iPSC-CMs with half-dose PKP2 expression do not exhibit these pathological phenotypes, suggesting that the haploinsufficiency of PKP2 is not sufficient to induce the above pathological phenotypes within 28 days after differentiation. In contrast, haploinsufficiency of PKP2 decreased contractility, which was evaluated using motion vector analysis, within 14 days of differentiation. As the monolayer protocol confers strong contraction to iPSC-CMs on culture plates immediately after differentiation[58,59], continuous tensile overload may facilitate the contractile phenotype among isogenic iPSC-CMs. A recent study used isogenic iPSC-CMs in which heterozygous or homozygous frameshift mutation was introduced into wild-type iPSC-CMs[53]. The authors reported that PKP2 deficiency decreased systolic force in three-dimensional cardiac microtissues. This further supported the functional relationship between PKP2 deficiency and contractile dysfunction. An experimental study using cardiac tissue-specific Pkp2 knockout mice demonstrated that the loss of Pkp2 increased the distance between the cell periphery and DES, an intermediate filament protein in cardiomyocytes[60]. As DES connects Z-discs of sarcomeres to sarcolemmal costameres, desmosomes, and nuclear envelope[11,61], further experimental studies focusing on these cellular networks are required to elucidate the pathogenesis of desmosome-related cardiomyopathy.
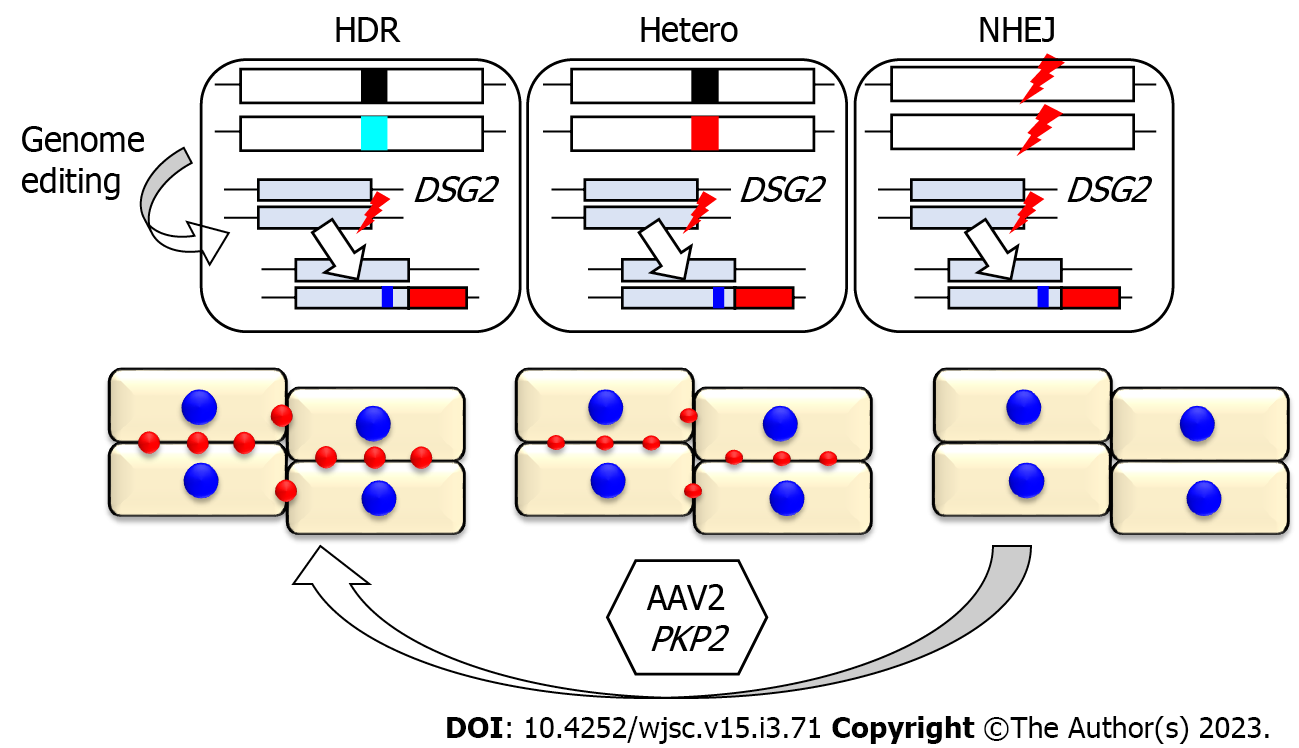
In the isogenic background, the haploinsufficiency of PKP2 did not affect the localization or expression levels of desmosomal proteins in iPSC-CMs as evidenced by the results of immunostaining or western blotting analyses. However, the desmosome area represented by dot distribution on the cell periphery in Hetero-iPSC-CMs was significantly lower than that in HDR-iPSC-CMs[54], suggesting that desmosome assembly is impaired by PKP2 haploinsufficiency. The impaired assembly of desmosomal proteins in human iPSC-CMs is supported by another study using isogenic iPSC-CMs. Fluorescence recovery after photobleaching experiments combined with lentivirus-mediated expression of fluorescent protein-tagged N-cadherin provided evidence that molecular stability of junctional N-cadherin is impaired by PKP2 deficiency[53]. To trace the molecular behavior of endogenous proteins in cardiomyocytes, fluorescent tagging of the structural proteins through genome editing is a powerful tool[62,63]. However, fluorescent tagging of endogenous desmosomal genes might affect desmosome structures or cell-to-cell integrity in iPSCs or iPSC-CMs. We previously identified a patient with DSG2-deficient cardiomyopathy due to a rare homozygous stop-gain mutation and demonstrated that complete loss of DSG2 in human iPSCs does not affect the differentiation or cellular morphology in iPSC-CMs[30]. These findings prompted us to use DSG2 as the target of endogenous tagging by fluorescent protein to trace desmosome dynamics in live human iPSC-CMs. Genome editing targeting DSG2 alleles was performed to establish the isogenic iPSC-CMs harboring identical two DSG2 alleles comprising intact and knocked-in tdTomato alleles under the adjusted PKP2 expression levels (Figure 2). The desmosome area (represented by desmoglein-2-tdTomato fusion protein) was significantly downregulated due to PKP2 haploinsufficiency. Adeno-associated virus (AAV), a small, nonenveloped virus with a linear, single-stranded DNA, is widely used for gene therapy targeting human diseases, including heart failure[64,65]. AAV-mediated gene replacement of PKP2 significantly restored the decreased contractility in Hetero-iPSC-CMs and NEHJ-iPSC-CMs, demonstrating the proof-of-concept for PKP2 gene therapy in human cells. Furthermore, time-lapse imaging using NHEJ-iPSC-CMs captured the recovery of desmosomes, which gradually assembled at the cell periphery after AAV-mediated PKP2 replacement (Figure 2). The established isogenic iPSCs harboring knocked-in tdTomato alleles allowed desmosome-imaging in living cells and provided distinct readouts for therapeutic development.
Several clinical trials using AAV-mediated gene replacement have been designed targeting cardiovascular disease[65,66]. A large-scale clinical trial was conducted as a randomized, multinational, double-blind, placebo-controlled phase 2 study targeting up to 250 patients with moderate-to-severe heart failure and reduced contractile function (CUPID2 trial)[67]. The study aimed to deliver sarcoplasmic reticulum Ca²+-ATPase (SERCA2a) into heart tissues via intracoronary injection. SERCA2a regulates cardiomyocyte contraction and relaxation by transporting Ca²+ from the cytosol into the sarcoplasmic reticulum during diastole[68]. The deficiency of SERCA2a is associated with heart failure progression[69,70]. Although promising results were achieved in preceding preclinical and clinical studies[71-73], gene replacement of SERCA2a did not improve the clinical course of patients with heart failure[74]. The two clinical trials of gene therapy targeting patients with heart failure conducted in the same period (AGENT-HF[75] and SERCA-LVAD[76]) were terminated due to the neutral result of the CUPID2 trial and the lack of functional benefit. The amount of vector DNA in heart tissues obtained from patients who received gene therapy and subsequently underwent heart transplantation or mechanical circulatory support device implantation was low, suggesting that only a small proportion of cardiomyocytes expressed AAV-delivered SERCA2a in the myocardium. Although these clinical trials demonstrate the difficulty of gene delivery targeting human heart tissues, they provide the evidence for the safety of cardiac gene therapy and a basis for the design of future gene therapy trials. Recent genetic analysis clarified a large number of genetic mutations that cause cardiomyopathies with advanced heart failure in a loss-of-function manner and can be targeted by specific gene replacement therapy[77,78]. In desmosome-related cardiomyopathy, most of the identified mutations in PKP2 are heterozygous[22,37,79,80]. However, in extremely rare cases, homozygous mutations of PKP2 cause lethal infantile heart failure with left ventricular non-compaction or hypoplastic left heart syndrome[81-83]. No effective therapies are available for these patients who require a novel therapeutic approach for desmosome-related cardiomyopathy. Proof-of-concept studies for structural and functional recovery using both human iPSC-CM models and in vivo models are required for future clinical application.
Although human iPSC-CMs are immature and do not fully recapitulate in vivo heart tissues[59], tissue engineering approaches[84,85] will promote the maturation of iPSC-CMs and provide a useful tool in combination with genome editing. The isogenic iPSC-CMs that we established represent a human disease model that recapitulates reduced contractility and impaired desmosome assembly and provides a convenient cellular platform for therapeutic screening to examine upstream molecular targets of desmosome-related cardiomyopathy.
The Department of Medical Therapeutics for Heart Failure was an endowment department supported by Actelion Pharmaceuticals Japan (2015–2020).
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cardiac and cardiovascular systems
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Campuzano O, Spain; Li SC, United States; Wang T, China S-Editor: Yan JP L-Editor: A P-Editor: Yan JP
| 1. | Tsutsui H, Ide T, Ito H, Kihara Y, Kinugawa K, Kinugawa S, Makaya M, Murohara T, Node K, Saito Y, Sakata Y, Shimizu W, Yamamoto K, Bando Y, Iwasaki YK, Kinugasa Y, Mizote I, Nakagawa H, Oishi S, Okada A, Tanaka A, Akasaka T, Ono M, Kimura T, Kosaka S, Kosuge M, Momomura SI. JCS/JHFS 2021 Guideline Focused Update on Diagnosis and Treatment of Acute and Chronic Heart Failure. J Card Fail. 2021;27:1404-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 103] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 2. | Levy D, Kenchaiah S, Larson MG, Benjamin EJ, Kupka MJ, Ho KK, Murabito JM, Vasan RS. Long-term trends in the incidence of and survival with heart failure. N Engl J Med. 2002;347:1397-1402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1514] [Cited by in RCA: 1504] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 3. | Roger VL, Weston SA, Redfield MM, Hellermann-Homan JP, Killian J, Yawn BP, Jacobsen SJ. Trends in heart failure incidence and survival in a community-based population. JAMA. 2004;292:344-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1172] [Cited by in RCA: 1197] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 4. | Jones NR, Roalfe AK, Adoki I, Hobbs FDR, Taylor CJ. Survival of patients with chronic heart failure in the community: a systematic review and meta-analysis. Eur J Heart Fail. 2019;21:1306-1325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 355] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 5. | Ono M, Yamaguchi O, Ohtani T, Kinugawa K, Saiki Y, Sawa Y, Shiose A, Tsutsui H, Fukushima N, Matsumiya G, Yanase M, Yamazaki K, Yamamoto K, Akiyama M, Imamura T, Iwasaki K, Endo M, Ohnishi Y, Okumura T, Kashiwa K, Kinoshita O, Kubota K, Seguchi O, Toda K, Nishioka H, Nishinaka T, Nishimura T, Hashimoto T, Hatano M, Higashi H, Higo T, Fujino T, Hori Y, Miyoshi T, Yamanaka M, Ohno T, Kimura T, Kyo S, Sakata Y, Nakatani T; JCS/JSCVS/JATS/JSVS Joint Working Group. JCS/JSCVS/JATS/JSVS 2021 Guideline on Implantable Left Ventricular Assist Device for Patients With Advanced Heart Failure. Circ J. 2022;86:1024-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 6. | Rosenbaum AN, Agre KE, Pereira NL. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol. 2020;17:286-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 141] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 7. | Marian AJ, Braunwald E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ Res. 2017;121:749-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 922] [Article Influence: 115.3] [Reference Citation Analysis (0)] |
| 8. | McNally EM, Mestroni L. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ Res. 2017;121:731-748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 546] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 9. | Corrado D, Basso C, Judge DP. Arrhythmogenic Cardiomyopathy. Circ Res. 2017;121:784-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 300] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 10. | Watkins H, Ashrafian H, Redwood C. Inherited cardiomyopathies. N Engl J Med. 2011;364:1643-1656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 418] [Cited by in RCA: 370] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 11. | Austin KM, Trembley MA, Chandler SF, Sanders SP, Saffitz JE, Abrams DJ, Pu WT. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat Rev Cardiol. 2019;16:519-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 183] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 12. | Kitaoka H, Tsutsui H, Kubo T, Ide T, Chikamori T, Fukuda K, Fujino N, Higo T, Isobe M, Kamiya C, Kato S, Kihara Y, Kinugawa K, Kinugawa S, Kogaki S, Komuro I, Hagiwara N, Ono M, Maekawa Y, Makita S, Matsui Y, Matsushima S, Sakata Y, Sawa Y, Shimizu W, Teraoka K, Tsuchihashi-Makaya M, Ishibashi-Ueda H, Watanabe M, Yoshimura M, Fukusima A, Hida S, Hikoso S, Imamura T, Ishida H, Kawai M, Kitagawa T, Kohno T, Kurisu S, Nagata Y, Nakamura M, Morita H, Takano H, Shiga T, Takei Y, Yuasa S, Yamamoto T, Watanabe T, Akasaka T, Doi Y, Kimura T, Kitakaze M, Kosuge M, Takayama M, Tomoike H; Japanese Circulation Society Joint Working Group. JCS/JHFS 2018 Guideline on the Diagnosis and Treatment of Cardiomyopathies. Circ J. 2021;85:1590-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 75] [Article Influence: 18.8] [Reference Citation Analysis (33)] |
| 13. | Khush KK, Cherikh WS, Chambers DC, Harhay MO, Hayes D Jr, Hsich E, Meiser B, Potena L, Robinson A, Rossano JW, Sadavarte A, Singh TP, Zuckermann A, Stehlik J; International Society for Heart and Lung Transplantation. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: Thirty-sixth adult heart transplantation report - 2019; focus theme: Donor and recipient size match. J Heart Lung Transplant. 2019;38:1056-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 672] [Article Influence: 112.0] [Reference Citation Analysis (0)] |
| 14. | Nakatani T, Fukushima N, Ono M, Saiki Y, Matsuda H, Nunoda S, Sawa Y, Isobe M. The Registry Report of Heart Transplantation in Japan (1999-2014). Circ J. 2016;80:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 15. | Giuliano K, Scheel P 3rd, Etchill E, Fraser CD 3rd, Suarez-Pierre A, Hsu S, Wittstein IS, Kasper EK, Florido R, Tandri H, Calkins H, Choi CW, Sharma K, Kilic A, Gilotra NA. Heart transplantation outcomes in arrhythmogenic right ventricular cardiomyopathy: a contemporary national analysis. ESC Heart Fail. 2022;9:988-997. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 16. | Coppini R, Ho CY, Ashley E, Day S, Ferrantini C, Girolami F, Tomberli B, Bardi S, Torricelli F, Cecchi F, Mugelli A, Poggesi C, Tardiff J, Olivotto I. Clinical phenotype and outcome of hypertrophic cardiomyopathy associated with thin-filament gene mutations. J Am Coll Cardiol. 2014;64:2589-2600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 17. | Marstrand P, Han L, Day SM, Olivotto I, Ashley EA, Michels M, Pereira AC, Wittekind SG, Helms A, Saberi S, Jacoby D, Ware JS, Colan SD, Semsarian C, Ingles J, Lakdawala NK, Ho CY; SHaRe Investigators. Hypertrophic Cardiomyopathy With Left Ventricular Systolic Dysfunction: Insights From the SHaRe Registry. Circulation. 2020;141:1371-1383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 146] [Article Influence: 29.2] [Reference Citation Analysis (33)] |
| 18. | Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, Feng Z, Müller S, Kayvanpour E, Vogel B, Sedaghat-Hamedani F, Lim WK, Zhao X, Fradkin D, Köhler D, Fischer S, Franke J, Marquart S, Barb I, Li DT, Amr A, Ehlermann P, Mereles D, Weis T, Hassel S, Kremer A, King V, Wirsz E, Isnard R, Komajda M, Serio A, Grasso M, Syrris P, Wicks E, Plagnol V, Lopes L, Gadgaard T, Eiskjær H, Jørgensen M, Garcia-Giustiniani D, Ortiz-Genga M, Crespo-Leiro MG, Deprez RH, Christiaans I, van Rijsingen IA, Wilde AA, Waldenstrom A, Bolognesi M, Bellazzi R, Mörner S, Bermejo JL, Monserrat L, Villard E, Mogensen J, Pinto YM, Charron P, Elliott P, Arbustini E, Katus HA, Meder B. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J. 2015;36:1123-135a. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 367] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 19. | Suwa Y, Higo S, Nakamoto K, Sera F, Kunimatsu S, Masumura Y, Kanzaki M, Mizote I, Mizuno H, Fujio Y, Hikoso S, Sakata Y. Old-Age Onset Progressive Cardiac Contractile Dysfunction in a Patient with Polycystic Kidney Disease Harboring a PKD1 Frameshift Mutation. Int Heart J. 2019;60:220-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Tobita T, Nomura S, Fujita T, Morita H, Asano Y, Onoue K, Ito M, Imai Y, Suzuki A, Ko T, Satoh M, Fujita K, Naito AT, Furutani Y, Toko H, Harada M, Amiya E, Hatano M, Takimoto E, Shiga T, Nakanishi T, Sakata Y, Ono M, Saito Y, Takashima S, Hagiwara N, Aburatani H, Komuro I. Genetic basis of cardiomyopathy and the genotypes involved in prognosis and left ventricular reverse remodeling. Sci Rep. 2018;8:1998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 21. | Towbin JA, McKenna WJ, Abrams DJ, Ackerman MJ, Calkins H, Darrieux FCC, Daubert JP, de Chillou C, DePasquale EC, Desai MY, Estes NAM 3rd, Hua W, Indik JH, Ingles J, James CA, John RM, Judge DP, Keegan R, Krahn AD, Link MS, Marcus FI, McLeod CJ, Mestroni L, Priori SG, Saffitz JE, Sanatani S, Shimizu W, van Tintelen JP, Wilde AAM, Zareba W. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy. Heart Rhythm. 2019;16:e301-e372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 515] [Article Influence: 85.8] [Reference Citation Analysis (0)] |
| 22. | Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med. 2008;5:258-267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 181] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 23. | Al-Jassar C, Bikker H, Overduin M, Chidgey M. Mechanistic basis of desmosome-targeted diseases. J Mol Biol. 2013;425:4006-4022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 24. | Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res. 2010;107:700-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 228] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 25. | Pugh TJ, Kelly MA, Gowrisankar S, Hynes E, Seidman MA, Baxter SM, Bowser M, Harrison B, Aaron D, Mahanta LM, Lakdawala NK, McDermott G, White ET, Rehm HL, Lebo M, Funke BH. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 275] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 26. | Garcia-Pavia P, Syrris P, Salas C, Evans A, Mirelis JG, Cobo-Marcos M, Vilches C, Bornstein B, Segovia J, Alonso-Pulpon L, Elliott PM. Desmosomal protein gene mutations in patients with idiopathic dilated cardiomyopathy undergoing cardiac transplantation: a clinicopathological study. Heart. 2011;97:1744-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 27. | Rigato I, Bauce B, Rampazzo A, Zorzi A, Pilichou K, Mazzotti E, Migliore F, Marra MP, Lorenzon A, De Bortoli M, Calore M, Nava A, Daliento L, Gregori D, Iliceto S, Thiene G, Basso C, Corrado D. Compound and digenic heterozygosity predicts lifetime arrhythmic outcome and sudden cardiac death in desmosomal gene-related arrhythmogenic right ventricular cardiomyopathy. Circ Cardiovasc Genet. 2013;6:533-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 194] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 28. | Gandjbakhch E, Redheuil A, Pousset F, Charron P, Frank R. Clinical Diagnosis, Imaging, and Genetics of Arrhythmogenic Right Ventricular Cardiomyopathy/Dysplasia: JACC State-of-the-Art Review. J Am Coll Cardiol. 2018;72:784-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 193] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 29. | Chen K, Rao M, Guo G, Duru F, Chen L, Chen X, Song J, Hu S. Recessive variants in plakophilin-2 contributes to early-onset arrhythmogenic cardiomyopathy with severe heart failure. Europace. 2019;21:970-977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 30. | Shiba M, Higo S, Kondo T, Li J, Liu L, Ikeda Y, Kohama Y, Kameda S, Tabata T, Inoue H, Nakamura S, Takeda M, Ito E, Takashima S, Miyagawa S, Sawa Y, Hikoso S, Sakata Y. Phenotypic recapitulation and correction of desmoglein-2-deficient cardiomyopathy using human-induced pluripotent stem cell-derived cardiomyocytes. Hum Mol Genet. 2021;30:1384-1397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 26] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Eshkind L, Tian Q, Schmidt A, Franke WW, Windoffer R, Leube RE. Loss of desmoglein 2 suggests essential functions for early embryonic development and proliferation of embryonal stem cells. Eur J Cell Biol. 2002;81:592-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 133] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 32. | Sayed N, Liu C, Wu JC. Translation of Human-Induced Pluripotent Stem Cells: From Clinical Trial in a Dish to Precision Medicine. J Am Coll Cardiol. 2016;67:2161-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 198] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 33. | Kondo T, Higo S, Shiba M, Kohama Y, Kameda S, Tabata T, Inoue H, Okuno S, Ogawa S, Nakamura S, Takeda M, Ito E, Li J, Liu L, Kuramoto Y, Lee JK, Takashima S, Miyagawa S, Sawa Y, Hikoso S, Sakata Y. Human-Induced Pluripotent Stem Cell-Derived Cardiomyocyte Model for TNNT2 Δ160E-Induced Cardiomyopathy. Circ Genom Precis Med. 2022;15:e003522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 34. | Tabata T, Masumura Y, Higo S, Kunimatsu S, Kameda S, Inoue H, Okuno S, Ogawa S, Takashima S, Watanabe M, Miyagawa S, Hikoso S, Sakata Y. Multiplexed measurement of cell type-specific calcium kinetics using high-content image analysis combined with targeted gene disruption. Biochem Biophys Res Commun. 2022;637:40-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 35. | Higo S, Hikoso S, Miyagawa S, Sakata Y. Genome Editing in Human Induced Pluripotent Stem Cells (hiPSCs). Methods Mol Biol. 2021;2320:235-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 36. | Padrón-Barthe L, Domínguez F, Garcia-Pavia P, Lara-Pezzi E. Animal models of arrhythmogenic right ventricular cardiomyopathy: what have we learned and where do we go? Basic Res Cardiol. 2017;112:50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Ohno S, Nagaoka I, Fukuyama M, Kimura H, Itoh H, Makiyama T, Shimizu A, Horie M. Age-dependent clinical and genetic characteristics in Japanese patients with arrhythmogenic right ventricular cardiomyopathy/dysplasia. Circ J. 2013;77:1534-1542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 38. | Groeneweg JA, Bhonsale A, James CA, te Riele AS, Dooijes D, Tichnell C, Murray B, Wiesfeld AC, Sawant AC, Kassamali B, Atsma DE, Volders PG, de Groot NM, de Boer K, Zimmerman SL, Kamel IR, van der Heijden JF, Russell SD, Jan Cramer M, Tedford RJ, Doevendans PA, van Veen TA, Tandri H, Wilde AA, Judge DP, van Tintelen JP, Hauer RN, Calkins H. Clinical Presentation, Long-Term Follow-Up, and Outcomes of 1001 Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy Patients and Family Members. Circ Cardiovasc Genet. 2015;8:437-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 361] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 39. | Novelli V, Malkani K, Cerrone M. Pleiotropic Phenotypes Associated With PKP2 Variants. Front Cardiovasc Med. 2018;5:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | El-Battrawy I, Zhao Z, Lan H, Cyganek L, Tombers C, Li X, Buljubasic F, Lang S, Tiburcy M, Zimmermann WH, Utikal J, Wieland T, Borggrefe M, Zhou XB, Akin I. Electrical dysfunctions in human-induced pluripotent stem cell-derived cardiomyocytes from a patient with an arrhythmogenic right ventricular cardiomyopathy. Europace. 2018;20:f46-f56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 41. | Buljubasic F, El-Battrawy I, Lan H, Lomada SK, Chatterjee A, Zhao Z, Li X, Zhong R, Xu Q, Huang M, Liao Z, Lang S, Cyganek L, Zhou X, Wieland T, Borggrefe M, Akin I. Nucleoside Diphosphate Kinase B Contributes to Arrhythmogenesis in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes from a Patient with Arrhythmogenic Right Ventricular Cardiomyopathy. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 42. | Ng R, Manring H, Papoutsidakis N, Albertelli T, Tsai N, See CJ, Li X, Park J, Stevens TL, Bobbili PJ, Riaz M, Ren Y, Stoddard CE, Janssen PM, Bunch TJ, Hall SP, Lo YC, Jacoby DL, Qyang Y, Wright N, Ackermann MA, Campbell SG. Patient mutations linked to arrhythmogenic cardiomyopathy enhance calpain-mediated desmoplakin degradation. JCI Insight. 2019;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 43. | Xia S, Wang X, Yue P, Li Y, Zhang D. Establishment of induced pluripotent stem cell lines from a family of an ARVC patient receiving heart transplantation in infant age carrying compound heterozygous mutations in DSP gene. Stem Cell Res. 2020;48:101977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 44. | Moreau A, Reisqs JB, Delanoe-Ayari H, Pierre M, Janin A, Deliniere A, Bessière F, Meli AC, Charrabi A, Lafont E, Valla C, Bauer D, Morel E, Gache V, Millat G, Nissan X, Faucherre A, Jopling C, Richard S, Mejat A, Chevalier P. Deciphering DSC2 arrhythmogenic cardiomyopathy electrical instability: From ion channels to ECG and tailored drug therapy. Clin Transl Med. 2021;11:e319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Reisqs JB, Moreau A, Charrabi A, Sleiman Y, Meli AC, Millat G, Briand V, Beauverger P, Richard S, Chevalier P. The PPARγ pathway determines electrophysiological remodelling and arrhythmia risks in DSC2 arrhythmogenic cardiomyopathy. Clin Transl Med. 2022;12:e748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 46. | Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, Gepstein L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet. 2013;6:557-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 47. | Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP, Chen HS. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 388] [Cited by in RCA: 412] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 48. | Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, Oh Y, Tan SH, Ng ML, Shim W, Wong P, Liew R. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2013;34:1122-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 49. | Dorn T, Kornherr J, Parrotta EI, Zawada D, Ayetey H, Santamaria G, Iop L, Mastantuono E, Sinnecker D, Goedel A, Dirschinger RJ, My I, Laue S, Bozoglu T, Baarlink C, Ziegler T, Graf E, Hinkel R, Cuda G, Kääb S, Grace AA, Grosse R, Kupatt C, Meitinger T, Smith AG, Laugwitz KL, Moretti A. Interplay of cell-cell contacts and RhoA/MRTF-A signaling regulates cardiomyocyte identity. EMBO J. 2018;37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 50. | Blazeski A, Lowenthal J, Wang Y, Teuben R, Zhu R, Gerecht S, Tomaselli G, Tung L. Engineered Heart Slice Model of Arrhythmogenic Cardiomyopathy Using Plakophilin-2 Mutant Myocytes. Tissue Eng Part A. 2019;25:725-735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 51. | Martewicz S, Luni C, Serena E, Pavan P, Chen HV, Rampazzo A, Elvassore N. Transcriptomic Characterization of a Human In Vitro Model of Arrhythmogenic Cardiomyopathy Under Topological and Mechanical Stimuli. Ann Biomed Eng. 2019;47:852-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 52. | Khudiakov A, Zaytseva A, Perepelina K, Smolina N, Pervunina T, Vasichkina E, Karpushev A, Tomilin A, Malashicheva A, Kostareva A. Sodium current abnormalities and deregulation of Wnt/β-catenin signaling in iPSC-derived cardiomyocytes generated from patient with arrhythmogenic cardiomyopathy harboring compound genetic variants in plakophilin 2 gene. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 53. | Zhang K, Cloonan PE, Sundaram S, Liu F, Das SL, Ewoldt JK, Bays JL, Tomp S, Toepfer CN, Marsiglia JDC, Gorham J, Reichart D, Eyckmans J, Seidman JG, Seidman CE, Chen CS. Plakophilin-2 truncating variants impair cardiac contractility by disrupting sarcomere stability and organization. Sci Adv. 2021;7:eabh3995. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 54. | Inoue H, Nakamura S, Higo S, Shiba M, Kohama Y, Kondo T, Kameda S, Tabata T, Okuno S, Ikeda Y, Li J, Liu L, Yamazaki S, Takeda M, Ito E, Takashima S, Miyagawa S, Sawa Y, Hikoso S, Sakata Y. Modeling reduced contractility and impaired desmosome assembly due to plakophilin-2 deficiency using isogenic iPS cell-derived cardiomyocytes. Stem Cell Reports. 2022;17:337-351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 55. | Kirchner F, Schuetz A, Boldt LH, Martens K, Dittmar G, Haverkamp W, Thierfelder L, Heinemann U, Gerull B. Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations. Circ Cardiovasc Genet. 2012;5:400-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 48] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 56. | Rasmussen TB, Nissen PH, Palmfeldt J, Gehmlich K, Dalager S, Jensen UB, Kim WY, Heickendorff L, Mølgaard H, Jensen HK, Baandrup UT, Bross P, Mogensen J. Truncating plakophilin-2 mutations in arrhythmogenic cardiomyopathy are associated with protein haploinsufficiency in both myocardium and epidermis. Circ Cardiovasc Genet. 2014;7:230-240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 57. | Corrado D, Perazzolo Marra M, Zorzi A, Beffagna G, Cipriani A, Lazzari M, Migliore F, Pilichou K, Rampazzo A, Rigato I, Rizzo S, Thiene G, Anastasakis A, Asimaki A, Bucciarelli-Ducci C, Haugaa KH, Marchlinski FE, Mazzanti A, McKenna WJ, Pantazis A, Pelliccia A, Schmied C, Sharma S, Wichter T, Bauce B, Basso C. Diagnosis of arrhythmogenic cardiomyopathy: The Padua criteria. Int J Cardiol. 2020;319:106-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 329] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 58. | Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11:855-860. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1016] [Cited by in RCA: 1192] [Article Influence: 108.4] [Reference Citation Analysis (0)] |
| 59. | Gintant G, Burridge P, Gepstein L, Harding S, Herron T, Hong C, Jalife J, Wu JC. Use of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes in Preclinical Cancer Drug Cardiotoxicity Testing: A Scientific Statement From the American Heart Association. Circ Res. 2019;125:e75-e92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 112] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 60. | Pérez-Hernández M, van Opbergen CJM, Bagwan N, Vissing CR, Marrón-Liñares GM, Zhang M, Torres Vega E, Sorrentino A, Drici L, Sulek K, Zhai R, Hansen FB, Christensen AH, Boesgaard S, Gustafsson F, Rossing K, Small EM, Davies MJ, Rothenberg E, Sato PY, Cerrone M, Jensen THL, Qvortrup K, Bundgaard H, Delmar M, Lundby A. Loss of Nuclear Envelope Integrity and Increased Oxidant Production Cause DNA Damage in Adult Hearts Deficient in PKP2: A Molecular Substrate of ARVC. Circulation. 2022;146:851-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 61. | Stroud MJ. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiomyopathy. Biophys Rev. 2018;10:1033-1051. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 62. | Ishizu T, Higo S, Masumura Y, Kohama Y, Shiba M, Higo T, Shibamoto M, Nakagawa A, Morimoto S, Takashima S, Hikoso S, Sakata Y. Targeted Genome Replacement via Homology-directed Repair in Non-dividing Cardiomyocytes. Sci Rep. 2017;7:9363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 63. | Kohama Y, Higo S, Masumura Y, Shiba M, Kondo T, Ishizu T, Higo T, Nakamura S, Kameda S, Tabata T, Inoue H, Motooka D, Okuzaki D, Takashima S, Miyagawa S, Sawa Y, Hikoso S, Sakata Y. Adeno-associated virus-mediated gene delivery promotes S-phase entry-independent precise targeted integration in cardiomyocytes. Sci Rep. 2020;10:15348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 64. | Tilemann L, Ishikawa K, Weber T, Hajjar RJ. Gene therapy for heart failure. Circ Res. 2012;110:777-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 107] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 65. | Ishikawa K, Weber T, Hajjar RJ. Human Cardiac Gene Therapy. Circ Res. 2018;123:601-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 66. | Chamberlain K, Riyad JM, Weber T. Cardiac gene therapy with adeno-associated virus-based vectors. Curr Opin Cardiol. 2017;32:275-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 67. | Greenberg B, Yaroshinsky A, Zsebo KM, Butler J, Felker GM, Voors AA, Rudy JJ, Wagner K, Hajjar RJ. Design of a phase 2b trial of intracoronary administration of AAV1/SERCA2a in patients with advanced heart failure: the CUPID 2 trial (calcium up-regulation by percutaneous administration of gene therapy in cardiac disease phase 2b). JACC Heart Fail. 2014;2:84-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 68. | Hasenfuss G, Pieske B. Calcium cycling in congestive heart failure. J Mol Cell Cardiol. 2002;34:951-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 315] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 69. | Kho C, Lee A, Hajjar RJ. Altered sarcoplasmic reticulum calcium cycling--targets for heart failure therapy. Nat Rev Cardiol. 2012;9:717-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 70. | Eisner D, Caldwell J, Trafford A. Sarcoplasmic reticulum Ca-ATPase and heart failure 20 years later. Circ Res. 2013;113:958-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 71. | del Monte F, Williams E, Lebeche D, Schmidt U, Rosenzweig A, Gwathmey JK, Lewandowski ED, Hajjar RJ. Improvement in survival and cardiac metabolism after gene transfer of sarcoplasmic reticulum Ca(2+)-ATPase in a rat model of heart failure. Circulation. 2001;104:1424-1429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 262] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 72. | Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, Yaroshinsky A, Zsebo KM, Dittrich H, Hajjar RJ; Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID) Investigators. Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation. 2011;124:304-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 579] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 73. | Cutler MJ, Wan X, Plummer BN, Liu H, Deschenes I, Laurita KR, Hajjar RJ, Rosenbaum DS. Targeted sarcoplasmic reticulum Ca2+ ATPase 2a gene delivery to restore electrical stability in the failing heart. Circulation. 2012;126:2095-2104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 74. | Greenberg B, Butler J, Felker GM, Ponikowski P, Voors AA, Desai AS, Barnard D, Bouchard A, Jaski B, Lyon AR, Pogoda JM, Rudy JJ, Zsebo KM. Calcium upregulation by percutaneous administration of gene therapy in patients with cardiac disease (CUPID 2): a randomised, multinational, double-blind, placebo-controlled, phase 2b trial. Lancet. 2016;387:1178-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 358] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 75. | Hulot JS, Salem JE, Redheuil A, Collet JP, Varnous S, Jourdain P, Logeart D, Gandjbakhch E, Bernard C, Hatem SN, Isnard R, Cluzel P, Le Feuvre C, Leprince P, Hammoudi N, Lemoine FM, Klatzmann D, Vicaut E, Komajda M, Montalescot G, Lompré AM, Hajjar RJ; AGENT-HF Investigators. Effect of intracoronary administration of AAV1/SERCA2a on ventricular remodelling in patients with advanced systolic heart failure: results from the AGENT-HF randomized phase 2 trial. Eur J Heart Fail. 2017;19:1534-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 76. | Lyon AR, Babalis D, Morley-Smith AC, Hedger M, Suarez Barrientos A, Foldes G, Couch LS, Chowdhury RA, Tzortzis KN, Peters NS, Rog-Zielinska EA, Yang HY, Welch S, Bowles CT, Rahman Haley S, Bell AR, Rice A, Sasikaran T, Johnson NA, Falaschetti E, Parameshwar J, Lewis C, Tsui S, Simon A, Pepper J, Rudy JJ, Zsebo KM, Macleod KT, Terracciano CM, Hajjar RJ, Banner N, Harding SE. Investigation of the safety and feasibility of AAV1/SERCA2a gene transfer in patients with chronic heart failure supported with a left ventricular assist device - the SERCA-LVAD TRIAL. Gene Ther. 2020;27:579-590. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 77. | Repetti GG, Toepfer CN, Seidman JG, Seidman CE. Novel Therapies for Prevention and Early Treatment of Cardiomyopathies. Circ Res. 2019;124:1536-1550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 78. | Hakui H, Kioka H, Miyashita Y, Nishimura S, Matsuoka K, Kato H, Tsukamoto O, Kuramoto Y, Takuwa A, Takahashi Y, Saito S, Ohta K, Asanuma H, Fu HY, Shinomiya H, Yamada N, Ohtani T, Sawa Y, Kitakaze M, Takashima S, Sakata Y, Asano Y. Loss-of-function mutations in the co-chaperone protein BAG5 cause dilated cardiomyopathy requiring heart transplantation. Sci Transl Med. 2022;14:eabf3274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 79. | Calkins H, Corrado D, Marcus F. Risk Stratification in Arrhythmogenic Right Ventricular Cardiomyopathy. Circulation. 2017;136:2068-2082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 147] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 80. | van Tintelen JP, Entius MM, Bhuiyan ZA, Jongbloed R, Wiesfeld AC, Wilde AA, van der Smagt J, Boven LG, Mannens MM, van Langen IM, Hofstra RM, Otterspoor LC, Doevendans PA, Rodriguez LM, van Gelder IC, Hauer RN. Plakophilin-2 mutations are the major determinant of familial arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation. 2006;113:1650-1658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 256] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 81. | Ramond F, Janin A, Di Filippo S, Chanavat V, Chalabreysse L, Roux-Buisson N, Sanlaville D, Touraine R, Millat G. Homozygous PKP2 deletion associated with neonatal left ventricle noncompaction. Clin Genet. 2017;91:126-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 82. | Katanyuwong P, Khongkraparn A, Wattanasirichaigoon D. A Novel Homozygous PKP2 Variant in Severe Neonatal Non-compaction and Concomitant Ventricular Septal Defect: A Case Report. Front Pediatr. 2021;9:801491. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 83. | Verhagen JMA, van den Born M, Kurul S, Asimaki A, van de Laar IMBH, Frohn-Mulder IME, Kammeraad JAE, Yap SC, Bartelings MM, van Slegtenhorst MA, von der Thüsen JH, Wessels MW. Homozygous Truncating Variant in PKP2 Causes Hypoplastic Left Heart Syndrome. Circ Genom Precis Med. 2018;11:e002397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 84. | Li J, Zhang L, Yu L, Minami I, Miyagawa S, Hörning M, Dong J, Qiao J, Qu X, Hua Y, Fujimoto N, Shiba Y, Zhao Y, Tang F, Chen Y, Sawa Y, Tang C, Liu L. Circulating re-entrant waves promote maturation of hiPSC-derived cardiomyocytes in self-organized tissue ring. Commun Biol. 2020;3:122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 85. | Li J, Minami I, Shiozaki M, Yu L, Yajima S, Miyagawa S, Shiba Y, Morone N, Fukushima S, Yoshioka M, Li S, Qiao J, Li X, Wang L, Kotera H, Nakatsuji N, Sawa Y, Chen Y, Liu L. Human Pluripotent Stem Cell-Derived Cardiac Tissue-like Constructs for Repairing the Infarcted Myocardium. Stem Cell Reports. 2017;9:1546-1559. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |