Published online Jul 26, 2022. doi: 10.4252/wjsc.v14.i7.490
Peer-review started: March 18, 2022
First decision: April 25, 2022
Revised: May 31, 2022
Accepted: July 8, 2022
Article in press: July 8, 2022
Published online: July 26, 2022
Processing time: 129 Days and 17.1 Hours
Stem cell fate determination is one of the central questions in stem cell biology, and although its regulation has been studied at genomic and proteomic levels, a variety of biological activities in cells occur at the metabolic level. Metabolomics studies have established the metabolome during stem cell differentiation and have revealed the role of metabolites in stem cell fate determination. While metabolism is considered to play a biological regulatory role as an energy source, recent studies have suggested the nexus between metabolism and epigenetics because several metabolites function as cofactors and substrates in epigenetic mechanisms, including histone modification, DNA methylation, and microRNAs. Additionally, the epigenetic modification is sensitive to the dynamic metabolites and consequently leads to changes in transcription. The nexus between metabolism and epigenetics proposes a novel stem cell-based therapeutic strategy through manipulating metabolites. In the present review, we summarize the possible nexus between metabolic and epigenetic regulation in stem cell fate determination, and discuss the potential preventive and therapeutic strategies via targeting metabolites.
Core Tip: Stem cell fate can be regulated by metabolites. Recent studies have suggested that there is a nexus between metabolism and epigenetics, as several metabolites could function as cofactors and substrates in epigenetic mechanisms. We review many basic and preclinical studies, and the results support this view. This finding may provide a clue to further studies on the co-effects of metabolism and epigenetics in cell fate determination.
- Citation: Liu Y, Cui DX, Pan Y, Yu SH, Zheng LW, Wan M. Metabolic-epigenetic nexus in regulation of stem cell fate. World J Stem Cells 2022; 14(7): 490-502
- URL: https://www.wjgnet.com/1948-0210/full/v14/i7/490.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i7.490
Stem cells are specialized cells with a capacity for prolonged self-renewal and production of various lineage cells, which contribute to the development, maintenance and repair of organs, such as teeth, hair follicles, and liver. These long-lived cells produce proliferating progenitors that differentiate into functional cells. Disorder of this procedure results in hyperplasia, hypoplasia or dysfunction of the organs[1]. How such cell fate determination is regulated is one of the central questions in stem cell biology. High-throughput sequencing has been conducted to establish gene expression profiles of both embryonic and adult stem cells, which helps address the crucial genes in stem cell fate regulation[2]. Epigenetic mechanisms, including histone modification, DNA methylation, and microRNAs (miRNAs), have uncovered the post-transcriptional regulation associated with stem cell fate[3]. Parallel proteomics studies have expanded our understanding of stem cell biology through constructing protein expression profiles of various stem cell populations. Despite these findings, the molecular network that regulates stem cell fate, maintaining pluripotency or initiating differentiation, is not completely understood due to the expression differences between mRNA and protein, the inconsistency between protein expression and its function, or the discordance between gene expression and cellular phenotype[4].
Although genomics and proteomics discuss the biological events at the gene and protein levels, respectively, several biological activities in cells occur at the metabolic level, including cell signaling, energy transfer, and intercellular communication[5]. To establish the metabolome, the collection of all metabolites at a specific time, metabolomics has been developed as one of the important components in system biology. Metabolomics is considered to be a prospective approach in various areas of researches, such as development, pathology, diagnosis, and environmental science, since it elaborates what happens in cells[6]. To address what occurs during regulation of stem cell fate determination, metabolomic research has been conducted to construct metabolic profiles of embryonic stem cells and differentiated neurons and cardiomyocytes in mice. Stem cells are characterized by highly unsaturated metabolites that regulate cell differentiation through oxidative reactions, suggesting the vital role of metabolism in stem cell fate determination. Metabolism is considered to function as a major energy source during the process[5,7,8]. Recent studies have demonstrated that lipid metabolism provided 90% of acetyl-CoA in histone acetylation. S-adenosylmethionine (SAM), one of the methionine metabolism metabolites, functions as a methyl donor in histone as well as DNA methylation[9]. Additionally, the epigenetic modification could be sensitive to the dynamic change in the metabolites, leading to changes in transcription. These findings provide compelling evidence that establishes the nexus exists between metabolism and epigenetics and propose a novel stem cell-based therapeutic strategy through manipulating metabolites[10-12].
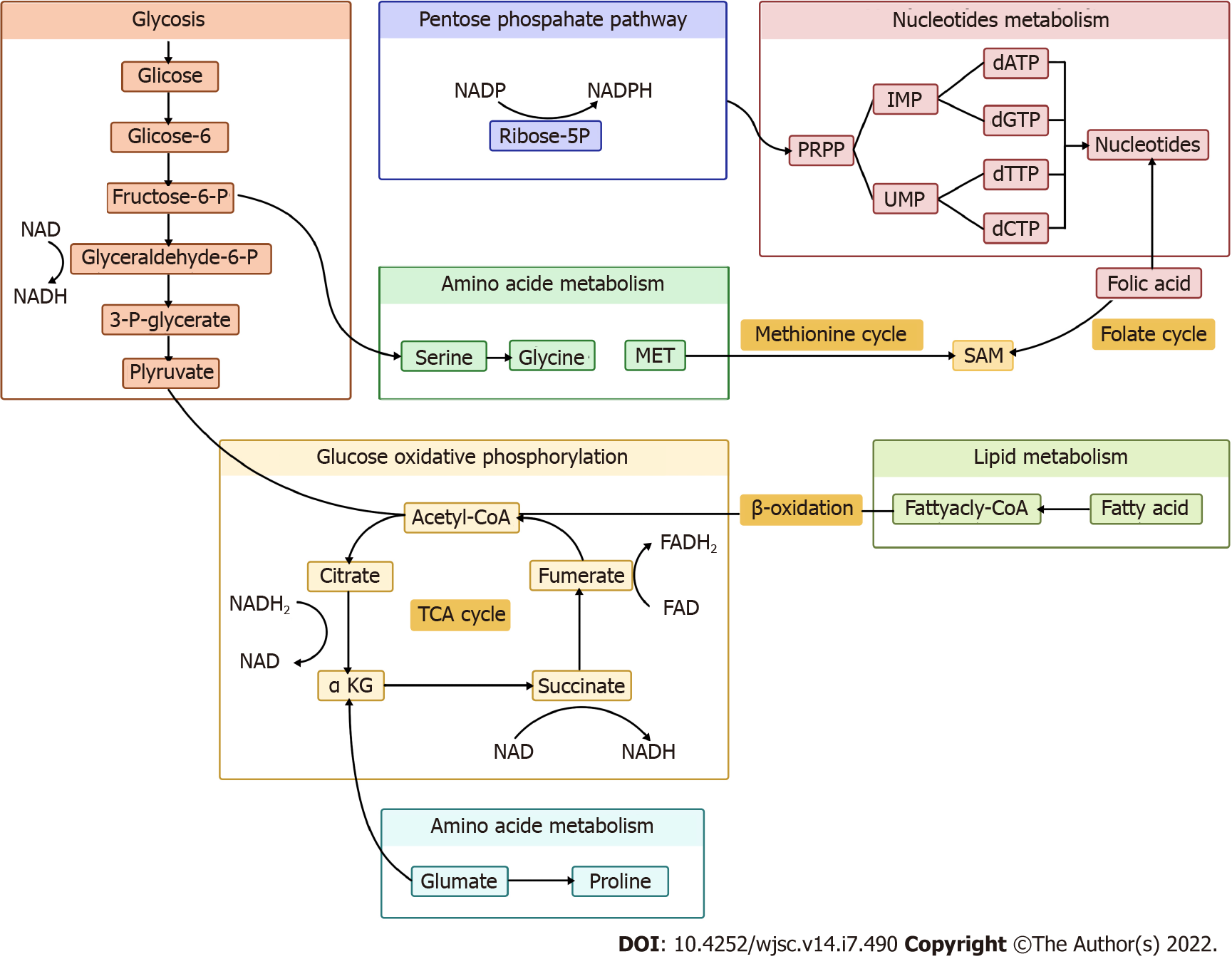
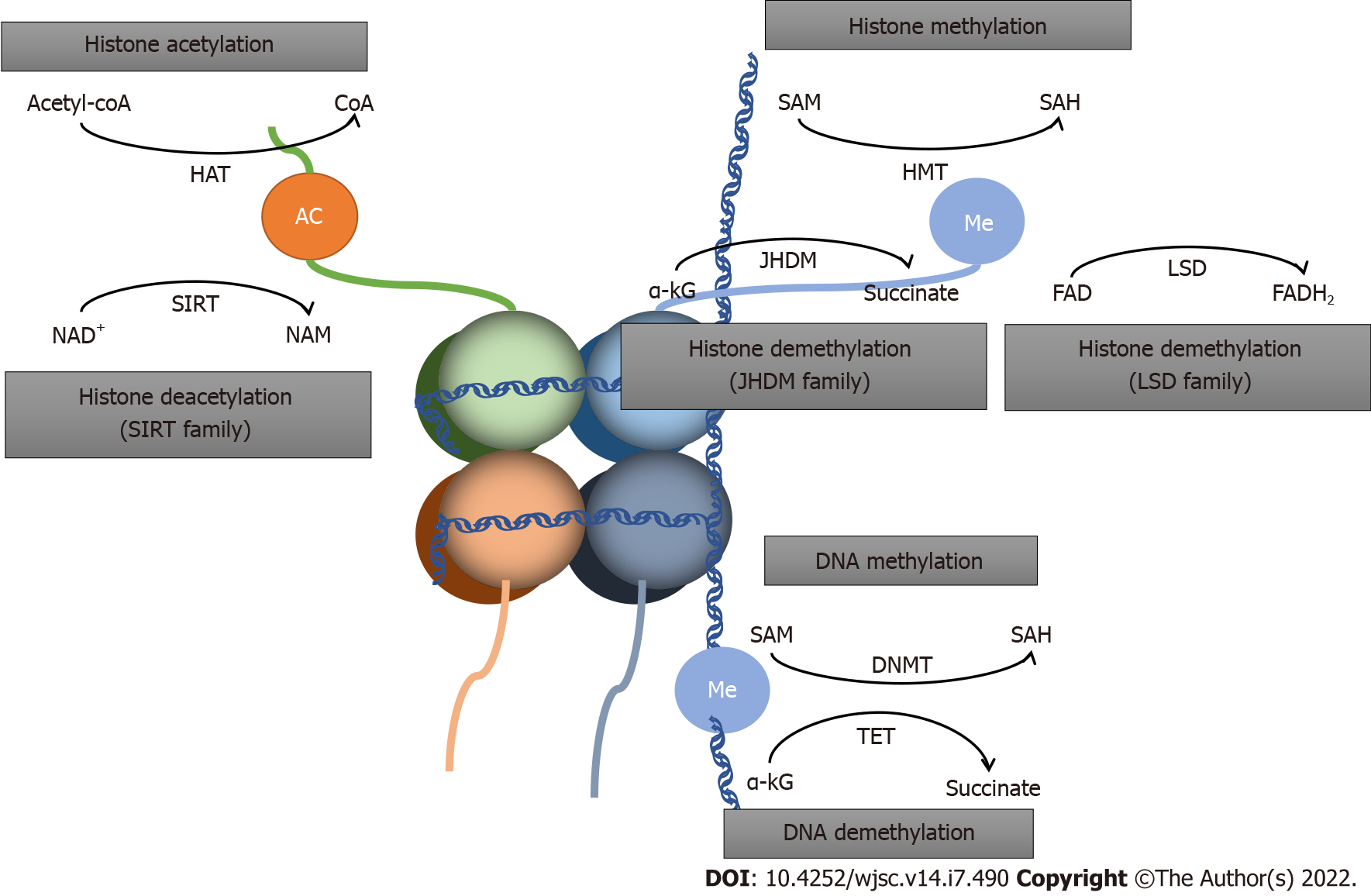
In the present review, we summarize the nexus between metabolic and epigenetic regulation in stem cell fate determination, along with potential preventive and therapeutic strategies targeting metabolites (Figures 1 and 2).
Lipids, crucial in maintaining cellular homeostasis, is attached to epigenetic reprogramming of homeostasis[13]. Acetyl-CoA from lipid metabolism could promote histone acetylation and drive cellular growth. Hence, acetyl-CoA is a crucial indicator for cell growth and development. Furthermore, acetyl-CoA reduces the production of b-hydroxybutyrate, an inhibitor of histone deacetylases (HDACs), which functions as antiproliferative and prodifferentiative properties[14]. The relation between lipid metabolism and epigenetic modification of gene expression in different stem cells has been reported by several studies[15].
Lipid metabolism contributes 90% of acetyl. Chromatin structure opening occurs when histone acetylation is present, activating stem cell transcription[16]. This suggests that lipid availability regulates the pluripotency of stem cells and promotes cell differentiation[17]. Itokazu group’s research on the importance of gangliosides in neural stem cells (NSCs) found that when the cellular histone deacetylase activity was inhibited by fatty acids, the levels of acetylated histone H3 and H4 on the GM2/GD2 synthase gene increased, promoting neuronal differentiation of NSCs[18]. Ardah et al[19] and Boddeke et al[20] also showed that the increased level of saturated fatty acids promoted NSC differentiation into neurons. Murray et al[21] reported that butyrate promoted myogenic differentiation of satellite cells.
Cornacchia et al[22] showed that the level of H3K27Ac, H3K9Ac and H4K8Ac was elevated by activation of histone acetylation in human pluripotent stem cells, while histone deacetylases (HDAC), sirtuin 1 (SIRT1) and HDAC1were limited. Similar evidence has also been reported in animal studies[23,24]. The level of H3K27ac decreased in the presence of low fatty acid metabolism in the gonads, leading to male differentiation-specific signal inhibition[25]. Acetyl-CoA production can be regulated by acetyl-CoA carboxylase , a rate-limiting enzyme whose activation limits the production of acetyl-CoA, thus promoting stem cell pluripotency. This is a traditional pathway in human as well as mouse embryonic stem cells (ESCs)[26,27]. These results show that lipid metabolism affects stem cell differentiation through histone acetylation modification.
Amino acids are one of the most fundamental substrates in cells, and are essential for metabolism of proteins, lipids and nucleotides. Previous reports have demonstrated that amino acid metabolism affected maintenance of stem cell pluripotency. In this review, we highlight the amino acids that influence stem cells critically.
Glutamine is the most abundant amino acid in metabolism, and is especially active in synthesis of nucleotides and fatty acids[28,29]. Glutamine changes α-ketoglutarate (α-KG) through deamination[30], which is a critical substrate for modification of proteins and DNA by demethylases. The mechanism is that α-KG acts as a substrate for Jumonji-C and ten-eleven translocation (TET), which regulate demethylase interaction with histone and DNA, respectively. Demethylases are essential for stem cell pluripotency acquisition and maintenance. Many studies have highlighted the role of glutamine metabolism in the maintenance and differentiation of stem cells.
DNA methylation correlates with the repression of expression. α-KG positively regulates demethylation of DNA and promotes stem cell differentiation[31,32], and is key to the determination of stem cells fate as an appropriate balance between H3K9me2 acquisition and H3K27me3 depletion. Tischler et al[33], Xing et al[34] and Zylicz et al[35] came to similar conclusions in experiment on mouse primordial germ cell–like cells (PGCLCs). Okabe et al[36] have reported that histone H3K9me3 demethylation induced by an increase in α-KG activates transcription, leading to steatoblast cellular differentiation. Glutamine also regulates fetal oocyte differentiation through DNA demethylation enzyme TET1[37].
As DNA demethylation can lead to higher levels of 5-hydroxymethylcytosine, several recent studies have reported that α-KG fluctuations influence ESC differentiation[38]. The self-renewal of ESCs decreases with deficiency of glutamine, but can recover with α-KG supplementation[39]. Hepatic stellate cell (HSC) and effector T cell differentiation is also promoted by α-KG. A surprising finding is that α-KG can suppress tumor initiation and influence progression. These effects are inhibited by succinate and fumarate, providing a possible therapy for cancer[40].
Singh et al[41] have suggested that α-KG induced cell death, with degradation of hypoxia-inducible facor-1α and suppression of histone H3 (Lys 27) acetylation. The exact mechanism of histone acetylation regulated by α-KG still needs to be explored. Morris et al[42] has shown that α-KG was an effector of p53-mediated tumor suppression, whose accumulation in p53-deficient tumors can drive tumor cell differentiation and inhibit malignant progression. Ascorbate has a positive effect on HSC differentiation and suppresses leukemogenesis[43].
All these studies above highlight the importance of glutamine in cell fate determination.
Methionine is an essential amino acid that plays an irreplaceable role in the synthesis of SAM. Methionine in the normal diet promotes production of SAM, which serves as a methyl donor for methyltransferases of histones and DNA[44]. The fluctuation of methionine and SAM levels regulates H3K4me3 formation and maintains the undifferentiated state of human ESCs/induced pluripotent stem cells (iPSCs)[45]. Kosti et al[46] have reported that limited methionine level was associated with neuronal differentiation, along with reduction of H3K27me3. Tang et al[47] have also provided evidence that reduced conversion of methionine to SAM lead to reduced ESC pluripotency. Zhang et al[48] have also found similar evidence that SAM played an important role in the differentiation of B cells into plasmablasts, and SAM deficiency was accompanied by induction of H3K27me3. The theory may be an attractive option for improving therapeutic effectiveness in patients with systemic lupus erythematosus.
Fluctuation in the methionine cycle is related to cancer epigenetics. The increase in H3K4me3 and H3K27me3 level in cells treated with methionine in cancer stem cells parallels the increase in SAM to some extent[49,50]. This may provide a new therapy for cancer[51].
Taken together, these findings show that methionine has an important influence on stem cell fates.
Proline is a nonessential amino acid derived from glutamine metabolism. Pyrroline-5-carboxylate (P5C) is an intermediate product of both proline biosynthesis and catabolism. P5C is converted to proline by P5C reductase (Pycr1). Emerging evidence indicates that L-proline influences the epigenetic landscape of stem cells by regulating histones and DNA methylation[52,53]. L-Proline regulates H3K9 methylation and activates reprogramming of stem cells. Supplementation with L-proline increases DNA 5-methylcytosine and reduces of 5-hydroxy-methylcytosine, which promotes DNA methylation. It has recently emerged that hypermethylation lead to α-KG depletion, limiting the activity of TETs and Jumonji, and resulting in increased DNA and histone methylation .A study on mouse embryonic stem cell has shown that L-proline influenced the balance between self-renewal and differentiation[54]. Proline availability increases DNA and histone methylation, and is an essential procedure in embryonic-stem-to-mesenchymal like transition[55].
Proline is one of the most important amino acids in stem cell fate determination because of its epigenetic effects.
Glycine takes part in one-carbon metabolism as a methyl group provider through the glycine cleavage system[56]. The glycine cleavage system is a multienzyme complex consisting of four individual components: glycine decarboxylase, amino methyltransferase, glycine cleavage system protein H, and dihydrolipoamide dehydrogenase[57]. It has been revealed that glycine influenced stem cell pluripotency by controlling the synthesis of SAM, thus promoting H3K4me3 modification, and open euchromatin[58]. This process is present in human and mouse PSCs[59].
Noncoding RNA (ncRNA) is RNA that does not encode a protein. ncRNA is transcribed from the genome and exerts its effects at the RNA level. Global ncRNA abundance influences cell fate determination and differentiation, and is important in embryonic development and its dysregulation causes cancer[60-62].
There are reports suggesting that long noncoding RNA (lncRNA) lnc13728 positively regulates expression of zinc finger BED-type containing 3 to promote the adipo-genic differentiation of human adipose-derived mesenchymal stem cells[63]. lncRNA has effects on hematopoietic cells in hematopoiesis regulation and the early stage of cell fate determination. Wu et al[64] have reported that, in hematopoietic stem cells and in differentiated lineage progenitors, lncRNA expression is given priority.
Griffiths and his colleagues have demonstrated that miRNA181a inhibition activated the early latent neurogenic gene to restore CA1 neurons, providing a positive clinical outcome in survivors of forebrain ischemia[65]. Zhang et al[66] have shown that miR-124 inhibited pancreatic progenitor cell proliferation to maintain a quiescent state, thus determining the fate of pancreatic progenitor cells. In cancer cells, miRNA might be a preferential pathway in cell reprograming. It has been reported that glucose transporter type 1 (GLUT1), GLUT3 and GLUT4 were overexpressed in most cancers. miR-122 regulates lipid levels in liver. miR-185 and miR-342 inhibit migration and invasion of prostate cancer cells, which could be a therapeutic option for prostate cancer[63]. He et al[67] have shown that miR-146a from exosomes had an effect on β-cell dedifferentiation, which provide a new therapy for type 2 diabetes[68]. High expression of miR-130a can increase osteogenic differentiation of bone marrow mesenchymal stem cells, which could be a potential therapy for age-related bone loss[69,70]. Huang et al[71] have reported that miR-330-5p negatively regulated differentiation of mesenchymal stem cells.
In summary, ncRNA plays an essential role in stem cell fate determination and could act as a breakthrough point in disease therapy. However, we still have a long way to go to understand the whole regulatory network of ncRNA.
Glucose and oxygen are important regulatory elements that help direct stem cell fate. In the undifferentiated state, stem cells, and their artificially reprogrammed equivalent iPSCs, are characterized by limited oxidative capacity and active anaerobic glycolysis. The importance of optimizing glucose metabolism during nuclear reprogramming by epigenetic regulation has been demonstrated in several studies.
Glycolysis is defined as a cytosolic redox reaction that transform a single glucose molecule into two pyruvate molecules accompanied by generation of two net ATP and two reduced NADH molecules. Although glycolysis is not as energetically efficient as complete oxidation, this pathway can occur in the absence of oxygen and enables a fast rate of ATP production, which may also be the reason why some highly proliferating cell types typically utilize glycolysis.
High glycolytic flux could be frequently observed in various stem cell populations and is critical for the acquisition and maintenance of cell pluripotency[72]. Li et al[73] have shown that GLIS family zinc finger 1 could directly bind to and open chromatin structure at glycolysis-related genes to promote glycolysis. Higher glycolytic flux subsequently upregulates cellular acetyl-CoA and lactate levels, leading to increased acetylation of H3K27 and pluripotency gene loci.
Glycolytic flux can be influenced by several factors, including epigenetic regulators and environmental conditions. For example, NAD-dependent histone deacetylase SIRT6 has been proved to act as a key regulator of glucose homeostasis, and its absence favors the metabolic profile of anaerobic glycolysis, which may activate gene reprogramming and pluripotency maintenance[74]. The epigenetic modifications are essential for the cell fate decisions in NSCs as well. High glucose levels increase H3K14 acetylation level, which can lead to premature neurogenetic differentiation of NSCs, providing a promising target for intervention in fetal neurodevelopment deficits[75]. Protein glycosylation is one of the most diverse and complicated co- and post-translational modifications, regulating self-renewal, pluripotency, and differentiation of stem cells through epigenetic mechanisms by histone modification and DNA methylation[76]. Glycolytic flux can also be regulated by oxygen. Glycolysis increases at 5% oxygen and acetylation of H3K9 and H3K27 is elevated, while H3K27 trimethylation is downregulated, leading to a more open chromatin structure and altered fate of human PSCs[77,78].
In summary, glycolysis is the dominant metabolic phenotype that controls stem cell fate.
Glucose oxidative phosphorylation is another critical pathway for maintaining bioenergetic homeostasis as a bridge between the tricarboxylic acid (TCA) cycle and ATP synthesis. Oxidative phosphorylation is a more efficient pathway for ATP production compared to glycolysis, producing 36 ATP molecules per glucose. Oxidative phosphorylation promotes stem cell differentiation. Uittenbogaard et al[79] have provided evidence that enhancing oxidative phosphorylation can trigger neuronal differentiation by generating H3K27ac. Oxidative phosphorylation also mediates hematopoiesis stem cell differentiation toward definitive hematopoiesis through actyl-CoA metabolism[80].
Several TCA-cycle-related metabolic intermediates like NADH, FADH, fumarate and succinate are reported to contribute to epigenetic regulation of transcription and be connected with stem cell fate.
NADH: NAD+ is a coenzyme that serves as a co-substrate for sirtuins, an HDAC family, and catalyzes deacetylation of histone lysine; a crucial protein post-translational modification[81-83].
NAD/NADH ratio can dictate the fate and function of different cell types. Increased NAD+ production is required for cell differentiation[84]. Bmal1 regulates primary myoblast proliferation and differentiation through increasing cytosolic NAD+. Reduced NAD+ level prevents the differentiation of preadipocytes[85]. Okabe et al[36] have confirmed that high NAD+ levels upregulated the TCA cycle, increasing α-KG and contributing to histone H3K9 demethylation and transcriptional activation. Zhu et al[85] have demonstrated that increasing cytosolic NAD levels could restore hypoxic cell proliferation and myofiber formation in Bmal1-deficient myoblasts, influencing oxygen-dependent myoblast cell fate. The effect of NAD/NADH ratio on stem cell fate is caused by generation of L-2-hydroxyglutaric acid, an analog of α-KG that regulates histone and DNA methylation by competitive inhibition of Jumonji-domain histone demethylase (JHDM) and TETs. There are reports revealing that increased NAD+ levels delay aging-related phenotypes, which may provide new therapeutic option for type 2 diabetes and heart failure[86,87]. Besides, NAD+ is a cosubstrate of Sirtuins, potentially regulating T cells, and could provide a therapeutic option for immune-related diseases[88].
In summary, NAD+ plays a key role in a diverse array of biological processes.
FADH: FAD, the oxidized form of FADH2, is a cofactor of human lysine-specific demethylase-1 (LSD1), and plays a pivotal role during early embryonic development and differentiation of ESCs and cancer stem cells[89-92]. LSD1 catalyzes the demethylation of mono- and dimethylated K4 or K9 on histone H3 via the FAD-dependent enzymatic oxidation[93]. Recent studies have found that LSD1 inhibition can enhance death in rhabdomyosarcoma cells[94]. Decreased expression of LSD1 is involved in the programmed oocyte death by autophagy in perinatal mice through promotion of H3K4me2 expression[95]. FAD also regulates NSC proliferation through modulation of histone methylation by affecting the action of LSD1. In addition, LSD1 is highly expressed in a few aggressive cancer types and is closely related with differentiation, proliferation, migration and invasion of cancer cells and poor prognosis.
Succinate: Succinate accumulation can decrease α-KG/succinate ratio, leading to inhibition of TET and JHDM enzymes and delayed differentiation of primed human PSCs. This effect can be reversed when the α-KG/succinate ratio increases[32,86]. Accumulation of succinate, resulting in genetic and epigenetic changes like histone hypermethylation, may lead to transformation of normal cells to cancerous cells[96,97] . Wong et al’s study in colorectal cancer cells showed that promoting accumulation of succinate upregulated DNA methylation and stem cell features[98]. AA6 is a novel compound succinic acid, identified as an inhibitor of α-KG dehydrogenase, which can increase the α-KG level in diabetic human cardiac mesenchymal cells and in the heart of high-fat diet, leading to DNA demethylation, and has beneficial effects of cardiac mesenchymal stem cells protection in diabetes[99].
Fumarate: Fumarate is reported to inhibit α-KG-dependent dioxygenases involved in DNA and histone demethylation. Laukka et al[100] have shown that fumarate downregulates global 5-hydroxymethylcytosine level in neuroblastoma cells via TET inhibition. Furthermore, Sharda et al[101] have reported that fumarate promotes monomer-to-dimer transition of malic enzyme 2 to enhance mitobiogenesis, linking metabolism to mitobiogenesis. Aberrant accumulation of fumarate may mediate epigenetic reprogramming. Some studies have reported the link between fumarate accumulation, epigenetic changes, and tumorigenesis. Accumulation of fumarate, inhibiting Tet-mediated demethylation, induces epithelial-to-mesenchymal transition; a phenotypic switch associated with cancer initiation, invasion and metastasis[102]. This implies that fumarate accumulation contributes to the aggressive features tumors[103].
The pentose phosphate pathway (PPP) is another glucose metabolism pathway, divided into oxidative and nonoxidative arms, producing NADH and ribose-5-phosphate and/or xylulose-5-phosphate that influence the regulation of transcription[104]. NADPH production in the pathway is involved in folate metabolism[105]. Previous studies have reported that regulation of the PPP resulted in iPSC reprogramming[106]. The PPP actively provides energy and metabolic intermediates for proliferation and pluripotency in cancer cells, ESCs and iPSCs[107,108]. Intracellular pH increase selectively activates catalysis, enhancing PPP flux, leading to nucleotide upregulation, increased NADPH/NADP+ ratio, and cell proliferation[109].
It remains to be elucidated whether PPP is linked to stem cell epigenetic remodeling.
The structure of scaffolds can affect stem cell metabolism. Three-dimensional graphene foam has better properties than two-dimensional foam for NSC differentiation. However, the possible mechanism needs to be explored[110].
Vitamin C: Vitamin C is a crucial micronutrient that may be involved in stem cell pluripotency by activating H3K36 and H3K9 demethylases through Jumonji-C function[111]. A study using human PGCLCs also indicated the pathway[112,113]. Micronutrients influence stem cells specification.
Folic acid: Folic acid is first metabolized to dihydrofolate and then to tetrahydrofolate, taking part in DNA synthesis, influencing DNA and histone methylation[105]. Several studies have elucidated the role of folate metabolism in regulating of the epigenetic landscape of stem cells[114,115]. Li et al[116] have shown that folic acid deficiency in NSCs decreased cell proliferative capacity but increased apoptosis. Kasulanati et al[117] in a study of ESCs have provided more evidence for the effect of folic acid on PSC pluripotency. Pei et al[118] in a study of mouse ESCs have demonstrated that under folate deficiency conditions, H2AK119ub1 increases, and expression of neural tube closure-associated genes decreases. This suggests a possible mechanism for neural tube defects. Xie et al[119] have shown that folate inhibition can activate histone modification of monomethylation at lysine 4 of histone H3 transcription, suggesting that epigenetic regulation varies for different histone modifications.
Crosslinking: Horitani et al[120] have reported that glucose along with triglyceride increased metabolic stress spikes in mice, resulting in demethylation of H3K27me3, and expression of senescence-like phenotypes in bone marrow stem/progenitor cells. This could provide a therapeutic method for patients with cardiovascular disease and type 2 diabetes.
This review summarizes the recent studies about the metabolic–epigenetic nexus and provides compelling evidence that metabolism regulates stem cell fate determination through epigenetic mechanisms, such as histone acetylation, histone methylation and DNA methylation, in a variety of physical and pathological phenomena. The latest studies have also suggested that potential manipulation of metabolites held great promise in developing novel preventive, diagnostic and therapeutic strategies for a variety of diseases, which still requires further study prior to application in clinic settings.
There are still some essential questions. For example, what is the outcome of the regulation of metabolism on the epigenetic and transcriptional procedures of stem cells. The interplay of metabolism and epigenetics also brings out the complexity in environmental exposures studies, as the method of cell metabolism and potential transgenerational inheritance has been changed[15,121].
Furthermore, there were still some limitations. Most studies have attached importance to the level of enzymatic activity in cells, and we must accept that there is a difference between the measured and actual values[15]. All researches were conducted under experimental and not physiological conditions, and it is not hard to conclude that there might be some variation.
In summary, when it comes to the mechanism of stem cell fate determination, there is indeed interplay between metabolism and epigenetics. We need more accurate data acquisition and more realistic simulation as well as more specific mechanisms. The development of new technologies makes it easier to measure cellular metabolic status, and the accumulation of past studies supports our further exploration in this field.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Portillo R, Czech Republic; Portius D, Germany; Wei W, China S-Editor: Zhang H L-Editor: A P-Editor: Zhang H
| 1. | Ohara TE, Colonna M, Stappenbeck TS. Adaptive differentiation promotes intestinal villus recovery. Dev Cell. 2022;57:166-179.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 2. | Neves de Oliveira BH, Dalmaz C, Zeidán-Chuliá F. Network-Based Identification of Altered Stem Cell Pluripotency and Calcium Signaling Pathways in Metastatic Melanoma. Med Sci (Basel). 2018;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 3. | Tian Q, Gao S, Zhou X, Zheng L, Zhou Y. Histone Acetylation in the Epigenetic Regulation of Bone Metabolism and Related Diseases. Stem Cells Int. 2021;2021:8043346. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 4. | Dvir S, Argoetti A, Lesnik C, Roytblat M, Shriki K, Amit M, Hashimshony T, Mandel-Gutfreund Y. Uncovering the RNA-binding protein landscape in the pluripotency network of human embryonic stem cells. Cell Rep. 2021;35:109198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 5. | Döhla J, Kuuluvainen E, Gebert N, Amaral A, Englund JI, Gopalakrishnan S, Konovalova S, Nieminen AI, Salminen ES, Torregrosa Muñumer R, Ahlqvist K, Yang Y, Bui H, Otonkoski T, Käkelä R, Hietakangas V, Tyynismaa H, Ori A, Katajisto P. Metabolic determination of cell fate through selective inheritance of mitochondria. Nat Cell Biol. 2022;24:148-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 59] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 6. | Yang Y, Fan TW, Lane AN, Higashi RM. Chloroformate derivatization for tracing the fate of Amino acids in cells and tissues by multiple stable isotope resolved metabolomics (mSIRM). Anal Chim Acta. 2017;976:63-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 7. | Sun Z, Zhao J, Yu H, Zhang C, Li H, Zeng Z, Zhang J. Metabolomics in Stem Cell Biology Research. Methods Mol Biol. 2019;1975:321-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Tencerova M, Rendina-Ruedy E, Neess D, Færgeman N, Figeac F, Ali D, Danielsen M, Haakonsson A, Rosen CJ, Kassem M. Metabolic programming determines the lineage-differentiation fate of murine bone marrow stromal progenitor cells. Bone Res. 2019;7:35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Ferrari A, Longo R, Silva R, Mitro N, Caruso D, De Fabiani E, Crestani M. Epigenome modifiers and metabolic rewiring: New frontiers in therapeutics. Pharmacol Ther. 2019;193:178-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | D'Aniello C, Cermola F, Patriarca EJ, Minchiotti G. Metabolic-Epigenetic Axis in Pluripotent State Transitions. Epigenomes. 2019;3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Verdikt R, Allard P. Metabolo-epigenetics: the interplay of metabolism and epigenetics during early germ cells development. Biol Reprod. 2021;105:616-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Li L, Chen K, Wu Y, Xiang G, Liu X. Epigenome-Metabolome-Epigenome signaling cascade in cell biological processes. J Genet Genomics. 2022;49:279-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Tsogtbaatar E, Landin C, Minter-Dykhouse K, Folmes CDL. Energy Metabolism Regulates Stem Cell Pluripotency. Front Cell Dev Biol. 2020;8:87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 140] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 14. | Prasad KN. Butyric acid: a small fatty acid with diverse biological functions. Life Sci. 1980;27:1351-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 165] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Fawal MA, Davy A. Impact of Metabolic Pathways and Epigenetics on Neural Stem Cells. Epigenet Insights. 2018;11:2516865718820946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 16. | McDonnell E, Crown SB, Fox DB, Kitir B, Ilkayeva OR, Olsen CA, Grimsrud PA, Hirschey MD. Lipids Reprogram Metabolism to Become a Major Carbon Source for Histone Acetylation. Cell Rep. 2016;17:1463-1472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 257] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 17. | Johnson MT, Mahmood S, Patel MS. Intermediary metabolism and energetics during murine early embryogenesis. J Biol Chem. 2003;278:31457-31460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Itokazu Y, Wang J, Yu RK. Gangliosides in Nerve Cell Specification. Prog Mol Biol Transl Sci. 2018;156:241-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 19. | Ardah MT, Parween S, Varghese DS, Emerald BS, Ansari SA. Saturated fatty acid alters embryonic cortical neurogenesis through modulation of gene expression in neural stem cells. J Nutr Biochem. 2018;62:230-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Balasubramaniyan V, Boddeke E, Bakels R, Küst B, Kooistra S, Veneman A, Copray S. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939-951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 111] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 21. | Murray RL, Zhang W, Liu J, Cooper J, Mitchell A, Buman M, Song J, Stahl CH. Tributyrin, a Butyrate Pro-Drug, Primes Satellite Cells for Differentiation by Altering the Epigenetic Landscape. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 22. | Cornacchia D, Zhang C, Zimmer B, Chung SY, Fan Y, Soliman MA, Tchieu J, Chambers SM, Shah H, Paull D, Konrad C, Vincendeau M, Noggle SA, Manfredi G, Finley LWS, Cross JR, Betel D, Studer L. Lipid Deprivation Induces a Stable, Naive-to-Primed Intermediate State of Pluripotency in Human PSCs. Cell Stem Cell. 2019;25:120-136.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 23. | Palmer NO, Fullston T, Mitchell M, Setchell BP, Lane M. SIRT6 in mouse spermatogenesis is modulated by diet-induced obesity. Reprod Fertil Dev. 2011;23:929-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 24. | Hayashi Y, Matsui Y. Metabolic Control of Germline Formation and Differentiation in Mammals. Sex Dev. 2022;1-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 25. | Xu Y, Xie J. Etomoxir regulates the differentiation of male germ cells by specifically reducing H3K27ac level. BMC Dev Biol. 2021;21:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 26. | Liu K, Cao J, Shi X, Wang L, Zhao T. Cellular metabolism and homeostasis in pluripotency regulation. Protein Cell. 2020;11:630-640. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Wang L, Zhang T, Wang L, Cai Y, Zhong X, He X, Hu L, Tian S, Wu M, Hui L, Zhang H, Gao P. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017;36:1330-1347. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 28. | Smith RJ, Wilmore DW. Glutamine nutrition and requirements. JPEN J Parenter Enteral Nutr. 1990;14:94S-99S. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 113] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 29. | Curi R, Newsholme P, Procopio J, Lagranha C, Gorjão R, Pithon-Curi TC. Glutamine, gene expression, and cell function. Front Biosci. 2007;12:344-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 30. | Baksh SC, Finley LWS. Metabolic Coordination of Cell Fate by α-Ketoglutarate-Dependent Dioxygenases. Trends Cell Biol. 2021;31:24-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 31. | Zhang Z, He C, Zhang L, Zhu T, Lv D, Li G, Song Y, Wang J, Wu H, Ji P, Liu G. Alpha-ketoglutarate affects murine embryo development through metabolic and epigenetic modulations. Reproduction. 2019;158:123-133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | TeSlaa T, Chaikovsky AC, Lipchina I, Escobar SL, Hochedlinger K, Huang J, Graeber TG, Braas D, Teitell MA. α-Ketoglutarate Accelerates the Initial Differentiation of Primed Human Pluripotent Stem Cells. Cell Metab. 2016;24:485-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 210] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 33. | Tischler J, Gruhn WH, Reid J, Allgeyer E, Buettner F, Marr C, Theis F, Simons BD, Wernisch L, Surani MA. Metabolic regulation of pluripotency and germ cell fate through α-ketoglutarate. EMBO J. 2019;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 74] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 34. | Xing M, Wang N, Zeng H, Zhang J. α-ketoglutarate promotes the specialization of primordial germ cell-like cells through regulating epigenetic reprogramming. J Biomed Res. 2020;35:36-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 35. | Zylicz JJ, Dietmann S, Günesdogan U, Hackett JA, Cougot D, Lee C, Surani MA. Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife. 2015;4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 89] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 36. | Okabe K, Nawaz A, Nishida Y, Yaku K, Usui I, Tobe K, Nakagawa T. NAD+ Metabolism Regulates Preadipocyte Differentiation by Enhancing α-Ketoglutarate-Mediated Histone H3K9 Demethylation at the PPARγ Promoter. Front Cell Dev Biol. 2020;8:586179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 37. | Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, Zhang K, Zhang Y. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443-447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 210] [Cited by in RCA: 235] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 38. | El-Habr EA, Dubois LG, Burel-Vandenbos F, Bogeas A, Lipecka J, Turchi L, Lejeune FX, Coehlo PL, Yamaki T, Wittmann BM, Fareh M, Mahfoudhi E, Janin M, Narayanan A, Morvan-Dubois G, Schmitt C, Verreault M, Oliver L, Sharif A, Pallud J, Devaux B, Puget S, Korkolopoulou P, Varlet P, Ottolenghi C, Plo I, Moura-Neto V, Virolle T, Chneiweiss H, Junier MP. A driver role for GABA metabolism in controlling stem and proliferative cell state through GHB production in glioma. Acta Neuropathol. 2017;133:645-660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 39. | Vardhana SA, Arnold PK, Rosen BP, Chen Y, Carey BW, Huangfu D, Carmona Fontaine C, Thompson CB, Finley LWS. Glutamine independence is a selectable feature of pluripotent stem cells. Nat Metab. 2019;1:676-687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 40. | Klysz D, Tai X, Robert PA, Craveiro M, Cretenet G, Oburoglu L, Mongellaz C, Floess S, Fritz V, Matias MI, Yong C, Surh N, Marie JC, Huehn J, Zimmermann V, Kinet S, Dardalhon V, Taylor N. Glutamine-dependent α-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci Signal. 2015;8:ra97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 393] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 41. | Singh K, Krug L, Basu A, Meyer P, Treiber N, Vander Beken S, Wlaschek M, Kochanek S, Bloch W, Geiger H, Maity P, Scharffetter-Kochanek K. Alpha-Ketoglutarate Curbs Differentiation and Induces Cell Death in Mesenchymal Stromal Precursors with Mitochondrial Dysfunction. Stem Cells. 2017;35:1704-1718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 42. | Morris JP 4th, Yashinskie JJ, Koche R, Chandwani R, Tian S, Chen CC, Baslan T, Marinkovic ZS, Sánchez-Rivera FJ, Leach SD, Carmona-Fontaine C, Thompson CB, Finley LWS, Lowe SW. α-Ketoglutarate links p53 to cell fate during tumour suppression. Nature. 2019;573:595-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 214] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 43. | Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, Grillo I, Bakogianni S, Ndiaye-Lobry D, Martín MT, Guillamot M, Banh RS, Xu M, Figueroa ME, Dickins RA, Abdel-Wahab O, Park CY, Tsirigos A, Neel BG, Aifantis I. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell. 2017;170:1079-1095.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 534] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 44. | Liu G, Yang W, Zhang X, Peng T, Zou Y, Zhang T, Wang H, Liu X, Tao LZ. Cystathionine beta-lyase is crucial for embryo patterning and the maintenance of root stem cell niche in Arabidopsis. Plant J. 2019;99:536-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | Van Winkle LJ, Ryznar R. One-Carbon Metabolism Regulates Embryonic Stem Cell Fate Through Epigenetic DNA and Histone Modifications: Implications for Transgenerational Metabolic Disorders in Adults. Front Cell Dev Biol. 2019;7:300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 46. | Kosti A, de Araujo PR, Li WQ, Guardia GDA, Chiou J, Yi C, Ray D, Meliso F, Li YM, Delambre T, Qiao M, Burns SS, Lorbeer FK, Georgi F, Flosbach M, Klinnert S, Jenseit A, Lei X, Sandoval CR, Ha K, Zheng H, Pandey R, Gruslova A, Gupta YK, Brenner A, Kokovay E, Hughes TR, Morris QD, Galante PAF, Tiziani S, Penalva LOF. The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation. Genome Biol. 2020;21:195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 47. | Tang S, Fang Y, Huang G, Xu X, Padilla-Banks E, Fan W, Xu Q, Sanderson SM, Foley JF, Dowdy S, McBurney MW, Fargo DC, Williams CJ, Locasale JW, Guan Z, Li X. Methionine metabolism is essential for SIRT1-regulated mouse embryonic stem cell maintenance and embryonic development. EMBO J. 2017;36:3175-3193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 48. | Zhang M, Iwata S, Hajime M, Ohkubo N, Todoroki Y, Miyata H, Ueno M, Hao H, Zhang T, Fan J, Nakayamada S, Yamagata K, Tanaka Y. Methionine Commits Cells to Differentiate Into Plasmablasts Through Epigenetic Regulation of BTB and CNC Homolog 2 by the Methyltransferase EZH2. Arthritis Rheumatol. 2020;72:1143-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 49. | Altundag Ö, Canpinar H, Çelebi-Saltik B. Methionine affects the expression of pluripotency genes and protein levels associated with methionine metabolism in adult, fetal, and cancer stem cells. J Cell Biochem. 2022;123:406-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 50. | Wang Z, Yip LY, Lee JHJ, Wu Z, Chew HY, Chong PKW, Teo CC, Ang HY, Peh KLE, Yuan J, Ma S, Choo LSK, Basri N, Jiang X, Yu Q, Hillmer AM, Lim WT, Lim TKH, Takano A, Tan EH, Tan DSW, Ho YS, Lim B, Tam WL. Methionine is a metabolic dependency of tumor-initiating cells. Nat Med. 2019;25:825-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 246] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 51. | Jung J, Kim LJ, Wang X, Wu Q, Sanvoranart T, Hubert CG, Prager BC, Wallace LC, Jin X, Mack SC, Rich JN. Nicotinamide metabolism regulates glioblastoma stem cell maintenance. JCI Insight. 2017;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 52. | Washington JM, Rathjen J, Felquer F, Lonic A, Bettess MD, Hamra N, Semendric L, Tan BS, Lake JA, Keough RA, Morris MB, Rathjen PD. L-Proline induces differentiation of ES cells: a novel role for an amino acid in the regulation of pluripotent cells in culture. Am J Physiol Cell Physiol. 2010;298:C982-C992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 53. | Casalino L, Comes S, Lambazzi G, De Stefano B, Filosa S, De Falco S, De Cesare D, Minchiotti G, Patriarca EJ. Control of embryonic stem cell metastability by L-proline catabolism. J Mol Cell Biol. 2011;3:108-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 54. | Shparberg RA, Glover HJ, Morris MB. Modeling Mammalian Commitment to the Neural Lineage Using Embryos and Embryonic Stem Cells. Front Physiol. 2019;10:705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 55. | D'Aniello C, Patriarca EJ, Phang JM, Minchiotti G. Proline Metabolism in Tumor Growth and Metastatic Progression. Front Oncol. 2020;10:776. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 56. | Tian S, Feng J, Cao Y, Shen S, Cai Y, Yang D, Yan R, Wang L, Zhang H, Zhong X, Gao P. Glycine cleavage system determines the fate of pluripotent stem cells via the regulation of senescence and epigenetic modifications. Life Sci Alliance. 2019;2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 57. | Narisawa A, Komatsuzaki S, Kikuchi A, Niihori T, Aoki Y, Fujiwara K, Tanemura M, Hata A, Suzuki Y, Relton CL, Grinham J, Leung KY, Partridge D, Robinson A, Stone V, Gustavsson P, Stanier P, Copp AJ, Greene ND, Tominaga T, Matsubara Y, Kure S. Mutations in genes encoding the glycine cleavage system predispose to neural tube defects in mice and humans. Hum Mol Genet. 2012;21:1496-1503. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 58. | Kidder BL, Hu G, Zhao K. KDM5B focuses H3K4 methylation near promoters and enhancers during embryonic stem cell self-renewal and differentiation. Genome Biol. 2014;15:R32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 59. | Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M, Shinoda G, Xie W, Cahan P, Wang L, Ng SC, Tintara S, Trapnell C, Onder T, Loh YH, Mikkelsen T, Sliz P, Teitell MA, Asara JM, Marto JA, Li H, Collins JJ, Daley GQ. LIN28 Regulates Stem Cell Metabolism and Conversion to Primed Pluripotency. Cell Stem Cell. 2016;19:66-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 252] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 60. | Wang Y, Hussein AM, Somasundaram L, Sankar R, Detraux D, Mathieu J, Ruohola-Baker H. microRNAs Regulating Human and Mouse Naïve Pluripotency. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Essig K, Kronbeck N, Guimaraes JC, Lohs C, Schlundt A, Hoffmann A, Behrens G, Brenner S, Kowalska J, Lopez-Rodriguez C, Jemielity J, Holtmann H, Reiche K, Hackermüller J, Sattler M, Zavolan M, Heissmeyer V. Roquin targets mRNAs in a 3'-UTR-specific manner by different modes of regulation. Nat Commun. 2018;9:3810. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 62. | Beilharz TH, See MM, Boag PR. 3'-UTRs and the Control of Protein Expression in Space and Time. Adv Exp Med Biol. 2019;1203:133-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 63. | Alamoudi AA, Alnoury A, Gad H. miRNA in tumour metabolism and why could it be the preferred pathway for energy reprograming. Brief Funct Genomics. 2018;17:157-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 64. | Wu Z, Gao S, Zhao X, Chen J, Keyvanfar K, Feng X, Kajigaya S, Young NS. Long noncoding RNAs of single hematopoietic stem and progenitor cells in healthy and dysplastic human bone marrow. Haematologica. 2019;104:894-906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 65. | Griffiths BB, Ouyang YB, Xu L, Sun X, Giffard RG, Stary CM. Postinjury Inhibition of miR-181a Promotes Restoration of Hippocampal CA1 Neurons after Transient Forebrain Ischemia in Rats. eNeuro. 2019;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 66. | Zhang Z, Zhai W, Liang J, Chen Z, Ma M, Zhao Y, Liang Y, Li X, Teng CB. Mutual inhibitions between epidermal growth factor receptor signaling and miR-124a control pancreatic progenitor proliferation. J Cell Physiol. 2019;234:12978-12988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 67. | He Q, Song J, Cui C, Wang J, Hu H, Guo X, Yang M, Wang L, Yan F, Liang K, Liu Z, Liu F, Sun Z, Dong M, Hou X, Chen L. Mesenchymal stem cell-derived exosomal miR-146a reverses diabetic β-cell dedifferentiation. Stem Cell Res Ther. 2021;12:449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 68. | Soltani A, Jafarian A, Allameh A. The Predominant microRNAs in β-cell Clusters for Insulin Regulation and Diabetic Control. Curr Drug Targets. 2020;21:722-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 69. | Lin Z, He H, Wang M, Liang J. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif. 2019;52:e12688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 131] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 70. | Chesi A, Wagley Y, Johnson ME, Manduchi E, Su C, Lu S, Leonard ME, Hodge KM, Pippin JA, Hankenson KD, Wells AD, Grant SFA. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat Commun. 2019;10:1260. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 71. | Huang W, Li K, Liu A, Yang Z, Hu C, Chen D, Wang H. miR3305p inhibits H2O2induced adipogenic differentiation of MSCs by regulating RXRγ. Int J Mol Med. 2018;42:2042-2052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 72. | Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park DJ, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348:1472-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 365] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 73. | Li L, Chen K, Wang T, Wu Y, Xing G, Chen M, Hao Z, Zhang C, Zhang J, Ma B, Liu Z, Yuan H, Long Q, Zhou Y, Qi J, Zhao D, Gao M, Pei D, Nie J, Ye D, Pan G, Liu X. Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat Metab. 2020;2:882-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 183] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 74. | Sanhueza Salas LF, García-Venzor A, Beltramone N, Capurro C, Toiber D, Silberman DM. Metabolic Imbalance Effect on Retinal Müller Glial Cells Reprogramming Capacity: Involvement of Histone Deacetylase SIRT6. Front Genet. 2021;12:769723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 75. | Ji S, Zhou W, Li X, Liu S, Wang F, Zhao T, Ji G, Du J, Hao A. Maternal hyperglycemia disturbs neocortical neurogenesis via epigenetic regulation in C57BL/6J mice. Cell Death Dis. 2019;10:211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 76. | Sheikh MA, Emerald BS, Ansari SA. Stem cell fate determination through protein O-GlcNAcylation. J Biol Chem. 2021;296:100035. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 77. | Lees JG, Cliff TS, Gammilonghi A, Ryall JG, Dalton S, Gardner DK, Harvey AJ. Oxygen Regulates Human Pluripotent Stem Cell Metabolic Flux. Stem Cells Int. 2019;2019:8195614. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 78. | Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, Bomze D, Elena-Herrmann B, Scherf T, Nissim-Rafinia M, Kempa S, Itskovitz-Eldor J, Meshorer E, Aberdam D, Nahmias Y. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 514] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 79. | Uittenbogaard M, Brantner CA, Chiaramello A. Epigenetic modifiers promote mitochondrial biogenesis and oxidative metabolism leading to enhanced differentiation of neuroprogenitor cells. Cell Death Dis. 2018;9:360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 80. | Oburoglu L, Mansell E, Canals I, Sigurdsson V, Guibentif C, Soneji S, Woods NB. Pyruvate metabolism guides definitive lineage specification during hematopoietic emergence. EMBO Rep. 2022;23:e54384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 81. | Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol. 2022;23:329-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 474] [Article Influence: 158.0] [Reference Citation Analysis (0)] |
| 82. | Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 720] [Cited by in RCA: 999] [Article Influence: 90.8] [Reference Citation Analysis (0)] |
| 83. | Williams EO, Taylor AK, Bell EL, Lim R, Kim DM, Guarente L. Sirtuin 1 Promotes Deacetylation of Oct4 and Maintenance of Naive Pluripotency. Cell Rep. 2016;17:809-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 84. | Chiarugi A, Dölle C, Felici R, Ziegler M. The NAD metabolome--a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12:741-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 501] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 85. | Zhu P, Hamlish NX, Thakkar AV, Steffeck AWT, Rendleman EJ, Khan NH, Waldeck NJ, DeVilbiss AW, Martin-Sandoval MS, Mathews TP, Chandel NS, Peek CB. BMAL1 drives muscle repair through control of hypoxic NAD+ regeneration in satellite cells. Genes Dev. 2022;36:149-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 86. | Chakrabarty RP, Chandel NS. Mitochondria as Signaling Organelles Control Mammalian Stem Cell Fate. Cell Stem Cell. 2021;28:394-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 266] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 87. | Lopaschuk GD, Karwi QG, Tian R, Wende AR, Abel ED. Cardiac Energy Metabolism in Heart Failure. Circ Res. 2021;128:1487-1513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 753] [Article Influence: 188.3] [Reference Citation Analysis (0)] |
| 88. | Hamaidi I, Kim S. Sirtuins are crucial regulators of T cell metabolism and functions. Exp Mol Med. 2022;54:207-215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 41] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 89. | Sun G, Alzayady K, Stewart R, Ye P, Yang S, Li W, Shi Y. Histone demethylase LSD1 regulates neural stem cell proliferation. Mol Cell Biol. 2010;30:1997-2005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 193] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 90. | Gao L, Alumkal J. Epigenetic regulation of androgen receptor signaling in prostate cancer. Epigenetics. 2010;5:100-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 91. | Tu WJ, McCuaig RD, Tan AHY, Hardy K, Seddiki N, Ali S, Dahlstrom JE, Bean EG, Dunn J, Forwood J, Tsimbalyuk S, Smith K, Yip D, Malik L, Prasanna T, Milburn P, Rao S. Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch. Front Immunol. 2020;11:1228. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 92. | Li Z, Ding L, Li Z, Wang Z, Suo F, Shen D, Zhao T, Sun X, Wang J, Liu Y, Ma L, Zhao B, Geng P, Yu B, Zheng Y, Liu H. Development of the triazole-fused pyrimidine derivatives as highly potent and reversible inhibitors of histone lysine specific demethylase 1 (LSD1/KDM1A). Acta Pharm Sin B. 2019;9:794-808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 93. | Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1269] [Cited by in RCA: 1371] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 94. | Schulz-Fincke J, Hau M, Barth J, Robaa D, Willmann D, Kürner A, Haas J, Greve G, Haydn T, Fulda S, Lübbert M, Lüdeke S, Berg T, Sippl W, Schüle R, Jung M. Structure-activity studies on N-Substituted tranylcypromine derivatives lead to selective inhibitors of lysine specific demethylase 1 (LSD1) and potent inducers of leukemic cell differentiation. Eur J Med Chem. 2018;144:52-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 95. | He M, Zhang T, Zhu Z, Qin S, Wang H, Zhao L, Zhang X, Hu J, Wen J, Cai H, Xin Q, Guo Q, Lin L, Zhou B, Zhang H, Xia G, Wang C. LSD1 contributes to programmed oocyte death by regulating the transcription of autophagy adaptor SQSTM1/p62. Aging Cell. 2020;19:e13102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 96. | Burnichon N, Brière JJ, Libé R, Vescovo L, Rivière J, Tissier F, Jouanno E, Jeunemaitre X, Bénit P, Tzagoloff A, Rustin P, Bertherat J, Favier J, Gimenez-Roqueplo AP. SDHA is a tumor suppressor gene causing paraganglioma. Hum Mol Genet. 2010;19:3011-3020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 506] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 97. | Nazar E, Khatami F, Saffar H, Tavangar SM. The Emerging Role of Succinate Dehyrogenase Genes (SDHx) in Tumorigenesis. Int J Hematol Oncol Stem Cell Res. 2019;13:72-82. [PubMed] |
| 98. | Wong CC, Xu J, Bian X, Wu JL, Kang W, Qian Y, Li W, Chen H, Gou H, Liu D, Yat Luk ST, Zhou Q, Ji F, Chan LS, Shirasawa S, Sung JJ, Yu J. In Colorectal Cancer Cells With Mutant KRAS, SLC25A22-Mediated Glutaminolysis Reduces DNA Demethylation to Increase WNT Signaling, Stemness, and Drug Resistance. Gastroenterology. 2020;159:2163-2180.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 115] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 99. | Spallotta F, Cencioni C, Atlante S, Garella D, Cocco M, Mori M, Mastrocola R, Kuenne C, Guenther S, Nanni S, Azzimato V, Zukunft S, Kornberger A, Sürün D, Schnütgen F, von Melchner H, Di Stilo A, Aragno M, Braspenning M, van Criekinge W, De Blasio MJ, Ritchie RH, Zaccagnini G, Martelli F, Farsetti A, Fleming I, Braun T, Beiras-Fernandez A, Botta B, Collino M, Bertinaria M, Zeiher AM, Gaetano C. Stable Oxidative Cytosine Modifications Accumulate in Cardiac Mesenchymal Cells From Type2 Diabetes Patients: Rescue by α-Ketoglutarate and TET-TDG Functional Reactivation. Circ Res. 2018;122:31-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 100. | Laukka T, Mariani CJ, Ihantola T, Cao JZ, Hokkanen J, Kaelin WG Jr, Godley LA, Koivunen P. Fumarate and Succinate Regulate Expression of Hypoxia-inducible Genes via TET Enzymes. J Biol Chem. 2016;291:4256-4265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 244] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 101. | Wang YP, Sharda A, Xu SN, van Gastel N, Man CH, Choi U, Leong WZ, Li X, Scadden DT. Malic enzyme 2 connects the Krebs cycle intermediate fumarate to mitochondrial biogenesis. Cell Metab. 2021;33:1027-1041.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 50] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 102. | De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1763] [Cited by in RCA: 1969] [Article Influence: 164.1] [Reference Citation Analysis (0)] |
| 103. | Sciacovelli M, Frezza C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J. 2017;284:3132-3144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 229] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 104. | Krüger A, Grüning NM, Wamelink MM, Kerick M, Kirpy A, Parkhomchuk D, Bluemlein K, Schweiger MR, Soldatov A, Lehrach H, Jakobs C, Ralser M. The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid Redox Signal. 2011;15:311-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 105. | Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, Rabinowitz JD. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1:404-415. [PubMed] |
| 106. | Ishida T, Nakao S, Ueyama T, Harada Y, Kawamura T. Metabolic remodeling during somatic cell reprogramming to induced pluripotent stem cells: involvement of hypoxia-inducible factor 1. Inflamm Regen. 2020;40:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 107. | Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA 4th, Ramalho-Santos J, Van Houten B, Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 499] [Cited by in RCA: 535] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 108. | Walvekar AS, Laxman S. Methionine at the Heart of Anabolism and Signaling: Perspectives From Budding Yeast. Front Microbiol. 2019;10:2624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 109. | Man CH, Mercier FE, Liu N, Dong W, Stephanopoulos G, Jiang L, Jung Y, Lin CP, Leung AYH, Scadden DT. Proton export alkalinizes intracellular pH and reprograms carbon metabolism to drive normal and malignant cell growth. Blood. 2022;139:502-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 110. | Fang Q, Zhang Y, Chen X, Li H, Cheng L, Zhu W, Zhang Z, Tang M, Liu W, Wang H, Wang T, Shen T, Chai R. Three-Dimensional Graphene Enhances Neural Stem Cell Proliferation Through Metabolic Regulation. Front Bioeng Biotechnol. 2019;7:436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 111. | Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, Guo L, Zhu J, Zhao X, Peng T, Zhang Y, Chen S, Li X, Li D, Wang T, Pei D. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 390] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 112. | Li Z, Fang F, Zhao Q, Li H, Xiong C. Supplementation of vitamin C promotes early germ cell specification from human embryonic stem cells. Stem Cell Res Ther. 2019;10:324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 113. | Zhitkovich A. Nuclear and Cytoplasmic Functions of Vitamin C. Chem Res Toxicol. 2020;33:2515-2526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 114. | Zhang Y, Kato H, Sato H, Yamaza H, Hirofuji Y, Han X, Masuda K, Nonaka K. Folic acid-mediated mitochondrial activation for protection against oxidative stress in human dental pulp stem cells derived from deciduous teeth. Biochem Biophys Res Commun. 2019;508:850-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 115. | Srivastava AC, Thompson YG, Singhal J, Stellern J, Srivastava A, Du J, O'Connor TR, Riggs AD. Elimination of human folypolyglutamate synthetase alters programming and plasticity of somatic cells. FASEB J. 2019;33:13747-13761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 116. | Li Z, Li W, Zhou D, Zhao J, Ma Y, Huang L, Dong C, Wilson JX, Huang G. Alleviating Oxidative Damage-Induced Telomere Attrition: a Potential Mechanism for Inhibition by Folic Acid of Apoptosis in Neural Stem Cells. Mol Neurobiol. 2022;59:590-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 117. | Kasulanati S, Venkatesan V. Understanding pluripotency under folic acid deficiency using embryonic stem cells as an in vitro model. Med Hypotheses. 2018;111:24-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 118. | Pei P, Cheng X, Yu J, Shen J, Li X, Wu J, Wang S, Zhang T. Folate deficiency induced H2A ubiquitination to lead to downregulated expression of genes involved in neural tube defects. Epigenetics Chromatin. 2019;12:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 119. | Xie Q, Li C, Song X, Wu L, Jiang Q, Qiu Z, Cao H, Yu K, Wan C, Li J, Yang F, Huang Z, Niu B, Jiang Z, Zhang T. Folate deficiency facilitates recruitment of upstream binding factor to hot spots of DNA double-strand breaks of rRNA genes and promotes its transcription. Nucleic Acids Res. 2017;45:2472-2489. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 120. | Horitani K, Iwasaki M, Kishimoto H, Wada K, Nakano M, Park H, Adachi Y, Motooka D, Okuzaki D, Shiojima I. Repetitive spikes of glucose and lipid induce senescence-like phenotypes of bone marrow stem cells through H3K27me3 demethylase-mediated epigenetic regulation. Am J Physiol Heart Circ Physiol. 2021;321:H920-H932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 121. | Cantor JR, Abu-Remaileh M, Kanarek N, Freinkman E, Gao X, Louissaint A Jr, Lewis CA, Sabatini DM. Physiologic Medium Rewires Cellular Metabolism and Reveals Uric Acid as an Endogenous Inhibitor of UMP Synthase. Cell. 2017;169:258-272.e17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 422] [Article Influence: 52.8] [Reference Citation Analysis (0)] |