Published online Jan 26, 2022. doi: 10.4252/wjsc.v14.i1.41
Peer-review started: March 16, 2021
First decision: May 5, 2021
Revised: May 13, 2021
Accepted: December 21, 2021
Article in press: December 21, 2021
Published online: January 26, 2022
Processing time: 309 Days and 19.6 Hours
The transforming growth factor (TGF)-β signaling pathway controls many cellular processes, including proliferation, differentiation, and apoptosis. Abnormalities in the TGF-β signaling pathway and its components are closely related to the occurrence of many human diseases, including cancer. Mothers against deca
Core Tip: Mothers against decapentaplegic homolog 4 (Smad4) is regarded as a tumor suppressor. Recent studies have shown that Smad4 plays a tumor-promoting role in specific types of cancer, rather than a tumor-suppressing role. Smad4 also correlates with the stem cells fate and drug resistance of cancer cells. Elucidating the specific role of Smad4 is of positive guiding significance for understanding the mechanism of tumorigenesis and cancer treatment. In this review, we focus on the multiple roles of Smad4 in tumorigenesis, stem cells, and drug resistance, and provide novel insights into the prospect of Smad4 in combination therapy.
- Citation: Dai CJ, Cao YT, Huang F, Wang YG. Multiple roles of mothers against decapentaplegic homolog 4 in tumorigenesis, stem cells, drug resistance, and cancer therapy. World J Stem Cells 2022; 14(1): 41-53
- URL: https://www.wjgnet.com/1948-0210/full/v14/i1/41.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i1.41
The transforming growth factor (TGF)-β signaling pathway controls several cell behaviors, including proliferation, inflammation, differentiation, and apoptosis[1,2]. Abnormalities of the TGF-β signaling pathway and its components are related to many human diseases such as fibrosis[3], immune diseases[4], and cancer[5]. Although TGF-β signals are mainly transmitted to the nucleus through the TGF-β/mothers against decapentaplegic homolog (Smad) signaling pathway[6], TGF-β superfamily ligands often interact with other signaling pathways, including JNK/p38, PI3K/AKT, ERK/MAPK, and integrin signaling pathways, regulating various cellular responses in a non-Smad-dependent manner[7-9]. The function of TGF-β in tumorigenesis is frequently described as a double-edged sword, but the precise mechanism for this phenomenon is still unclear[5,10,11]. In addition, TGF-β plays a role in the deve
Cancer has become a major threat to human health worldwide. In 2020 alone, 19.3 million new cases were diagnosed, and nearly 10 million people died of cancer[17]. Unfortunately, our current understanding of the mechanism of tumorigenesis is not enough to completely overcome cancer[18]. Smad4, a key component of the canonical TGF-β signaling pathway, exhibits varying degrees of inactivation among cancers[19], and its expression is significantly correlated with tumor development and prognosis in cancer patients[20,21]. Therefore, Smad4 is potently regarded as a tumor suppressor. Moreover, considering the critical role of Smad4 in tumor progression, Smad4 can be an attractive target for cancer treatment[22]. Additional novel options for cancer therapy were revealed by demonstrating that many RNAs directly/indirectly targeting Smad4, including miRNAs, circular (circ) RNAs, and long noncoding (lnc) RNAs, are dysregulated during cancer progression[23-25]. In the past decade, it has been found that Smad4 seems to play a tumor-promoting role in certain types of cancer, such as hepatocellular carcinoma[26,27]. In addition, an increasing number of studies demonstrated a close association of Smad4 with stem cell fate[28] and drug resistance of cancer cells[29]. Elucidating the specific role of Smad4 is important for understanding the mechanism of tumorigenesis and cancer treatment. Here, we review the identification and characteristics of Smad4, and the canonical TGF-β signaling pathway, and summarize the multiple regulatory functions of Smad4 in tumorigenesis, stem cell fate, and drug resistance. In addition, we provide new insights into the prospects of Smad4-targeting cancer therapy and its challenges in the future.
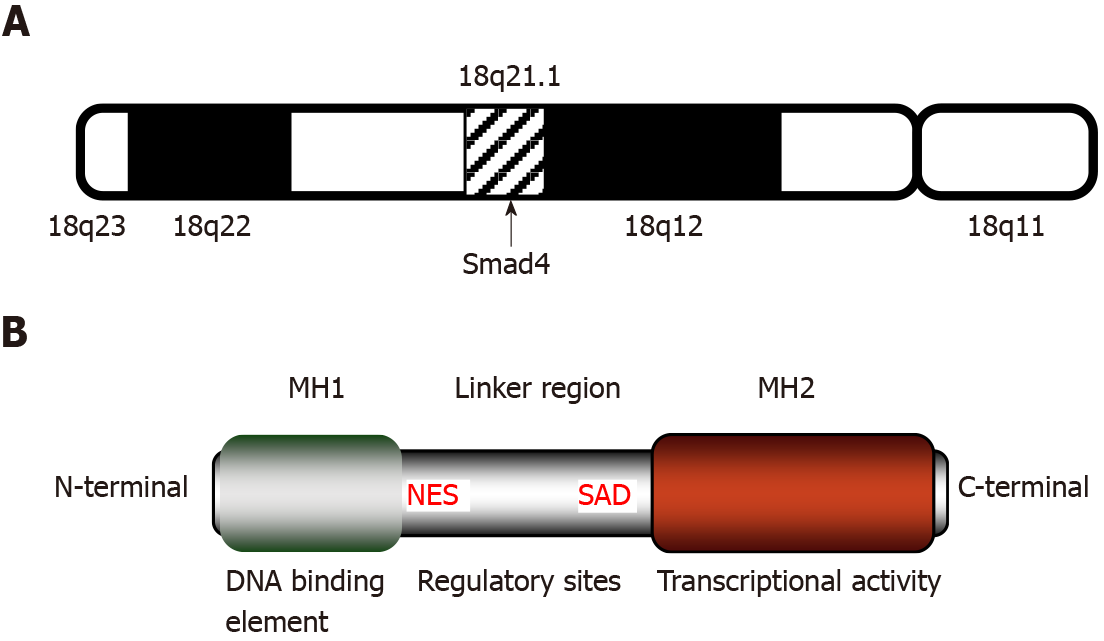
Smad4, also known as deleted in pancreatic cancer locus 4 (DPC4), was initially found as a tumor suppressor candidate gene in human pancreatic carcinoma in 1996[30]. The term “Smad” was a combination of the sma gene of Caenorhabditis elegans and the mad gene of Drosophila melanogaster[31]. Subsequently, Smad4 mutations were found in additional types of tumors, such as gastrointestinal carcinoid[32], prostate cancer[33], squamous cell carcinoma[34], and lung cancer[35]. The gene coding for Smad4 is located at human chromosome locus 18q21.1 (Figure 1A) and is composed of 12 exons and 10 introns[36]. The 12th exon was named exon 0 because it was discovered after the identification of 11 exons and is located upstream of exon 1[32].
Smad4 protein consists of 552 amino acids and has a molecular weight of 60 kDa[36]. It is composed of the N-terminal mad homology domain 1 (MH1), the middle linker region including nuclear export signal and Smad activation domain (SAD), as well as the C-terminal MH2[37]. The MH1 domain of Smad4 is involved in DNA binding by recognizing the Smad-binding site of DNA. The SAD of the linker region and MH2 domain are responsible for transcriptional activity. Additionally, the MH2 domain interacts with the MH1 domain of other Smads, enabling the transduction of various signaling pathways, including the TGF-β signaling (Figure 1B).
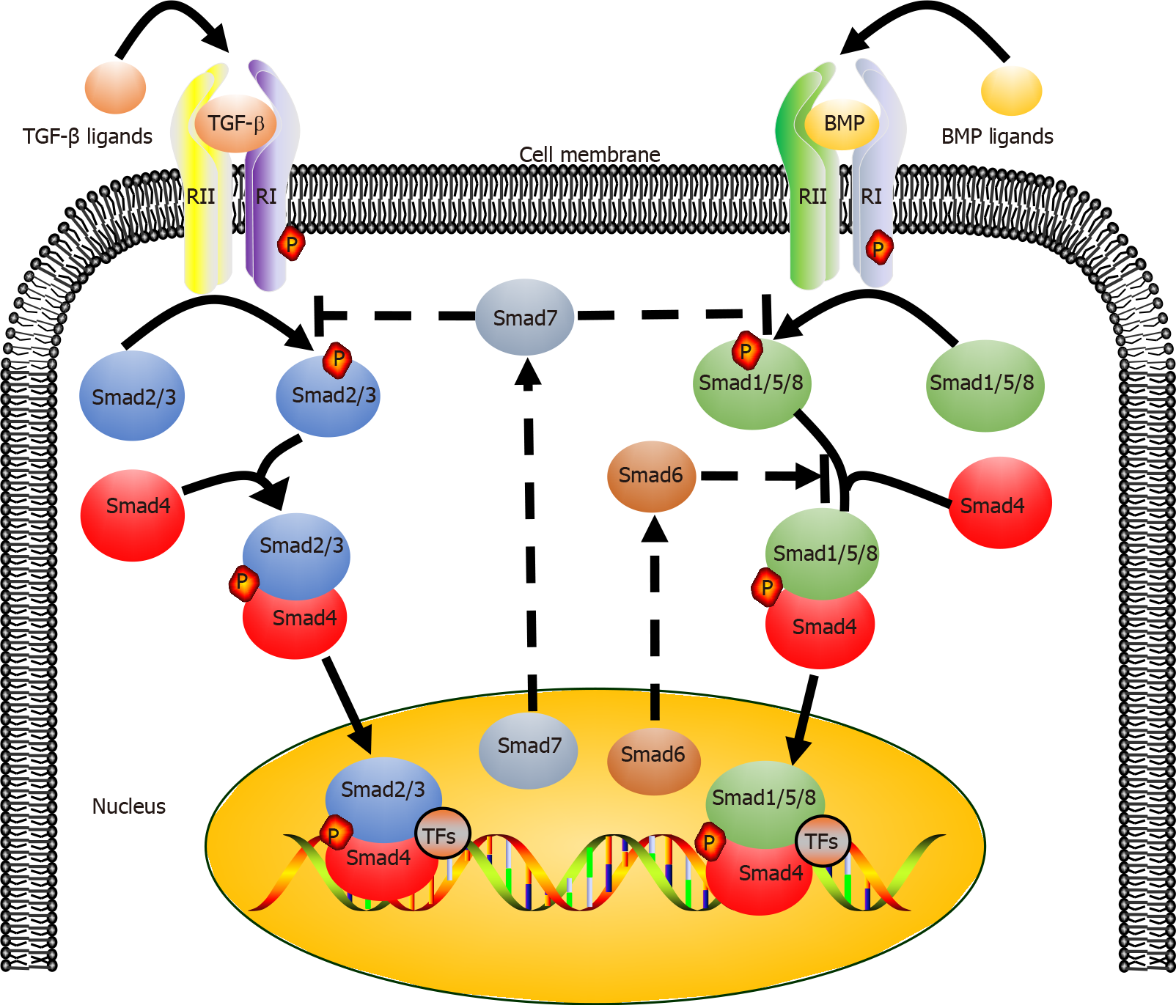
The canonical TGF-β signaling pathway is an uncomplicated linear cascade, which involves TGF-β superfamily ligands, receptors, and signal transducers (Figure 2)[38]. At present, there are 33 known TGF-β ligands encoded by mammalian genomes, including activin, nodal, TGF-βs, bone morphogenetic proteins (BMPs), and growth differentiation factors (GDFs)[39,40]. According to the difference in structure and function, these polypeptides can be subdivided into two families: TGF-βs (TGF-β, GDF, nodal, and activin) and BMPs (BMP2, BMP4, and BMP7). The TGF-β type I and type II receptors (TβRI and TβRII) are composed of several pairs of serine/threonine protein kinases[41,42].
The eight known Smads are divided into three categories, including a common Smad (Co-Smad, Smad4), two inhibitory Smads (I-Smads, Smad6, and Smad7), and receptor-regulated Smads (R-Smads, Smad2/3 transducing TGF-β signaling, and Smad1/5/8 transducing BMP signaling). Co-Smad and Smad4/DPC4 bind to R-Smads, forming two different complexes, Smad4/Smad2/3 and Smad4/Smad1/5/8. Subsequently, the heteromeric complexes are translocated into the nucleus, where they interact with transcriptional factors and bind to regulatory elements of target genes, affecting their expression[43]. Smad6 and Smad7 can both inhibit the transcriptional activity of target genes, thus blocking TGF-β signal transmission. It must be empha
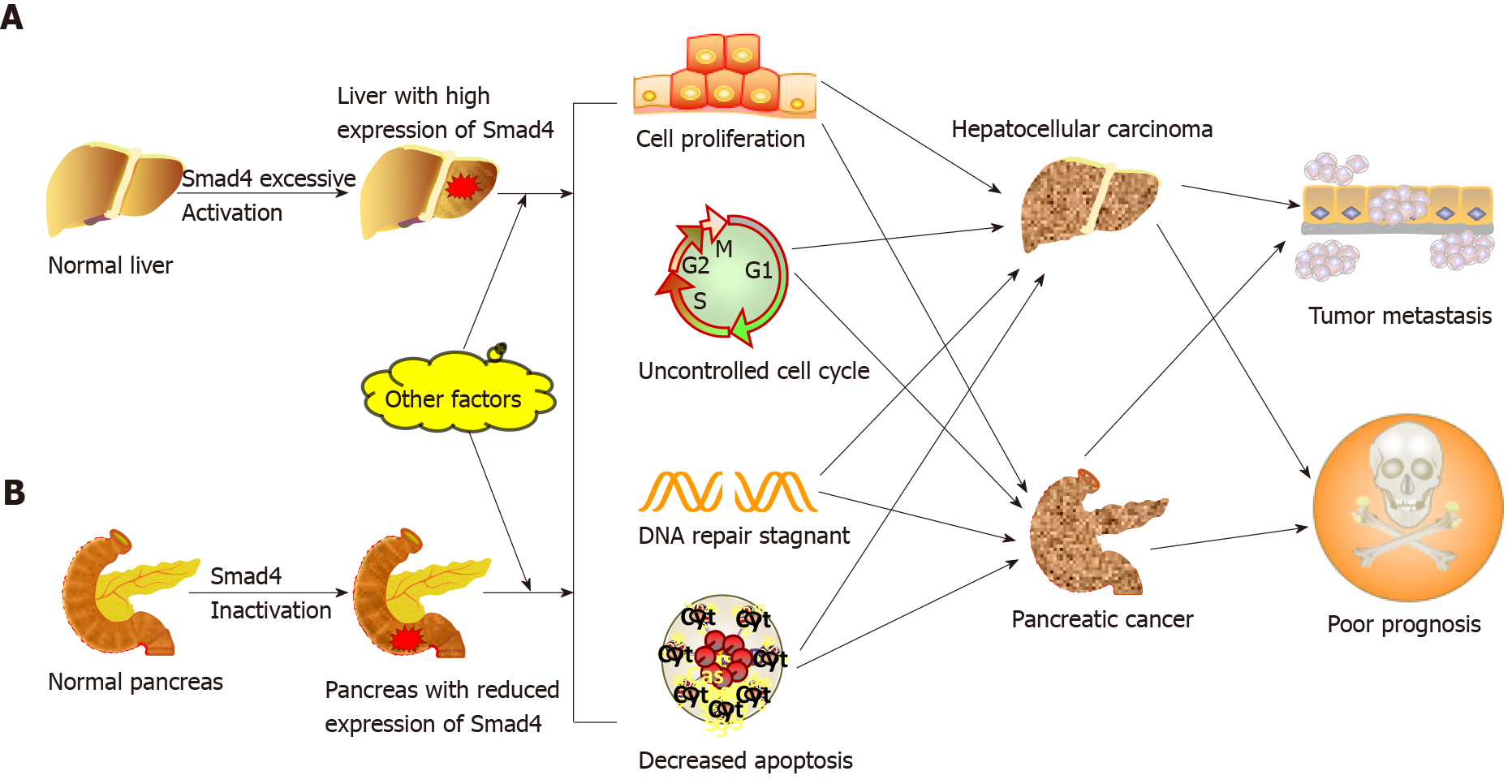
Due to the multiple interactions of environmental chemicals, genes, and endogenous signals, the process of carcinogenesis is extremely complex[46]. As a tumor suppressor gene, Smad4 exerts its inhibitory effect on tumor cells primarily via the canonical TGF-β signaling pathway[47,48]. The mechanism of this inhibition, which prevents carcinogenesis, involves the role of Smad4 in inhibiting the tumor-promoting activity of proinflammatory cytokines, inducing the cell cycle arrest, and promoting apoptosis through activating the TGF-β/BMP/Smad4 axis. However, once the Smad4 gene is mutated, TGF-β cannot induce G1 or G2 cell cycle arrest and switch from tumor suppressor to tumor promoter, leading to tumor growth and metastasis[10,49]. In addition, Smad4 is indispensable for the tumor suppressor function of TGF-β[50]. So far, there are many contradictory results and conclusions about the role of Smad4 in tumorigenesis. Here, we focus on pancreatic cancer and hepatocellular carcinoma (HCC) to discuss the role of Smad4 in cancer progression (Figure 3).
It is recognized that Smad4 acts as a tumor suppressor in pancreatic cancer. Smad4 mutation or deletion is found in > 50% of pancreatic cancer and is associated with the proliferation and metastasis of tumor cells[51,52]. The alterations in the Smad4 gene mainly include deletion, frameshift mutation, point mutation, amplification, and translocation[19,22]. Furthermore, Smad4 gene mutation is associated with stages of pancreatic cancer. A study by Notta et al[53] showed that the inactivation rate of Smad4 in mid-advanced pancreatic cancer was higher than it in early pancreatic cancer. Therefore, Smad4 dysfunction may be considered an advanced event in pancreatic cancer. Mechanically, the behavior that Smad4 deletion accelerates the progression of pancreatic cancer may be related to the increased expression of HNF4G, PAR-4, and PGK-1[51,54,55]. Additionally, Smad4 mutation in mice does not directly contribute to the formation of pancreatic tumors[56], indicating that a single Smad4 mutation is not sufficient to initiate pancreatic carcinogenesis. Thus, mutations in Smad4 are likely to cooperate with mutations in other genes in promoting pancreatic cancer progression. For example, Izeradjene et al[57] revealed that the formation of mucinous cystic neoplasms is induced by synergistic effects of Kras-G12D and Smad4 mutations. The role of Smad4 in other human cancers, such as colorectal cancer (CRC)[58], gastric cancer[59], ovarian cancer[60] and head and neck squamous cell carcinoma (HNSCC)[61], is similar to that in pancreatic cancer. Downregulation of Smad4 is considered an early event of HNSCC, which is different from pancreatic cancer.
An interesting fact regarding HCC is that the role of Smad4 in HCC differs significantly from that in cancers mentioned above. The nuclear level of Smad4 in liver cancer tissue is markedly higher than that in the adjacent noncancerous tissue[26]. The ability of liver cancer cells to form colonies and migrate was significantly reduced after Smad4 gene knockout in mouse models. Yuan et al[62] showed that ubiquitin-specific proteases promote HCC cell migration and invasion by deubiquitinating and stabilizing Smad4 protein. These findings seem to indicate that Smad4 plays a tumor-promoting rather than tumor-suppressing role in HCC. However, the underlying mechanism of Smad4 in the pathogenesis of HCC remains elusive. We propose that this difference may be explained by the fact that the cellular behaviors of the TGF-β/Smad4 signaling pathway varies with types of cells, their extracellular matrix, TGF-β concentration, and tumor microenvironment. These possibilities warrant further investigation. It is certain that Smad4 plays multiple regulatory roles in carcinogenesis, and whether it has a tumor-promoting or tumor-suppressing function may depend on the microenvironment of tumor cells and surrounding stromal cells.
Besides tumorigenesis, Smad4 is involved in the self-renewal and pluripotency of human stem cells. It has been reported that several miRNAs targeting Smad4 negatively regulate the differentiation of human mesenchymal stem cells (hMSCs)[63,64], but the underlying mechanisms of stem cell differentiation are poorly understood. The Smad4/TAZ axis might be involved in this process. TAZ protein, a transcriptional coactivator with PDZ-binding motif, can bind to Smad4 protein and translocate to the nucleus, where it enhances the expression of osteogenic genes, promoting the osteogenic differentiation of hMSCs[65]. These findings suggest that Smad4 is essential for stem cell differentiation. However, Avery et al[28] demonstrated that human embryonic stem cells (hESCs) remain undifferentiated after Smad4 gene knockdown, indicating that the single event of Smad4 inactivation does not induce stem cell differentiation. Similarly, Smad4 inactivation does not directly induce self-renewal of stem cells. For instance, Smad4 mutation has no effect on hESC self-renewal but is essential for their differentiation into cardiac mesodermal precursors[66].
BMP signaling is also indispensable for regulation of the proliferation of stem cells and maintenance of metabolic homeostasis[67]. BMP binds with leukemia inhibitory factor to maintain the self-renewal of ESCs[68]. Subsequent studies have documented that stem cell fate decisions induced by BMP may be related to Smad4. The BMP/Smad axis regulates the proliferation and differentiation of alveolar stem cells. BMP suppresses proliferation of alveolar type 2 epithelial cells (AT2s), while ant
Cancer stem cells (CSCs) have many characteristics similar to stem cells. However, in contrast to stem cells, they exhibit tumorigenicity and invasiveness[70]. The gradual accumulation of genetic mutations in stem cells during the life of the organism is associated with the development of cancer[71], and results in tumor heterogeneity[72]. Therefore, elucidating the relationship between Smad4 and CSC fate may provide a clinical diagnostic index or potential therapeutic target for cancer. Xia et al[73] found that cyclin D1 interacts with Smad2/3 and Smad4, activating the cyclin D1/Smad pathway and upregulating the expression of stemness genes in liver CSCs. This result implies that the use of Smad inhibitors may be an effective strategy for targeting liver CSCs. Wen et al[74] showed that blocking the TGF-β/Smad/EMT pathway inhibited the self-renewal and metastasis of ovarian CSCs. In another study[75], Smad4 muta
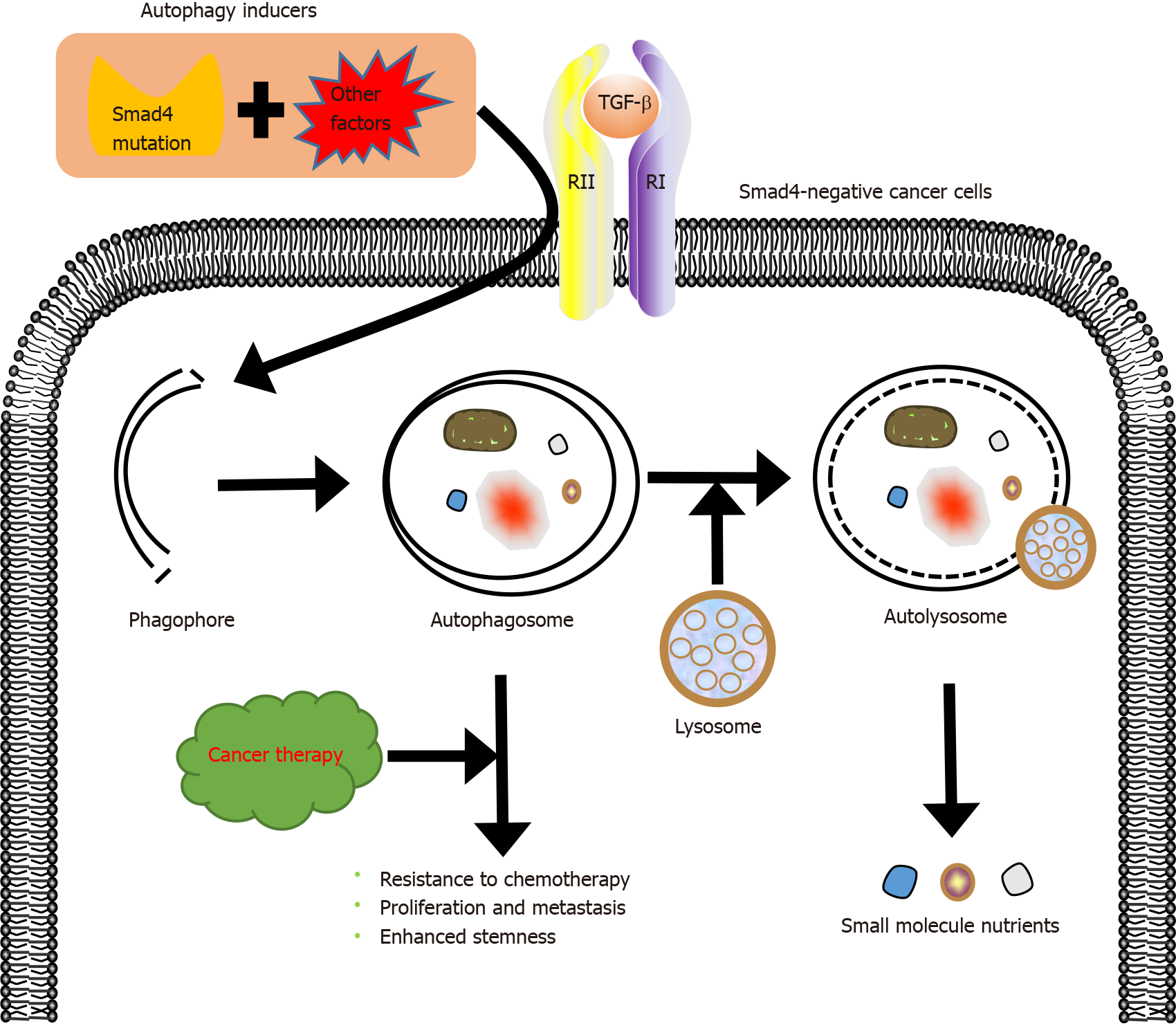
Drug resistance is the main cause of chemotherapy failure and tumor recurrence in cancer patients[76]. Drug resistance has been shown to be related to the activation of autophagy[77], a highly conserved catabolic process in which large cellular structures are degraded. Autophagy contributes to cell survival by recycling constituents of cellular structures[78]. Activation of autophagy not only mediates the resistance to chemotherapy but also induces autophagy-mediated cell death, which helps to eliminate tumor cells[52,79]. For instance, autophagy protects breast cancer cells from epirubicin-induced apoptosis and promotes the development of epirubicin resistance[80]. Peptidylarginine deiminase IV, a protein involved in many pathological processes, induces the resistance of HCC to chemotherapy by activating autophagy[81]. Autophagy may promote the development of multidrug resistance. Fortunately, inducing autophagy-mediated cell death can help overcome the resistance to chemotherapy, and there are many clinically available drugs that can regulate autophagy, such as chloroquine or hydroxychloroquine[82]. These autophagy regu
Smad4 inactivation not only involves the development of drug resistance in cancer but also contributes to the crosstalk between TGF-β and other signaling pathways, accelerating this process. Zhang et al[86] demonstrated that Smad4 deletion induces the resistance of colon cancer cells to 5-fluorouracil (5-FU)-based therapy by activating the AKT pathway. Moreover, Smad4 deficiency inhibits 5-FU-mediated apoptosis[87]. Besides, the inactivation of Smad4 makes tumors resistant to other chemotherapeutic drugs. Smad4 mutation contributes to platinum resistance in non-small cell lung carcinoma[35] and to cetuximab resistance in HNSCC[29]. Targeting the rapamycin-insensitive companion of mTOR, a component of mammalian target of rapamycin complex 2, increases the sensitivity of Smad4-negative colon cancer to irinotecan[88]. Recent studies have demonstrated that Smad4 deficiency correlates with the development of resistance to chemotherapy and radiotherapy[89]. Therefore, Smad4 modifications may be a critical marker predicting the resistance of tumor cells to chemoradiation therapy[90]. In our view, elucidating the specific role of Smad4 in resistance of tumors to drugs and developing rational therapeutic applications of autophagy will help to improve the outcomes of treatment of drug-resistant tumors.
Given the key role of Smad4 in tumorigenesis, Smad4 is expected to be an attractive therapeutic target for tumors resistant to radiotherapy and chemotherapy. Many miRNAs, such as miR-224[91], miR-34a[92], and miR-205[93], are essential regulators of TGF-β-induced tumor suppression by affecting the TGF-β/Smad signaling pathway. Therefore, Smad4-targeting miRNAs may become novel therapeutic agents for cancer treatment. Other RNAs, including circRNAs[94] and lncRNAs[95], regulate the level of Smad4 protein by targeting Smad4-targeting miRNAs directly or indirectly. Proteins, such as ALK[96] and tripartite motif 47[97] that promote, respectively, Smad4 phosphorylation and ubiquitination, diminish the tumor suppressor effect of Smad4, indicating that suppressor molecules or enzymes that inhibit Smad4 activity may be a new option for the treatment of Smad4-negative cancers.
Many inhibitors targeting the TGF-β signaling pathway are being developed clinically[98]. Molecules such as TGF-β antibodies, antisense oligonucleotides, and small molecule inhibitors of TGF-β receptor kinase activity show immense clinical potential[99]. Although inhibition of the TGF-β signaling pathway is one of the strategies for cancer treatment, the clinical outcomes of targeting TGF-β signaling and its superfamily members are not satisfactory, suggesting that single target-based therapies are not sufficient to inhibit tumors[10].
Combination therapy is an increasingly important part of anticancer therapies, and has more advantages than single-drug therapy[100-102]. The combination of Smad4 targeting and traditional/nontraditional therapies may become a novel choice for anticancer treatment. Mariathasan et al[103] showed that the combined application of TGF-β inhibitors together with anti-PD-L1 antibodies decreased the TGF-β level and promoted the infiltration of T-cells into tumor cells, thereby enhancing antitumor immune response and leading to tumor apoptosis. Kassardjian and Wang[104] found that Smad4-positive tumors had a better response to neoadjuvant therapy, and the lymph node metastasis rate of Smad4-positive tumors was significantly lower, suggesting that Smad4 plays an important role in neoadjuvant therapy. In our previous research[105], we combined oncolytic virus therapy and targeted gene therapy to design a new oncolytic adenovirus CD55-Smad4, which can stably produce Smad4 protein in vitro and in vivo. CD55-Smad4 significantly inhibited proliferation, metastasis, and stemness of CRC cells. All these show that the combination therapy targeting Smad4 has clinical potential. Therefore, we believe that combination of Smad4-targeted therapy with traditional therapies of surgery, radiotherapy, and chemotherapy, and with emerging treatments such as PD-LI inhibitors, chimeric antigen receptor cell therapy, as well as our targeting gene virotherapy will sig
Targeting abnormal signal transduction or abnormal metabolic pathway has always been the focus of anticancer research. Smad4 gene mutations and its abnormal expression have been confirmed to dysregulate the TGF-β signaling pathway, transforming its function from a tumor suppressor to a tumor promoter[106]. This signaling disorder accelerates tumor progression, but the precise mechanism by which Smad4 affects tumor development remains unclear. An increasing number of studies have shown that targeting oncogenic miRNAs, circRNAs, and other RNAs can inhibit TGF-β-induced tumorigenesis by directly or indirectly acting on Smad4, increasing susceptibility of tumors to chemoradiation and improving the survival rate of cancer patients. Although the possibility of regulating tumor progression by these RNAs shows clinical potential, the underlying mechanisms are poorly understood.
Fortunately, the vigorous development of genome editing technologies such as CRISPR-Cas9 systems and AAV gene vectors has enabled the transformation of Smad4-targeted therapies into clinical reality[107,108]. Furthermore, it is asserted that the presence of Smad4 protein promotes the cell-to-cell spread of the vaccinia virus, which enables combining Smad4 gene therapy with oncolytic virus therapy to effectively eliminate tumors[109]. In summary, although the specific mechanisms by which Smad4 affects carcinogenesis, metastasis, drug resistance, and other malignant features of tumors have not been completely clarified, Smad4-targeted therapies coupled with traditional therapies and emerging anticancer treatments have achieved good antitumor effects in animal models. Smad4 combination therapy shows potential for future clinical applications.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zhou P S-Editor: Wang JJ L-Editor: Wang TQ P-Editor: Wang JJ
| 1. | Blank U, Karlsson S. TGF-β signaling in the control of hematopoietic stem cells. Blood. 2015;125:3542-3550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 208] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 2. | Haque S, Morris JC. Transforming growth factor-β: A therapeutic target for cancer. Hum Vaccin Immunother. 2017;13:1741-1750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 219] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 3. | Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12:325-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1512] [Cited by in RCA: 2546] [Article Influence: 282.9] [Reference Citation Analysis (0)] |
| 4. | Batlle E, Massagué J. Transforming Growth Factor-β Signaling in Immunity and Cancer. Immunity. 2019;50:924-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 787] [Cited by in RCA: 1642] [Article Influence: 273.7] [Reference Citation Analysis (0)] |
| 5. | Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1726] [Cited by in RCA: 1776] [Article Influence: 74.0] [Reference Citation Analysis (0)] |
| 6. | Tzavlaki K, Moustakas A. TGF-β Signaling. Biomolecules. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 531] [Article Influence: 106.2] [Reference Citation Analysis (0)] |
| 7. | Luo K. Signaling Cross Talk between TGF-β/Smad and Other Signaling Pathways. Cold Spring Harb Perspect Biol. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 415] [Cited by in RCA: 437] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 8. | Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 788] [Cited by in RCA: 802] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 9. | Jahan N, Hannila SS. Transforming growth factor β-induced expression of chondroitin sulfate proteoglycans is mediated through non-Smad signaling pathways. Exp Neurol. 2015;263:372-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Colak S, Ten Dijke P. Targeting TGF-β Signaling in Cancer. Trends Cancer. 2017;3:56-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 752] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 11. | Wang Y, Xue Q, Zheng Q, Jin Y, Shen X, Yang M, Zhou X, Li Y. SMAD4 mutation correlates with poor prognosis in non-small cell lung cancer. Lab Invest. 2021;101:463-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Khalil H, Kanisicak O, Prasad V, Correll RN, Fu X, Schips T, Vagnozzi RJ, Liu R, Huynh T, Lee SJ, Karch J, Molkentin JD. Fibroblast-specific TGF-β-Smad2/3 signaling underlies cardiac fibrosis. J Clin Invest. 2017;127:3770-3783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 691] [Article Influence: 86.4] [Reference Citation Analysis (0)] |
| 13. | Tatler AL, John AE, Jolly L, Habgood A, Porte J, Brightling C, Knox AJ, Pang L, Sheppard D, Huang X, Jenkins G. Integrin αvβ5-mediated TGF-β activation by airway smooth muscle cells in asthma. J Immunol. 2011;187:6094-6107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 14. | Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu Rev Immunol. 2014;32:51-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 659] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 15. | Caja L, Dituri F, Mancarella S, Caballero-Diaz D, Moustakas A, Giannelli G, Fabregat I. TGF-β and the Tissue Microenvironment: Relevance in Fibrosis and Cancer. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 257] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 16. | Derynck R, Budi EH. Specificity, versatility, and control of TGF-β family signaling. Sci Signal. 2019;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 544] [Article Influence: 90.7] [Reference Citation Analysis (0)] |
| 17. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64637] [Article Influence: 16159.3] [Reference Citation Analysis (176)] |
| 18. | Qiu L, Hu X, Jing Q, Zeng X, Chan KM, Han J. Mechanism of cancer: Oncohistones in action. J Genet Genomics. 2018;45:227-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 19. | Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 Mutations in Pancreatic Cancer. Cancers (Basel). 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 190] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 20. | Ahmed S, Bradshaw AD, Gera S, Dewan MZ, Xu R. The TGF-β/Smad4 Signaling Pathway in Pancreatic Carcinogenesis and Its Clinical Significance. J Clin Med. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 125] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 21. | Rosic J, Dragicevic S, Miladinov M, Despotovic J, Bogdanovic A, Krivokapic Z, Nikolic A. SMAD7 and SMAD4 expression in colorectal cancer progression and therapy response. Exp Mol Pathol. 2021;123:104714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Qian Y, Gong Y, Fan Z, Luo G, Huang Q, Deng S, Cheng H, Jin K, Ni Q, Yu X, Liu C. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J Hematol Oncol. 2020;13:130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 223] [Cited by in RCA: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 23. | Yan W, Liu Z, Yang W, Wu G. miRNA expression profiles in Smad4-positive and Smad4-negative SW620 human colon cancer cells detected by next-generation small RNA sequencing. Cancer Manag Res. 2018;10:5479-5490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Ma C, Wang X, Yang F, Zang Y, Liu J, Xu X, Li W, Jia J, Liu Z. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020;19:157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 25. | Zhang C, Hao Y, Wang Y, Xu J, Teng Y, Yang X. TGF-β/SMAD4-Regulated LncRNA-LINP1 Inhibits Epithelial-Mesenchymal Transition in Lung Cancer. Int J Biol Sci. 2018;14:1715-1723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 26. | Hernanda PY, Chen K, Das AM, Sideras K, Wang W, Li J, Cao W, Bots SJ, Kodach LL, de Man RA, Ijzermans JN, Janssen HL, Stubbs AP, Sprengers D, Bruno MJ, Metselaar HJ, ten Hagen TL, Kwekkeboom J, Peppelenbosch MP, Pan Q. SMAD4 exerts a tumor-promoting role in hepatocellular carcinoma. Oncogene. 2015;34:5055-5068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 27. | Hiwatashi K, Ueno S, Sakoda M, Kubo F, Tateno T, Kurahara H, Mataki Y, Maemura K, Ishigami S, Shinchi H, Natsugoe S. Strong Smad4 expression correlates with poor prognosis after surgery in patients with hepatocellular carcinoma. Ann Surg Oncol. 2009;16:3176-3182. [PubMed] [DOI] [Full Text] |
| 28. | Avery S, Zafarana G, Gokhale PJ, Andrews PW. The role of SMAD4 in human embryonic stem cell self-renewal and stem cell fate. Stem Cells. 2010;28:863-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Ozawa H, Ranaweera RS, Izumchenko E, Makarev E, Zhavoronkov A, Fertig EJ, Howard JD, Markovic A, Bedi A, Ravi R, Perez J, Le QT, Kong CS, Jordan RC, Wang H, Kang H, Quon H, Sidransky D, Chung CH. SMAD4 Loss Is Associated with Cetuximab Resistance and Induction of MAPK/JNK Activation in Head and Neck Cancer Cells. Clin Cancer Res. 2017;23:5162-5175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 30. | Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1723] [Cited by in RCA: 1669] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 31. | Derynck R, Gelbart WM, Harland RM, Heldin CH, Kern SE, Massagué J, Melton DA, Mlodzik M, Padgett RW, Roberts AB, Smith J, Thomsen GH, Vogelstein B, Wang XF. Nomenclature: vertebrate mediators of TGFbeta family signals. Cell. 1996;87:173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 138] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Mamot C, Mild G, Reuter J, Laffer U, Metzger U, Terracciano L, Boulay JL, Herrmann R, Rochlitz C. Infrequent mutation of the tumour-suppressor gene Smad4 in early-stage colorectal cancer. Br J Cancer. 2003;88:420-423. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Horvath LG, Henshall SM, Kench JG, Turner JJ, Golovsky D, Brenner PC, O'Neill GF, Kooner R, Stricker PD, Grygiel JJ, Sutherland RL. Loss of BMP2, Smad8, and Smad4 expression in prostate cancer progression. Prostate. 2004;59:234-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 34. | Hernandez AL, Young CD, Wang JH, Wang XJ. Lessons learned from SMAD4 Loss in squamous cell carcinomas. Mol Carcinog. 2019;58:1648-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 35. | Tan X, Tong L, Li L, Xu J, Xie S, Ji L, Fu J, Liu Q, Shen S, Liu Y, Xiao Y, Gao F, Moses RE, Bardeesy N, Wang Y, Zhang J, Tang L, Wong K-K, Song D, Yang X, Liu J, Li X. Loss of Smad4 promotes aggressive lung cancer metastasis by de-repression of PAK3 via miRNA regulation. Nat Commun. 2021;12 (1):4853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 44] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 36. | Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 340] [Cited by in RCA: 346] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 37. | Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14:111-123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 221] [Cited by in RCA: 443] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 38. | Budi EH, Duan D, Derynck R. Transforming Growth Factor-β Receptors and Smads: Regulatory Complexity and Functional Versatility. Trends Cell Biol. 2017;27:658-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 224] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 39. | Morikawa M, Derynck R, Miyazono K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb Perspect Biol. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 972] [Article Influence: 108.0] [Reference Citation Analysis (0)] |
| 40. | Weiss A, Attisano L. The TGFbeta superfamily signaling pathway. Wiley Interdiscip Rev Dev Biol. 2013;2:47-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 406] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 41. | David CJ, Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19:419-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 615] [Article Influence: 87.9] [Reference Citation Analysis (0)] |
| 42. | Vander Ark A, Cao J, Li X. TGF-β receptors: In and beyond TGF-β signaling. Cell Signal. 2018;52:112-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 328] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 43. | Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783-2810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 1886] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 44. | Hata A, Lagna G, Massagué J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 543] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 45. | Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365-1375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1031] [Cited by in RCA: 1079] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 46. | Peters JM, Gonzalez FJ. The Evolution of Carcinogenesis. Toxicol Sci. 2018;165:272-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 47. | Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor {beta} (TGF-{beta}) target genes and distinguishes TGF-{beta}-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108-8125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 275] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 48. | Ke Y, Wang XJ. TGFβ Signaling in Photoaging and UV-Induced Skin Cancer. J Invest Dermatol. 2021;141:1104-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 82] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 49. | Hao Y, Baker D, Ten Dijke P. TGF-β-Mediated Epithelial-Mesenchymal Transition and Cancer Metastasis. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 363] [Cited by in RCA: 807] [Article Influence: 134.5] [Reference Citation Analysis (0)] |
| 50. | Li X, Li J, Cai Y, Peng S, Wang J, Xiao Z, Wang Y, Tao Y, Leng Q, Wu D, Yang S, Ji Z, Han Y, Li L, Gao X, Zeng C, Wen X. Hyperglycaemia-induced miR-301a promotes cell proliferation by repressing p21 and Smad4 in prostate cancer. Cancer Lett. 2018;418:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Liang C, Shi S, Qin Y, Meng Q, Hua J, Hu Q, Ji S, Zhang B, Xu J, Yu XJ. Localisation of PGK1 determines metabolic phenotype to balance metastasis and proliferation in patients with SMAD4-negative pancreatic cancer. Gut. 2020;69:888-900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 52. | Fei N, Wen S, Ramanathan R, Hogg ME, Zureikat AH, Lotze MT, Bahary N, Singhi AD, Zeh HJ, Boone BA. SMAD4 loss is associated with response to neoadjuvant chemotherapy plus hydroxychloroquine in patients with pancreatic adenocarcinoma. Clin Transl Sci. 2021;14:1822-1829. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 53. | Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, Denroche RE, Liang SB, Brown AM, Kim JC, Wang T, Simpson JT, Beck T, Borgida A, Buchner N, Chadwick D, Hafezi-Bakhtiari S, Dick JE, Heisler L, Hollingsworth MA, Ibrahimov E, Jang GH, Johns J, Jorgensen LG, Law C, Ludkovski O, Lungu I, Ng K, Pasternack D, Petersen GM, Shlush LI, Timms L, Tsao MS, Wilson JM, Yung CK, Zogopoulos G, Bartlett JM, Alexandrov LB, Real FX, Cleary SP, Roehrl MH, McPherson JD, Stein LD, Hudson TJ, Campbell PJ, Gallinger S. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature. 2016;538:378-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 400] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 54. | Wang C, Zhang T, Liao Q, Dai M, Guo J, Yang X, Tan W, Lin D, Wu C, Zhao Y. Metformin inhibits pancreatic cancer metastasis caused by SMAD4 deficiency and consequent HNF4G upregulation. Protein Cell. 2021;12:128-144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 55. | Mohd Faheem M, Rasool RU, Ahmad SM, Jamwal VL, Chakraborty S, Katoch A, Gandhi SG, Bhagat M, Goswami A. Par-4 mediated Smad4 induction in PDAC cells restores canonical TGF-β/ Smad4 axis driving the cells towards lethal EMT. Eur J Cell Biol. 2020;99:151076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 56. | Colvin EK, Scarlett CJ. A historical perspective of pancreatic cancer mouse models. Semin Cell Dev Biol. 2014;27:96-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 57. | Izeradjene K, Combs C, Best M, Gopinathan A, Wagner A, Grady WM, Deng CX, Hruban RH, Adsay NV, Tuveson DA, Hingorani SR. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell. 2007;11:229-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 279] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 58. | Ogawa R, Yamamoto T, Hirai H, Hanada K, Kiyasu Y, Nishikawa G, Mizuno R, Inamoto S, Itatani Y, Sakai Y, Kawada K. Loss of SMAD4 Promotes Colorectal Cancer Progression by Recruiting Tumor-Associated Neutrophils via the CXCL1/8-CXCR2 Axis. Clin Cancer Res. 2019;25:2887-2899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 59. | Leng A, Liu T, He Y, Li Q, Zhang G. Smad4/Smad7 balance: a role of tumorigenesis in gastric cancer. Exp Mol Pathol. 2009;87:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 60. | Wang J, Li Y, Zhou JH, Shen FR, Shi X, Chen YG. CircATRNL1 activates Smad4 signaling to inhibit angiogenesis and ovarian cancer metastasis via miR-378. Mol Oncol. 2021;15:1217-1233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 61. | Lin LH, Chang KW, Cheng HW, Liu CJ. SMAD4 Somatic Mutations in Head and Neck Carcinoma Are Associated With Tumor Progression. Front Oncol. 2019;9:1379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 62. | Yuan T, Chen Z, Yan F, Qian M, Luo H, Ye S, Cao J, Ying M, Dai X, Gai R, Yang B, He Q, Zhu H. Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein. Mol Oncol. 2020;14:197-210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 63. | Ma X, Fan C, Wang Y, Du Y, Zhu Y, Liu H, Lv L, Liu Y, Zhou Y. MiR-137 knockdown promotes the osteogenic differentiation of human adipose-derived stem cells via the LSD1/BMP2/SMAD4 signaling network. J Cell Physiol. 2020;235:909-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 64. | Gu Z, Long J, Li Y, Wang X, Wang H. MiR-125a-3p negatively regulates osteoblastic differentiation of human adipose derived mesenchymal stem cells by targeting Smad4 and Jak1. Am J Transl Res. 2019;11:2603-2615. [PubMed] |
| 65. | Park JS, Kim M, Song NJ, Kim JH, Seo D, Lee JH, Jung SM, Lee JY, Lee J, Lee YS, Park KW, Park SH. A Reciprocal Role of the Smad4-Taz Axis in Osteogenesis and Adipogenesis of Mesenchymal Stem Cells. Stem Cells. 2019;37:368-381. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 66. | Xu J, Gruber PJ, Chien KR. SMAD4 Is Essential for Human Cardiac Mesodermal Precursor Cell Formation. Stem Cells. 2019;37:216-225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 67. | Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, Hogan BLM. Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development. 2018;145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 213] [Cited by in RCA: 218] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 68. | Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1593] [Cited by in RCA: 1551] [Article Influence: 73.9] [Reference Citation Analysis (0)] |
| 69. | Qi Z, Li Y, Zhao B, Xu C, Liu Y, Li H, Zhang B, Wang X, Yang X, Xie W, Li B, Han JJ, Chen YG. BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat Commun. 2017;8:13824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 226] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 70. | Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1234] [Cited by in RCA: 1891] [Article Influence: 236.4] [Reference Citation Analysis (0)] |
| 71. | Vlashi E, Pajonk F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin Cancer Biol. 2015;31:28-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 231] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 72. | Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 331] [Cited by in RCA: 565] [Article Influence: 70.6] [Reference Citation Analysis (0)] |
| 73. | Xia W, Lo CM, Poon RYC, Cheung TT, Chan ACY, Chen L, Yang S, Tsao GSW, Wang XQ. Smad inhibitor induces CSC differentiation for effective chemosensitization in cyclin D1- and TGF-β/Smad-regulated liver cancer stem cell-like cells. Oncotarget. 2017;8:38811-38824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 74. | Wen H, Qian M, He J, Li M, Yu Q, Leng Z. Inhibiting of self-renewal, migration and invasion of ovarian cancer stem cells by blocking TGF-β pathway. PLoS One. 2020;15:e0230230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 75. | Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 837] [Article Influence: 83.7] [Reference Citation Analysis (0)] |
| 76. | Assaraf YG, Brozovic A, Gonçalves AC, Jurkovicova D, Linē A, Machuqueiro M, Saponara S, Sarmento-Ribeiro AB, Xavier CPR, Vasconcelos MH. The multi-factorial nature of clinical multidrug resistance in cancer. Drug Resist Updat. 2019;46:100645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 371] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 77. | Zamame Ramirez JA, Romagnoli GG, Kaneno R. Inhibiting autophagy to prevent drug resistance and improve anti-tumor therapy. Life Sci. 2021;265:118745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 78. | Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247:708-718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 282] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 79. | Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 430] [Cited by in RCA: 738] [Article Influence: 73.8] [Reference Citation Analysis (0)] |
| 80. | Sun WL, Chen J, Wang YP, Zheng H. Autophagy protects breast cancer cells from epirubicin-induced apoptosis and facilitates epirubicin-resistance development. Autophagy. 2011;7:1035-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 81. | Fan T, Zhang C, Zong M, Zhao Q, Yang X, Hao C, Zhang H, Yu S, Guo J, Gong R, Fan S, Wei L, Fan L. Peptidylarginine deiminase IV promotes the development of chemoresistance through inducing autophagy in hepatocellular carcinoma. Cell Biosci. 2014;4:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 82. | Amaravadi RK, Kimmelman AC, Debnath J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov. 2019;9:1167-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 670] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 83. | Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ, Coppes RP, Engedal N, Mari M, Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1423] [Cited by in RCA: 1456] [Article Influence: 208.0] [Reference Citation Analysis (0)] |
| 84. | Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, Kano MR, Sugimoto K, Miyazono K. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res. 2009;69:8844-8852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 239] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 85. | Liang C, Xu J, Meng Q, Zhang B, Liu J, Hua J, Zhang Y, Shi S, Yu X. TGFB1-induced autophagy affects the pattern of pancreatic cancer progression in distinct ways depending on SMAD4 status. Autophagy. 2020;16:486-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 86. | Zhang B, Zhang B, Chen X, Bae S, Singh K, Washington MK, Datta PK. Loss of Smad4 in colorectal cancer induces resistance to 5-fluorouracil through activating Akt pathway. Br J Cancer. 2014;110:946-957. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 87. | Papageorgis P, Cheng K, Ozturk S, Gong Y, Lambert AW, Abdolmaleky HM, Zhou JR, Thiagalingam S. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71:998-1008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 177] [Cited by in RCA: 175] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 88. | Wong CK, Lambert AW, Ozturk S, Papageorgis P, Lopez D, Shen N, Sen Z, Abdolmaleky HM, Győrffy B, Feng H, Thiagalingam S. Targeting RICTOR Sensitizes SMAD4-Negative Colon Cancer to Irinotecan. Mol Cancer Res. 2020;18:414-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 89. | Jiang D, Wang X, Wang Y, Philips D, Meng W, Xiong M, Zhao J, Sun L, He D, Li K. Mutation in BRAF and SMAD4 associated with resistance to neoadjuvant chemoradiation therapy in locally advanced rectal cancer. Virchows Arch. 2019;475:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 90. | Ezrova Z, Nahacka Z, Stursa J, Werner L, Vlcak E, Kralova Viziova P, Berridge MV, Sedlacek R, Zobalova R, Rohlena J, Boukalova S, Neuzil J. SMAD4 loss limits the vulnerability of pancreatic cancer cells to complex I inhibition via promotion of mitophagy. Oncogene. 2021;40:2539-2552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 91. | Zhou J, Hu M, Wang F, Song M, Huang Q, Ge B. miR-224 Controls Human Colorectal Cancer Cell Line HCT116 Proliferation by Targeting Smad4. Int J Med Sci. 2017;14:937-942. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 92. | Qi Y, Zhao A, Yang P, Jin L, Hao C. miR-34a-5p Attenuates EMT through targeting SMAD4 in silica-induced pulmonary fibrosis. J Cell Mol Med. 2020;24:12219-12224. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 93. | Chu P, Liang A, Jiang A, Zong L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol Lett. 2018;15:7571-7578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 94. | Liu J, Dai X, Guo X, Cheng A, Mac SM, Wang Z. Circ-OXCT1 Suppresses Gastric Cancer EMT and Metastasis by Attenuating TGF-β Pathway Through the Circ-OXCT1/miR-136/SMAD4 Axis. Onco Targets Ther. 2020;13:3987-3998. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 95. | Xiao T, Zou Z, Xue J, Syed BM, Sun J, Dai X, Shi M, Li J, Wei S, Tang H, Zhang A, Liu Q. LncRNA H19-mediated M2 polarization of macrophages promotes myofibroblast differentiation in pulmonary fibrosis induced by arsenic exposure. Environ Pollut. 2021;268:115810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 96. | Zhang Q, Xiao M, Gu S, Xu Y, Liu T, Li H, Yu Y, Qin L, Zhu Y, Chen F, Wang Y, Ding C, Wu H, Ji H, Chen Z, Zu Y, Malkoski S, Li Y, Liang T, Ji J, Qin J, Xu P, Zhao B, Shen L, Lin X, Feng XH. ALK phosphorylates SMAD4 on tyrosine to disable TGF-β tumour suppressor functions. Nat Cell Biol. 2019;21:179-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 97. | Liang Q, Tang C, Tang M, Zhang Q, Gao Y, Ge Z. TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J Exp Clin Cancer Res. 2019;38:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 98. | Huynh LK, Hipolito CJ, Ten Dijke P. A Perspective on the Development of TGF-β Inhibitors for Cancer Treatment. Biomolecules. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 139] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 99. | Syed V. TGF-β Signaling in Cancer. J Cell Biochem. 2016;117:1279-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 355] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 100. | Nastiuk KL, Krolewski JJ. Opportunities and challenges in combination gene cancer therapy. Adv Drug Deliv Rev. 2016;98:35-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 101. | Colli LM, Machiela MJ, Zhang H, Myers TA, Jessop L, Delattre O, Yu K, Chanock SJ. Landscape of Combination Immunotherapy and Targeted Therapy to Improve Cancer Management. Cancer Res. 2017;77:3666-3671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 102. | Martin NT, Bell JC. Oncolytic Virus Combination Therapy: Killing One Bird with Two Stones. Mol Ther. 2018;26:1414-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 103. | Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, Jhunjhunwala S, Banchereau R, Yang Y, Guan Y, Chalouni C, Ziai J, Şenbabaoğlu Y, Santoro S, Sheinson D, Hung J, Giltnane JM, Pierce AA, Mesh K, Lianoglou S, Riegler J, Carano RAD, Eriksson P, Höglund M, Somarriba L, Halligan DL, van der Heijden MS, Loriot Y, Rosenberg JE, Fong L, Mellman I, Chen DS, Green M, Derleth C, Fine GD, Hegde PS, Bourgon R, Powles T. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544-548. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3113] [Cited by in RCA: 3655] [Article Influence: 522.1] [Reference Citation Analysis (1)] |
| 104. | Kassardjian A, Wang HL. SMAD4-Expressing Pancreatic Ductal Adenocarcinomas Have Better Response to Neoadjuvant Therapy and Significantly Lower Lymph Node Metastasis Rates. Pancreas. 2020;49:1153-1160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 105. | Xiao B, Zhang L, Liu H, Fang H, Wang C, Huang B, Liu X, Zhou X, Wang Y. Oncolytic Adenovirus CD55-Smad4 Suppresses Cell Proliferation, Metastasis, and Tumor Stemness in Colorectal Cancer by Regulating Wnt/β-Catenin Signaling Pathway. Biomedicines. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 106. | Itatani Y, Kawada K, Sakai Y. Transforming Growth Factor-β Signaling Pathway in Colorectal Cancer and Its Tumor Microenvironment. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 179] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 107. | Hudry E, Vandenberghe LH. Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality. Neuron. 2019;101:839-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 240] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 108. | Maeder ML, Gersbach CA. Genome-editing Technologies for Gene and Cell Therapy. Mol Ther. 2016;24:430-446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 507] [Cited by in RCA: 458] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 109. | Gowripalan A, Abbott CR, McKenzie C, Chan WS, Karupiah G, Levy L, Newsome TP. Cell-to-cell spread of vaccinia virus is promoted by TGF-β-independent Smad4 signalling. Cell Microbiol. 2020;22:e13206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |