Published online Sep 26, 2022. doi: 10.4252/wjsc.v14.i9.714
Peer-review started: May 2, 2022
First decision: June 11, 2022
Revised: June 24, 2022
Accepted: August 15, 2022
Article in press: August 15, 2022
Published online: September 26, 2022
Processing time: 143 Days and 21.5 Hours
The effect of hypoxia on mesenchymal stem cells (MSCs) is an emerging topic in MSC biology. Although long non-coding RNAs (lncRNAs) and messenger RNAs (mRNAs) are reported to play a critical role in regulating the biological characteristics of MSCs, their specific expression and co-expression profiles in human placenta-derived MSCs (hP-MSCs) under hypoxia and the underlying mech
To reveal the specific expression profiles of lncRNAs in hP-MSCs under hypoxia and initially explored the possible mechanism of lncRNAs on hP-MSC biology.
Here, we used a multigas incubator (92.5% N2, 5% CO2, and 2.5% O2) to mimic the hypoxia condition and observed that hypoxic culture significantly promoted the proliferation potential of hP-MSCs. RNA sequencing technology was applied to identify the exact expression profiles of lncRNAs and mRNAs under hypoxia.
We identified 289 differentially expressed lncRNAs and 240 differentially expressed mRNAs between the hypoxia and normoxia groups. Among them, the lncRNA SNHG16 was upregulated under hypoxia, which was also validated by reverse transcription-polymerase chain reaction. SNHG16 was confirmed to affect hP-MSC proliferation rates using a SNHG16 knockdown model. SNHG16 overexpression could significantly enhance the proliferation capacity of hP-MSCs, activate the PI3K/AKT pathway, and upregulate the expression of cell cycle-related proteins.
Our results revealed the specific expression characteristics of lncRNAs and mRNAs in hypoxia-cultured hP-MSCs and that lncRNA SNHG16 can promote hP-MSC proliferation through the PI3K/AKT pathway.
Core Tip: This study revealed the specific expression and co-expressed profiles of long non-coding RNAs and messenger RNAs in human placenta-derived mesenchymal stem cells under hypoxia by RNA sequencing assays. Through the performance of a series of systemic bioinformatic analyses, the hypoxia-responsive long non-coding RNA SNHG16 that may play a role in proliferation was screened out. Furthermore, through the use of molecular biology experiments, SNHG16 was found to affect human placenta-derived mesenchymal stem cell proliferation rates and cell cycle progression by activating the PI3K/AKT pathway and upregulating the expression of the key cell cycle regulators.
- Citation: Feng XD, Zhou JH, Chen JY, Feng B, Hu RT, Wu J, Pan QL, Yang JF, Yu J, Cao HC. Long non-coding RNA SNHG16 promotes human placenta-derived mesenchymal stem cell proliferation capacity through the PI3K/AKT pathway under hypoxia. World J Stem Cells 2022; 14(9): 714-728
- URL: https://www.wjgnet.com/1948-0210/full/v14/i9/714.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i9.714
Recently, mesenchymal stem cells (MSCs) have gained much attention due to their therapeutic effects and potential applications in regenerative medicine[1]. MSCs have recently been shown to have therapeutic efficacy in various disease models and clinical diseases such as liver injury, coronavirus disease 2019, and Crohn’s disease[2-5]. MSCs have been reported to be present in bone marrow, pla
To improve the proliferation potential of hP-MSCs in vitro, most researchers use different methods to stimulate the microenvironment of MSCs in vivo[10]. Among the proposed approaches to mimic the natural cellular microenvironment, hypoxia has garnered enormous interest. Hypoxia has been observed in different tissue niches, including the placenta (1%-5% O2) where hP-MSCs reside[11]. Since the oxygen concentration (almost 21%) in the ex vivo culture system is much higher than the physiological oxygen concentration in the body, hypoxia could act as a physiological stimulus with a significant influence on cell fate. Numerous studies have reported that hypoxia can affect various biological properties of MSCs, such as proliferation capacity, multidirectional differentiation potential, migration, and apoptosis[12-14]. However, the underlying molecular mechanisms by which hypoxia regulates MSC biology remain unclear.
Long non-coding RNAs (lncRNAs) are RNAs longer than 200 nt with no protein-coding potential[15]. LncRNAs are the coordinators of the cellular biological regulatory network, participating in a variety of biological and pathological cellular processes such as cellular survival, proliferation, or migration through regulation of gene expression at transcriptional, post-transcriptional, or translational levels[16]. With advancements in gene sequencing technology, more and more lncRNAs related to cellular functions have been identified. However, the impact of hypoxia on the lncRNA expression profile of MSCs remains unclear. In addition, the roles of hypoxia-responsive lncRNAs remain to be explored. In this study, we investigated the effect of hypoxia on the proliferation potential of hP-MSCs and explored the role of lncRNAs in it.
The protocols for hP-MSC isolation and hypoxic culture were as previously described[7]. All protocols for the processing of human tissues and cells were approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang University (No. 2020-1088).
For the colony-forming unit-fibroblast assay, 1000 hP-MSCs were plated on six-well plates in triplicate and cultured in complete medium for 14 d under normoxic or hypoxic conditions with medium changes every 3 d. Colonies were fixed with paraformaldehyde and then stained with crystal violet for enumeration.
The corresponding cells were inoculated into 96-well cell culture plates at a density of 2000 cells per well. After 24, 48, 72 or 96-h culture, 10 μL of cell counting kit-8 reagent (Dojindo, Kumamoto, Japan) was added into each well to incubate for 2 h. The optical density value at 450 nm was measured using a microplate reader.
The cells were collected with trypsin and fixed with cold 70% ethanol for 2 h. Fixed cells were then treated with propidium iodide staining solution (Beyotime, Nanjing, China). The cells were finally analyzed by flow cytometry. The proportions of cell population in G0/G1, S, and G2/M phases of the cell cycle were fitted and calculated using ModFit software.
Total RNA of cells was obtained using Trizol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s protocol; the concentration of total RNA was quantified using a NanoDrop-2000 (Thermo Fisher Scientific, Waltham, MA, United States). cDNA was synthesized by reverse transcription reaction using a commercial lncRNA quantitative reverse transcription polymerase chain reaction (PCR) Starter Kit (RiboBio, Guangzhou, China). The final relative expression levels of genes were analyzed through the 2−ΔΔCt method using GAPDH as the internal control. Primers were as follows: GAPDH: (forward) 5’-ACAACTTTGGTATCGTGGAAGG-3’, (reverse) 5’-GCCATCACGCCACAGTTTC-3’; SNHG16: (forward) 5’-GTTGCCACCCACAACCATT-3’, and (reverse) 5’-GCGGAGACACCAGGAGAACT-3’.
The cellular protein was harvested using RIPA lysis buffer supplemented with protease and phosphatase inhibitor cocktail (Beyotime). The protein concentrations were detected using a BCA kit (Beyotime). The western blot was conducted as previously described[7]. The primary antibodies were anti-β-actin (Abcam, Cambridge, United Kingdom), anti-GAPDH (Abcam), anti-hypoxia-inducible factor 1α (HIF-1α) (Cell Signaling Technology, Danvers, MA, United States), anti-c-MYC (Abcam), anti-proliferating cell nuclear antigen (Abcam), anti-CDK2 (Abcam), anti-CDK4 (Abcam), anti-CDK6 (Abcam), anti-CyclinD1 (Abcam), anti-CyclinE1 (Abcam), anti-AKT (Abcam), and anti-phospho-AKT (Abcam).
Whole-transcriptome sequencing was quantitatively analyzed by Oebiotech (Shanghai, China). The libraries [including lncRNA and messenger RNA (mRNA)] were generated using TruSeq Stranded Total RNA with Ribo-Zero Gold (Illumina, San Diego, CA, United States) according to the manual. RNA was then sequenced on a HiSeq 2500 instrument (Illumina). Differential expression analysis of lncRNA and mRNA between the hypoxic and normoxic groups was conducted using the DESeq software package. The differentially expressed genes were identified with the criteria of fold change > 1.5 and P < 0.05. The Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed for differentially expressed mRNAs to explore their biological functions. Association analysis between lncRNAs and transcription factors and lncRNA–mRNA co-expression analysis were conducted to investigate lncRNA functions in cell biology.
Lentivirus-mediated short hairpin RNA for silencing SNHG16 in cells and recombinant lentivirus for SNHG16 overexpression were constructed by Genomeditech (Shanghai, China). Transfection was performed following the manufacturer's instructions. Lentiviruses were added to infect cells at a multiplicity of infection of 50:1.
All data were expressed as the mean ± SD. Statistical evaluation of two groups was conducted using Student’s t test; a P value < 0.05 was considered to indicate statistical significance.
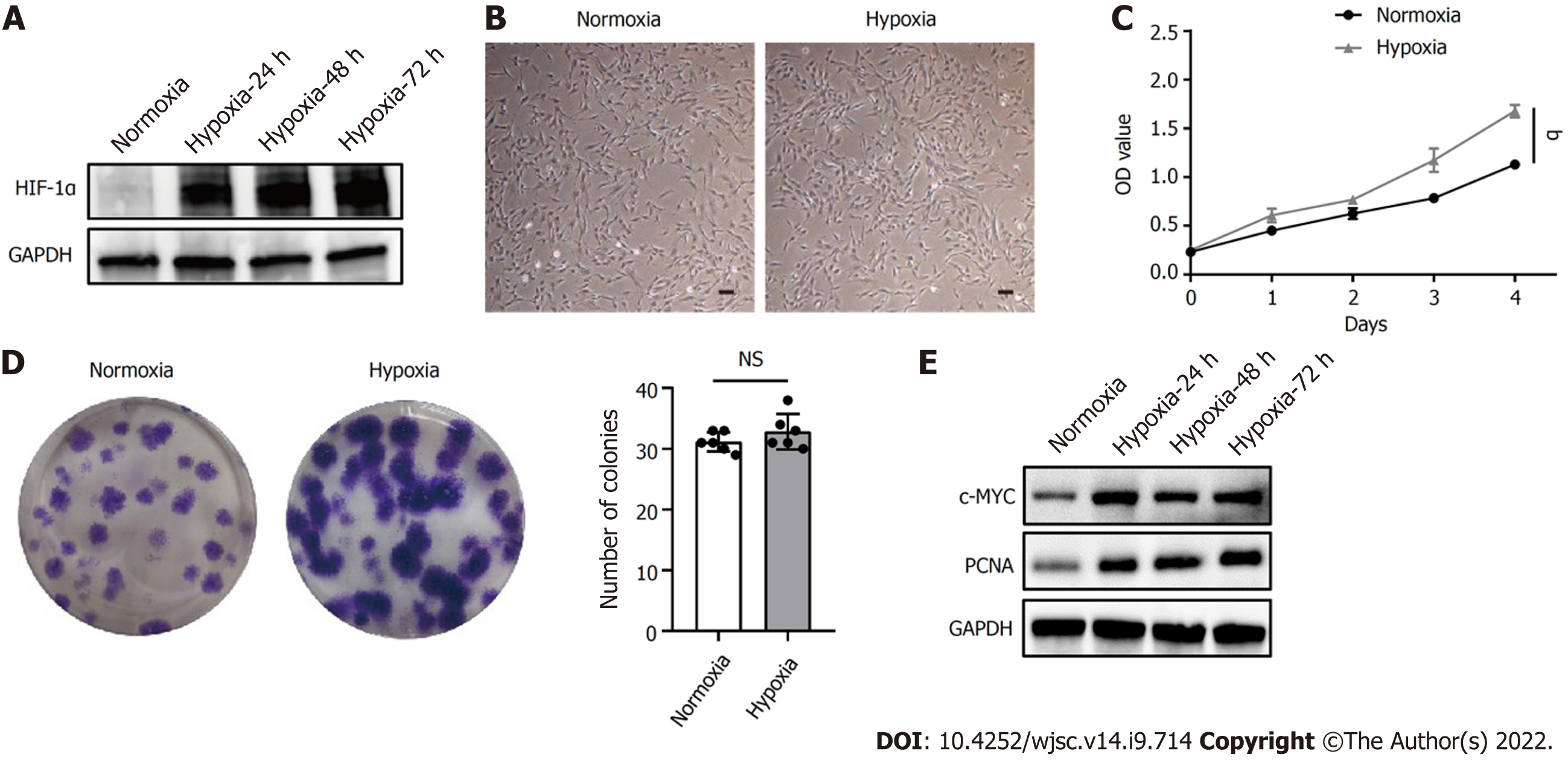
HIF-1α is a critical regulator of cellular adaptation to the hypoxic microenvironment. When the expression of HIF-1α protein under hypoxia was assessed by western blot, hypoxia induced the expression of HIF-1α in hP-MSCs and stabilized its expression level during cell growth (Figure 1A). In addition, hP-MSCs cultured under hypoxia appeared to be relatively small, with a spindle-shaped morphology (Figure 1B). The cell counting kit-8 assay showed that hP-MSCs had higher proliferation potential (P < 0.0001) when they were maintained under hypoxia (Figure 1C). Similarly, the colony-forming unit-fibroblast assay indicated that hypoxia enhanced the hP-MSC proliferation rate. Although the difference in the number of hP-MSC colonies between the hypoxia and normoxia groups was not significant (P = 0.249), the colony size of the hypoxia group was larger with darker staining, indicating a higher number of cells (Figure 1D). c-MYC and proliferating cell nuclear antigen are molecules closely related to cell proliferation and can be adopted to determine the status of cell proliferation. As expected, hypoxia significantly increased the expression of c-MYC and proliferating cell nuclear antigen, indicating that cells proliferated more rapidly under hypoxia (Figure 1E).
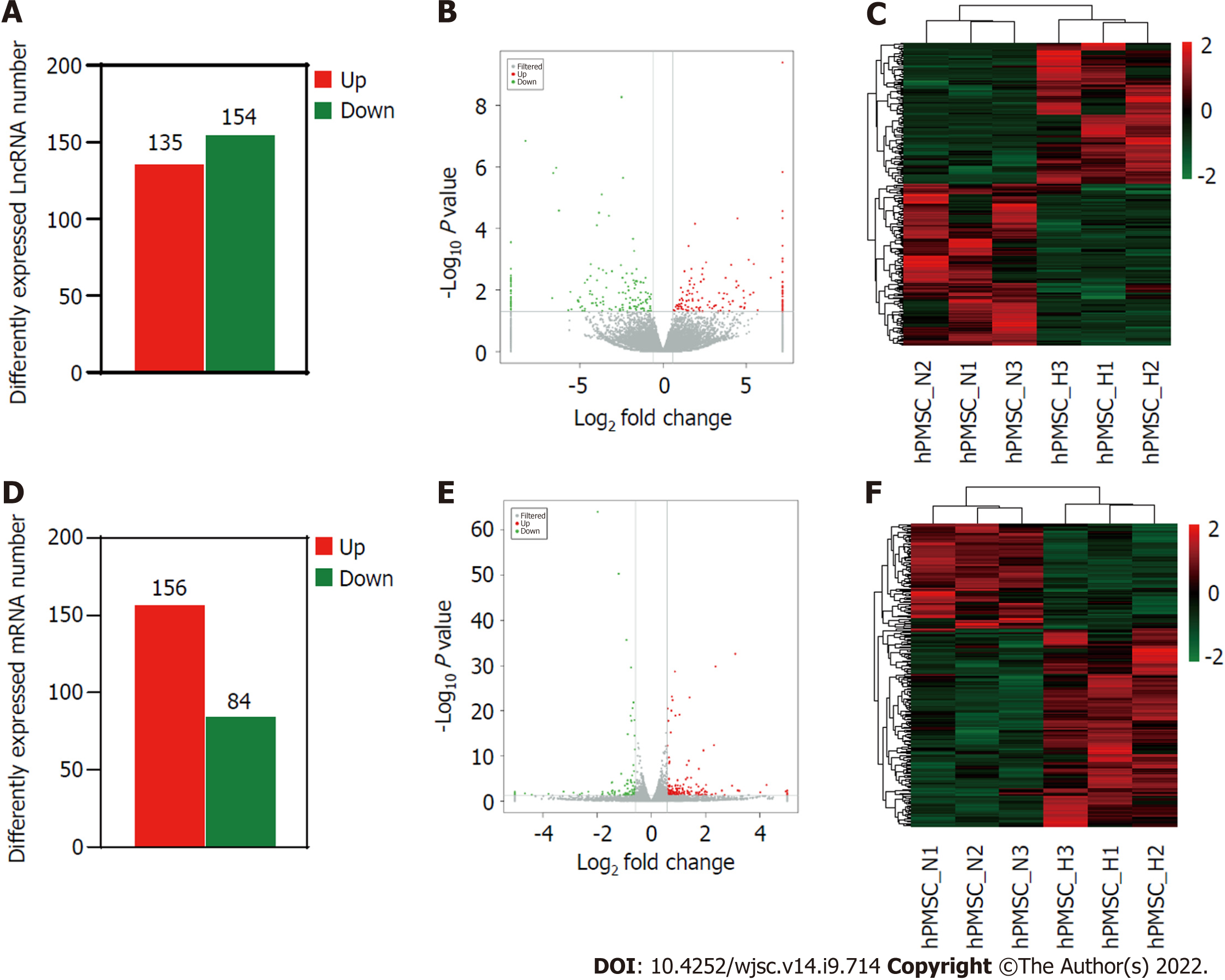
To further investigate the influence of hypoxia on hP-MSCs, whole-transcriptome sequencing was performed. First, six high-throughput sequenced transcriptomes were generated, containing over 650 million clean reads, among which three were from the normoxic group and three were from the hypoxic group. More than 96% of the raw reads were high-quality clean reads (Table 1). Ultimately, 10387 putative lncRNAs and 16041 mRNAs were identified. We further identified 289 differentially expressed lncRNAs (135 upregulated and 154 downregulated) and 240 differentially expressed mRNAs (156 upregulated and 84 downregulated) in the hypoxia group compared to normoxia group (Figure 2). Heatmap analysis clearly distinguished the hP-MSCs cultured under hypoxia from those cultured under normoxia. The top 20 differentially expressed lncRNAs and mRNAs are summarized in Tables 2 and 3.
| Summary | MSC_N1 | MSC_N2 | MSC_N3 | MSC_H1 | MSC_H2 | MSC_H3 |
| Raw reads (M) | 103.22 | 113.01 | 117.09 | 114.00 | 117.77 | 106.81 |
| Clean reads (M) | 99.95 | 109.86 | 113.73 | 110.04 | 113.72 | 103.86 |
| Clean reads rate (%) | 96.83 | 97.21 | 97.13 | 96.53 | 96.56 | 97.24 |
| Q30 (%) | 94.32 | 94.63 | 94.62 | 94.22 | 94.18 | 94.78 |
| GC (%) | 46.71 | 45.65 | 45.35 | 46.18 | 44.99 | 45.92 |
| Total mapped reads | 320502139 | 324502629 | ||||
| Uniquely mapped reads | 308534583 | 312982527 | ||||
| Upregulated lncRNA | Fold-change | Downregulated lncRNA | Fold-change |
| ENST00000480904 | 144.7046633 | TCONS_00024987 | 558.6529515 |
| ENST00000420168 | 88.58973499 | TCONS_00022901 | 305.9141231 |
| ENST00000447687 | 51.87793204 | ENST00000580684 | 101.5147448 |
| TCONS_00022897 | 43.87602707 | XR_951092.3 | 96.15131381 |
| NR_135828.1 | 43.01226344 | TCONS_00040744 | 85.07807992 |
| ENST00000652331 | 37.64067361 | NR_151707.1 | 75.91477831 |
| ENST00000615566 | 35.57471309 | XR_924538.2 | 51.52143558 |
| XR_001740831.1 | 30.29369126 | ENST00000533146 | 47.14143364 |
| XR_943245.2 | 29.4766044 | NR_046472.1 | 45.2199291 |
| ENST00000641463 | 29.12107436 | ENST00000379848 | 34.42041339 |
| ENST00000587838 | 28.77216197 | XR_002957073.1 | 33.6627267 |
| NR_027295.2 | 28.51478291 | NR_138037.1 | 31.71645024 |
| XR_001738493.2 | 26.08189868 | ENST00000424751 | 31.28206241 |
| ENST00000622955 | 24.98325918 | ENST00000476224 | 29.12601955 |
| ENST00000437589 | 23.11958922 | NR_152515.1 | 28.62941025 |
| NR_028397.1 | 22.10295425 | ENST00000608741 | 25.94241325 |
| NR_138259.1 | 21.16258012 | XR_001740695.2 | 25.76946681 |
| XR_947992.2 | 21.13351381 | NR_102280.1 | 25.69150571 |
| XR_930796.2 | 18.16731229 | ENST00000513626 | 23.42886779 |
| ENST00000542086 | 17.05330496 | NR_132369.1 | 21.10943719 |
| Upregulated mRNA | Fold-change | Downregulated mRNA | Fold-change |
| S100A1 | 32.28708068 | MMP13 | 32.7146796 |
| IL20 | 31.01976446 | RASAL3 | 25.22521191 |
| GUCY2D | 18.95647724 | ITGAM | 21.2376519 |
| PIK3R5 | 16.27652505 | FGF14 | 13.88641698 |
| KCNJ15 | 9.485618545 | KCNS2 | 9.402226087 |
| CA9 | 9.092268285 | DCT | 7.332486828 |
| AK4 | 8.577190243 | TMEM247 | 6.224926429 |
| CD99 | 7.853653254 | TNFRSF4 | 5.89336975 |
| VSIG2 | 6.545400641 | SOX7 | 5.002461264 |
| CKMT2 | 6.072884506 | TXNIP | 3.914446648 |
| FOLR1 | 5.195878685 | NBPF6 | 3.514713231 |
| C5orf46 | 5.180793591 | 3.476083157 | |
| PPFIA4 | 4.979164209 | LHX4 | 3.46662373 |
| TEC | 4.732591252 | CD14 | 3.005111835 |
| INHBB | 4.597149928 | HHIPL2 | 2.918560997 |
| GLDC | 4.140246682 | RASL11B | 2.770760137 |
| GPR146 | 4.107008037 | LMO3 | 2.767289903 |
| VASH2 | 4.051761622 | MYH11 | 2.728808288 |
| C4orf47 | 3.969880654 | DIRAS2 | 2.718905726 |
| SCHIP1 | 3.945719906 | AJM1 | 2.696612138 |
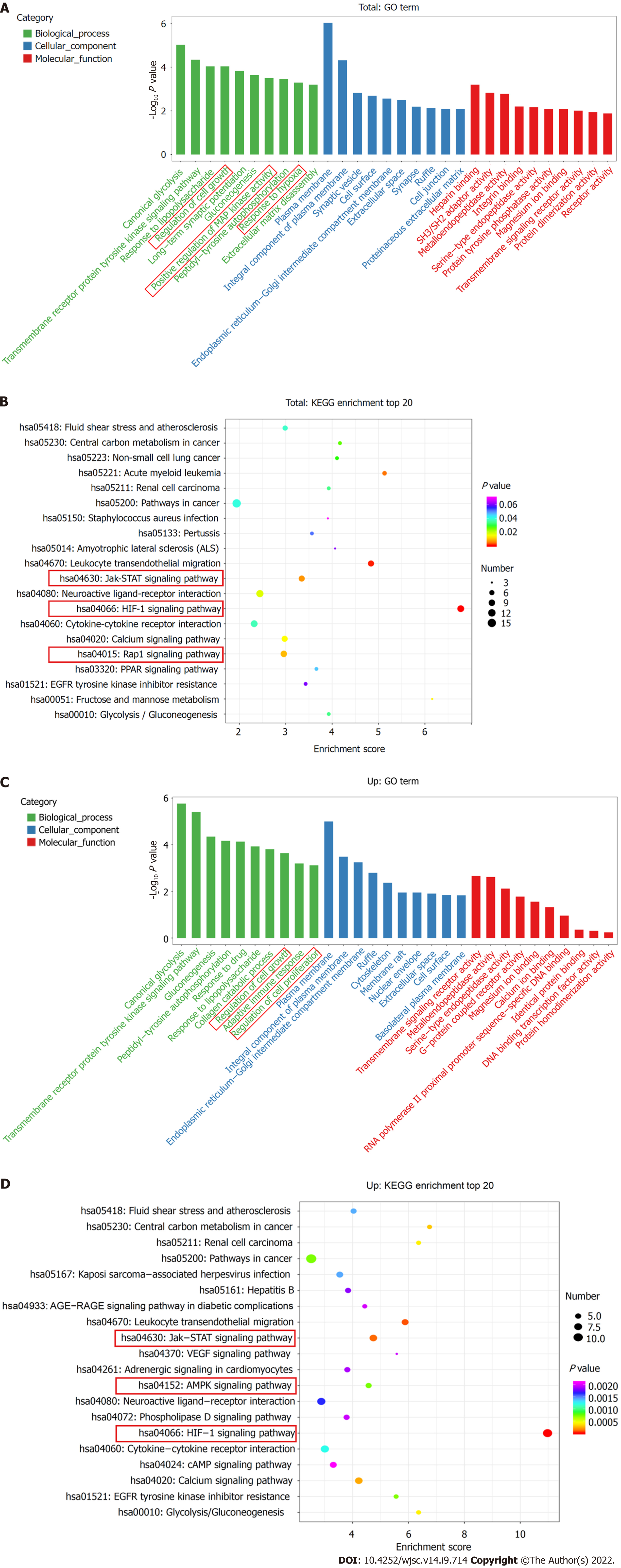
The top ten enriched GO terms in biological process, molecular function, and cellular component were determined. For biological process, the differentially expressed mRNAs were related to regulation of cell growth (GO: 0001558), positive regulation of MAP kinase activity (GO: 0043406), and response to hypoxia (GO: 0001666) (Figure 3A). KEGG pathway enrichment analysis revealed several significantly enriched pathways, such as the HIF-1 signaling pathway (KEGG: hsa04066), Jak-STAT signaling pathway (KEGG: hsa04630), and Rap1 signaling pathway (KEGG: has04015) (Figure 3B). We further performed GO and KEGG analyses on the upregulated and downregulated genes separately. Upregulated genes were involved in regulation of cell growth (GO: 0001558) and regulation of cell proliferation (GO: 0042127) (Figure 3C) and were enriched in the HIF-1 signaling pathway (KEGG: hsa04066), Jak-STAT signaling pathway (KEGG: hsa04630), and AMPK signaling pathway (KEGG: hsa04152) (Figure 3D). Thus, hypoxia mainly affected cell functions, such as proliferation, by upregulating the expression of certain genes through several signaling pathways.
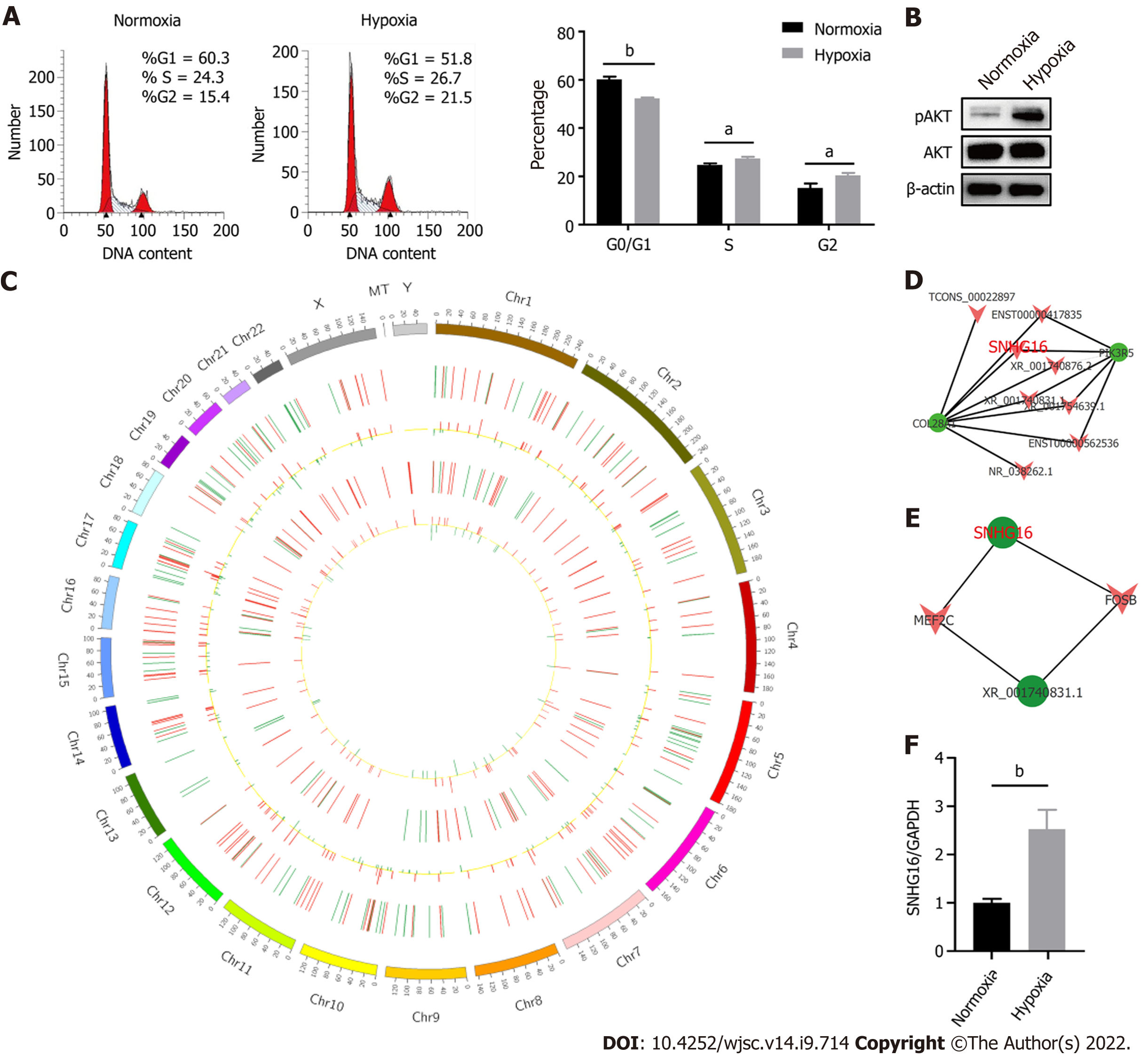
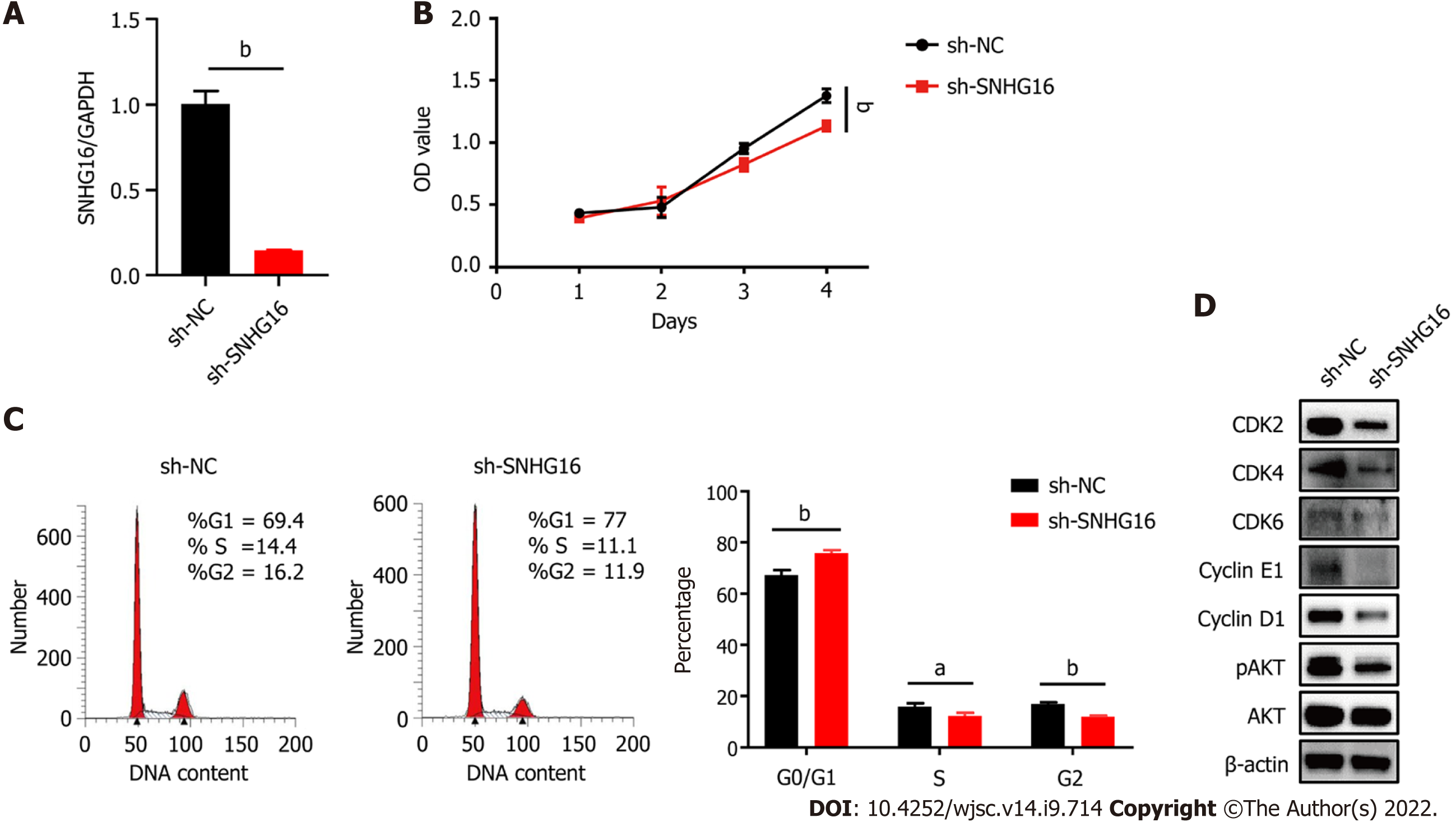
Hypoxia affected cell proliferation by regulating cell cycle progression as the percentages of S (P = 0.011) and G2/M phase cells (P = 0.014) were larger under the hypoxic condition (Figure 4A). At the same time, the PI3K/AKT pathway was activated under hypoxia (Figure 4B). The PI3K/AKT pathway is responsible for coordinating a diverse range of cell functions, including proliferation and survival. These findings suggest that hypoxia can activate the PI3K/AKT pathway and modulate the cell cycle. To explore whether there are specific hypoxia-responsive lncRNAs that play a role in hypoxia-promoted cell proliferation, association analysis between lncRNAs and transcription factors and lncRNA–mRNA co-expression analysis were performed. SNHG16 was related to the expression of PIK3R5, a gene encoding the regulatory subunit of the PI3K gamma complex (Figure 4C and D). SNHG16 was also correlated with FOSB, a key transcription factor in the cell cycle (Figure 4E). Quantitative reverse transcription PCR analysis confirmed that hypoxia induced the expression of SNHG16 (P = 0.003), consistent with the results of RNA sequencing (Figure 4F). Thus, SNHG16 is a potential promoter of hP-MSC proliferation under hypoxia.
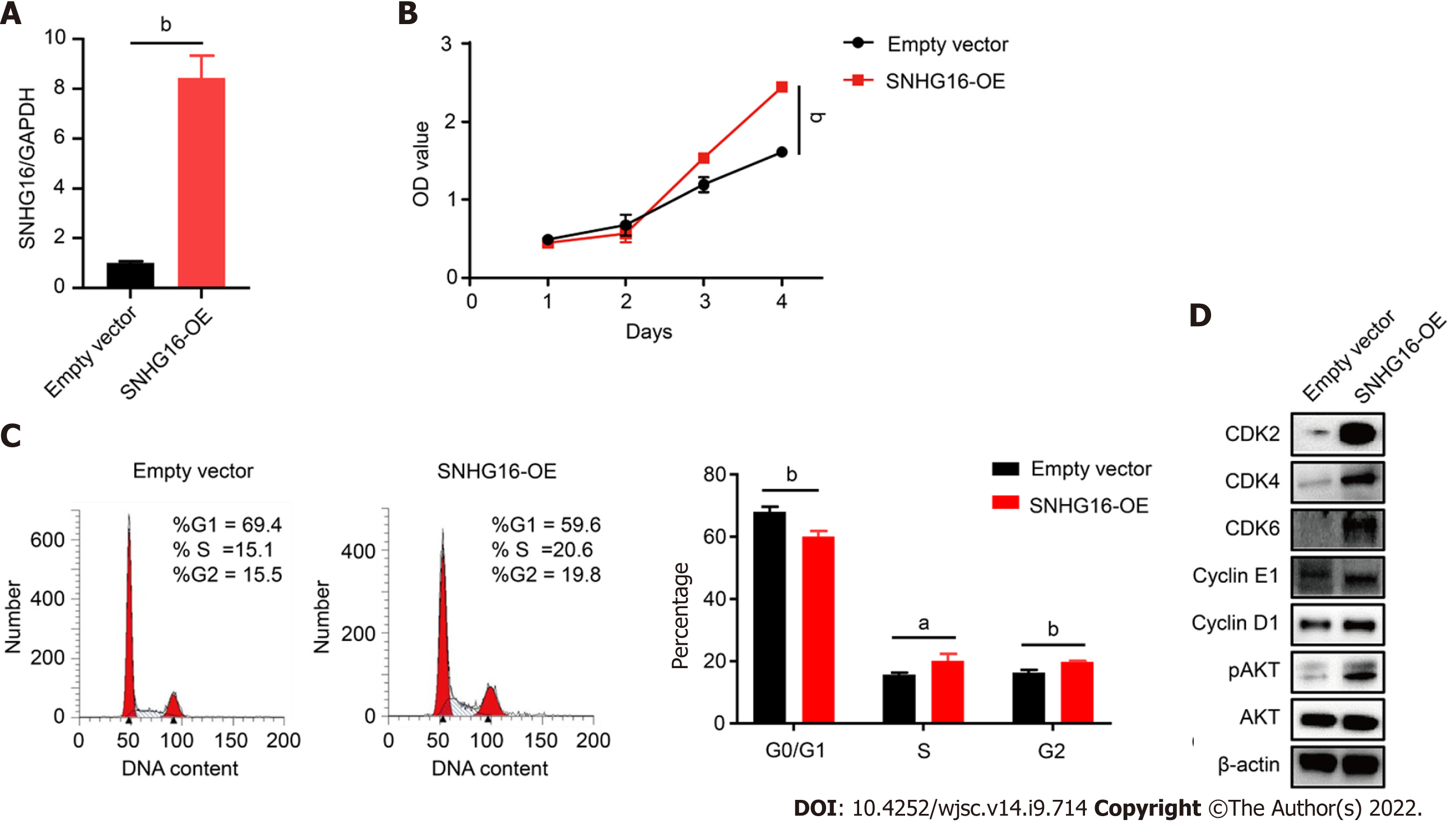
To further confirm the biological function of SNHG16 in hP-MSCs, short hairpin RNA was used to specifically knock down SNHG16, whereas lentivirus overexpressing SNHG16 was used to increase SNHG16 expression. By transfecting SNHG16 short hairpin RNA, we found that sh-SNHG16 sign
MSCs have great potential to cure a variety of diseases, as evidenced by the rapid growth in the number of published preclinical and clinical studies. However, MSCs are found in very small numbers in most adult tissues, such as bone marrow, placenta, adipose tissue, umbilical cord, amniotic fluid, and muscle[17,18]. To generate sufficient clinical therapeutic quantities, in vitro expansion is necessary[19]. Managing and modifying culture conditions during amplification of MSCs in vitro is critical for the manufacture of effective cell therapies, as these in vitro culture conditions affect the cell properties and cell behaviors after transplantation[20].
MSCs are widely located in the hypoxic microenvironment[21,22]. This physiological oxygen concentration is significantly lower than normoxic conditions typically used for MSC culture in the laboratory. Therefore, the application of physiological oxygen tension in stem cell research has attracted attention. Culturing MSCs under hypoxia has been consistently associated with increased cell proliferative rate, increased clonogenicity, decreased spontaneous differentiation, transcriptional alterations, and other cellular behaviors[11,23-25].
In the current study, we focused on the influence of hypoxia on hP-MSC proliferation ability. We found that hypoxic culture could facilitate hP-MSC proliferation, but enhanced clonogenicity under hypoxia was not observed in hP-MSCs. This finding provides a basis for exploring the underlying mechanism of the increased proliferation of hP-MSCs under hypoxic conditions.
Previous findings suggested that lncRNAs could exert regulatory function in MSC proliferation or differentiation. For example, Meng et al[26] revealed that lincRNA-p21 promotes the migration and survival capabilities of mouse bone marrow-derived MSCs via the HIF-1α/CXCR4 and CXCR7 axis under hypoxia[26]. In addition, LINC01119 negatively regulates osteogenic differentiation of human bone marrow-derived MSCs, while the lncRNAs LOC100126784 and POM121L9P improve the osteogenic differentiation of human bone marrow-derived MSCs[27,28]. However, the role of lncRNAs in hP-MSCs has rarely been reported.
Here, we employed RNA sequencing technology to obtain a comprehensive and systematic un
The results of the bioinformatic analysis were consistent with our experimental results. Combined with the individual analysis of upregulated genes, we found that hypoxia affected multiple cellular functions, mainly through upregulating the expression of certain genes. Moreover, hypoxia could mediate cell cycle progression and activate the PI3K/AKT pathway. Similarly, lncRNA–mRNA co-expression network analysis indicated that SNHG16, a hypoxia-responsive lncRNA, is associated with key genes in the cell cycle or PI3K/AKT pathway. Therefore, SNHG16 was selected as a potential promoter of the hP-MSC proliferative rate under hypoxia.
SNHG16 is a member of the SNHGs and is well-documented for its oncogenic properties in numerous types of malignancies[29]. SNHG16 is reported to be involved in multiple cell biological functions, including cell cycle progression, proliferation, and migration[30-32]. In our study, we found that hypoxic culture could induce the expression of SNHG16 in hP-MSCs. We further verified that SNHG16 could promote cell cycle progression and cell proliferation of hP-MSCs by using knockdown and overexpression models. Moreover, we demonstrated that overexpression of SNHG16 could increase the phosphorylation of AKT with a simultaneous elevation in the expression levels of G1 to S phase transition related proteins, including CDK2, CDK4, CDK6, cyclin E1, and cyclin D1.
However, how SNHG16 becomes integrated in the PI3K/AKT signaling pathway in the study remains unknown. There are some related articles on the mechanism by which SNHG16 regulates the AKT pathway in other models. For example, SNHG16 could activate the PI3K/AKT pathway through SNHG16/miR-338-3p/PLK4 axis in cisplatin-resistant neuroblastoma cells[33]. Moreover, SNHG16 was found to facilitate proliferative diabetes-related abnormalities in cell proliferation through regulating miR-7-5p/IRS1 to activate PI3K/AKT pathway in HG-stimulated hRMECs[31]. It can be seen that SNHG16 mainly acts as a competing endogenous RNA to participate in the regulation of the PI3K/AKT signaling pathway. Our follow-up studies will take this as a starting point to elucidate the detailed mechanism of SNHG16 regulation of the PI3K/AKT pathway.
In this study, we have shown that hypoxia enhanced hP-MSCs proliferation ability and could specifically alter the lncRNA and mRNA expression profile. Furthermore, we identified a hypoxia-responsive lncRNA, SNHG16, which may serve as a regulator of promoting hP-MSCs proliferation under hypoxia. Mechanically, SNHG16 was shown to activate the PI3K/AKT signaling pathway and upregulate the expression of key cell cycle regulators to induce cell cycle transition.
As the role of hypoxia on mesenchymal stem cells (MSCs) is an emerging topic of MSCs biology, increasing studies are devoted to researching the regulation mechanisms of hypoxia on the biological functions of MSCs. Long non-coding RNAs (lncRNAs) and messenger RNAs (mRNAs) are reported to possess a critical role in regulating MSC biological characteristics. Nonetheless, the specific expression and co-expressed profiles of lncRNAs and mRNAs in human placenta-derived MSCs (hP-MSCs) under hypoxia and underlying mechanism of lncRNAs on hP-MSCs biology are still unknown.
Although some studies have explored the effects of hypoxia on MSCs, the role of lncRNAs in them remains unclear.
In this study, we aimed to reveal the specific expression profiles of lncRNAs in hP-MSCs under hypoxia and initially explored the possible mechanism of lncRNAs on hP-MSCs biology.
Here, we used a multigas incubator (92.5% N2, 5%CO2 and 2.5% O2) to mimic a hypoxia condition and observed that hypoxic culture can significantly promote the proliferation potential of hP-MSCs. RNA sequencing technology was applied to identify the exact expression profiles of lncRNAs and mRNAs under hypoxia. After establishment of SNHG16-knockdown and SNHG16-overexpression hP-MSCs, the effect of SNHG16 on proliferation capacity of hP-MSCs was analyzed via cell counting kit-8 and cell cycle analysis. Finally, the underlying mechanism was analyzed by western blot.
We identified 289 differentially expressed lncRNAs and 240 differentially expressed mRNAs between hypoxia group and normoxia group. Among them, the lncRNA SNHG16 was upregulated under hypoxia, which was also validated by reverse transcription polymerase chain reaction. SNHG16 was confirmed to affect hP-MSCs proliferation rates by studying the SNHG16 knockdown model. SNHG16 overexpression could significantly enhance proliferation capacity of hP-MSCs, activate PI3K/AKT pathway, and upregulate the expression of cell cycle-related proteins.
Our results revealed the specific expression characteristics of lncRNAs and mRNAs in hypoxia-cultured hP-MSCs and identified that hypoxia-responsive lncRNA SNHG16 can promote hP-MSC proliferation through the PI3K/AKT pathway.
This study may contribute to understanding the role of noncoding RNAs in MSC biology.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: Asian Pacific Association for the Study of the Liver, No. MN-2162.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jabbarpour Z, Iran; Luo Y, China; Wang J, China S-Editor: Fan JR L-Editor: Filipodia P-Editor: Zhang YL
| 1. | El Agha E, Kramann R, Schneider RK, Li X, Seeger W, Humphreys BD, Bellusci S. Mesenchymal Stem Cells in Fibrotic Disease. Cell Stem Cell. 2017;21:166-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 317] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 2. | Lightner AL, Faubion WA. Mesenchymal Stem Cell Injections for the Treatment of Perianal Crohn's Disease: What We Have Accomplished and What We Still Need to Do. J Crohns Colitis. 2017;11:1267-1276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Hu C, Zhao L, Zhang L, Bao Q, Li L. Mesenchymal stem cell-based cell-free strategies: safe and effective treatments for liver injury. Stem Cell Res Ther. 2020;11:377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 4. | Zhu R, Yan T, Feng Y, Liu Y, Cao H, Peng G, Yang Y, Xu Z, Liu J, Hou W, Wang X, Li Z, Deng L, Wang S, Li J, Han Q, Li H, Shan G, Cao Y, An X, Yan J, Zhang Z, Qu X, Zhu J, Zhou S, Wang J, Zhang F, Gao J, Jin R, Xu D, Ma YQ, Huang T, Peng S, Zheng Z, Stambler I, Gilson E, Lim LW, Moskalev A, Cano A, Chakrabarti S, Ulfhake B, Su H, Xu H, Xu S, Wei F, Brown-Borg HM, Min KJ, Ellison-Hughes G, Caruso C, Jin K, Zhao RC. Mesenchymal stem cell treatment improves outcome of COVID-19 patients via multiple immunomodulatory mechanisms. Cell Res. 2021;31:1244-1262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 88] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 5. | Shi L, Wang L, Xu R, Zhang C, Xie Y, Liu K, Li T, Hu W, Zhen C, Wang FS. Mesenchymal stem cell therapy for severe COVID-19. Signal Transduct Target Ther. 2021;6:339. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 6. | Wang ZG, He ZY, Liang S, Yang Q, Cheng P, Chen AM. Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells. Stem Cell Res Ther. 2020;11:511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 163] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 7. | Feng XD, Zhu JQ, Zhou JH, Lin FY, Feng B, Shi XW, Pan QL, Yu J, Li LJ, Cao HC. Hypoxia-inducible factor-1α-mediated upregulation of CD99 promotes the proliferation of placental mesenchymal stem cells by regulating ERK1/2. World J Stem Cells. 2021;13:317-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Zhou J, Feng X, Zhu J, Feng B, Yao Q, Pan Q, Yu J, Yang J, Li L, Cao H. Mesenchymal stem cell treatment restores liver macrophages homeostasis to alleviate mouse acute liver injury revealed by single-cell analysis. Pharmacol Res. 2022;179:106229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 9. | Meng F, Xu R, Wang S, Xu Z, Zhang C, Li Y, Yang T, Shi L, Fu J, Jiang T, Huang L, Zhao P, Yuan X, Fan X, Zhang JY, Song J, Zhang D, Jiao Y, Liu L, Zhou C, Maeurer M, Zumla A, Shi M, Wang FS. Human umbilical cord-derived mesenchymal stem cell therapy in patients with COVID-19: a phase 1 clinical trial. Signal Transduct Target Ther. 2020;5:172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 236] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 10. | Samal JRK, Rangasami VK, Samanta S, Varghese OP, Oommen OP. Discrepancies on the Role of Oxygen Gradient and Culture Condition on Mesenchymal Stem Cell Fate. Adv Healthc Mater. 2021;10:e2002058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 11. | Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1249] [Cited by in RCA: 1144] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 12. | Li L, Jaiswal PK, Makhoul G, Jurakhan R, Selvasandran K, Ridwan K, Cecere R. Hypoxia modulates cell migration and proliferation in placenta-derived mesenchymal stem cells. J Thorac Cardiovasc Surg. 2017;154:543-552.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Hung SP, Ho JH, Shih YR, Lo T, Lee OK. Hypoxia promotes proliferation and osteogenic differentiation potentials of human mesenchymal stem cells. J Orthop Res. 2012;30:260-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 190] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 14. | Rosová I, Dao M, Capoccia B, Link D, Nolta JA. Hypoxic preconditioning results in increased motility and improved therapeutic potential of human mesenchymal stem cells. Stem Cells. 2008;26:2173-2182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 566] [Cited by in RCA: 537] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 15. | Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1868] [Article Influence: 116.8] [Reference Citation Analysis (0)] |
| 16. | Taniue K, Akimitsu N. The Functions and Unique Features of LncRNAs in Cancer Development and Tumorigenesis. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 145] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 17. | Liu J, Li P, Zhu J, Lin F, Zhou J, Feng B, Sheng X, Shi X, Pan Q, Yu J, Gao J, Li L, Cao H. Mesenchymal stem cell-mediated immunomodulation of recruited mononuclear phagocytes during acute lung injury: a high-dimensional analysis study. Theranostics. 2021;11:2232-2246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 18. | Phinney DG, Kopen G, Isaacson RL, Prockop DJ. Plastic adherent stromal cells from the bone marrow of commonly used strains of inbred mice: variations in yield, growth, and differentiation. J Cell Biochem. 1999;72:570-585. [PubMed] |
| 19. | Aust L, Devlin B, Foster SJ, Halvorsen YD, Hicok K, du Laney T, Sen A, Willingmyre GD, Gimble JM. Yield of human adipose-derived adult stem cells from liposuction aspirates. Cytotherapy. 2004;6:7-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 524] [Cited by in RCA: 507] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 20. | Elabd C, Ichim TE, Miller K, Anneling A, Grinstein V, Vargas V, Silva FJ. Comparing atmospheric and hypoxic cultured mesenchymal stem cell transcriptome: implication for stem cell therapies targeting intervertebral discs. J Transl Med. 2018;16:222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 21. | Mas-Bargues C, Sanz-Ros J, Román-Domínguez A, Inglés M, Gimeno-Mallench L, El Alami M, Viña-Almunia J, Gambini J, Viña J, Borrás C. Relevance of Oxygen Concentration in Stem Cell Culture for Regenerative Medicine. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 140] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 22. | Barry FP, Murphy JM. Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol. 2004;36:568-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1203] [Cited by in RCA: 1173] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 23. | Adesida AB, Mulet-Sierra A, Jomha NM. Hypoxia mediated isolation and expansion enhances the chondrogenic capacity of bone marrow mesenchymal stromal cells. Stem Cell Res Ther. 2012;3:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 24. | Studer L, Csete M, Lee SH, Kabbani N, Walikonis J, Wold B, McKay R. Enhanced proliferation, survival, and dopaminergic differentiation of CNS precursors in lowered oxygen. J Neurosci. 2000;20:7377-7383. [PubMed] |
| 25. | Das R, Jahr H, van Osch GJ, Farrell E. The role of hypoxia in bone marrow-derived mesenchymal stem cells: considerations for regenerative medicine approaches. Tissue Eng Part B Rev. 2010;16:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 225] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 26. | Meng SS, Xu XP, Chang W, Lu ZH, Huang LL, Xu JY, Liu L, Qiu HB, Yang Y, Guo FM. LincRNA-p21 promotes mesenchymal stem cell migration capacity and survival through hypoxic preconditioning. Stem Cell Res Ther. 2018;9:280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 27. | Gao H, Dong H, Zheng J, Jiang X, Gong M, Hu L, He J, Wang Y. LINC01119 negatively regulates osteogenic differentiation of mesenchymal stem cells via the Wnt pathway by targeting FZD4. Stem Cell Res Ther. 2022;13:43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Xu Y, Xin R, Sun H, Long D, Li Z, Liao H, Xue T, Zhang Z, Kang Y, Mao G. Long Non-coding RNAs LOC100126784 and POM121L9P Derived From Bone Marrow Mesenchymal Stem Cells Enhance Osteogenic Differentiation via the miR-503-5p/SORBS1 Axis. Front Cell Dev Biol. 2021;9:723759. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Zimta AA, Tigu AB, Braicu C, Stefan C, Ionescu C, Berindan-Neagoe I. An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front Oncol. 2020;10:389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 30. | Jiang X, Ru Q, Li P, Ge X, Shao K, Xi L, Xu B, Wang Q, Huang S. LncRNA SNHG16 induces proliferation and fibrogenesis via modulating miR-141-3p and CCND1 in diabetic nephropathy. Gene Ther. 2020;27:557-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Cai F, Jiang H, Li Y, Li Q, Yang C. Upregulation of long non-coding RNA SNHG16 promotes diabetes-related RMEC dysfunction via activating NF-κB and PI3K/AKT pathways. Mol Ther Nucleic Acids. 2021;24:512-527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 32. | Xia W, Liu Y, Cheng T, Xu T, Dong M, Hu X. Extracellular Vesicles Carry lncRNA SNHG16 to Promote Metastasis of Breast Cancer Cells via the miR-892b/PPAPDC1A Axis. Front Cell Dev Biol. 2021;9:628573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 33. | Xu Z, Sun Y, Wang D, Sun H, Liu X. SNHG16 promotes tumorigenesis and cisplatin resistance by regulating miR-338-3p/PLK4 pathway in neuroblastoma cells. Cancer Cell Int. 2020;20:236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |