Published online Aug 26, 2021. doi: 10.4252/wjsc.v13.i8.1134
Peer-review started: February 18, 2021
First decision: March 30, 2021
Revised: April 19, 2021
Accepted: July 5, 2021
Article in press: July 5, 2021
Published online: August 26, 2021
Processing time: 182 Days and 6.2 Hours
Fibromyalgia (FM) and myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) are devastating metabolic neuroimmune diseases that are difficult to diagnose because of the presence of numerous symptoms and a lack of specific biomarkers. Despite patient heterogeneity linked to patient subgroups and variation in disease severity, anomalies are found in the blood and plasma of these patients when compared with healthy control groups. The seeming specificity of these “plasma factors”, as recently reported by Ron Davis and his group at Stanford University, CA, United States, and observations by our group, have led to the proposal that induced pluripotent stem cells (iPSCs) may be used as metabolic sensors for FM and ME/CFS, a hypothesis that is the basis for this in-depth review.
To identify metabolic signatures in FM and/or ME/CFS supporting the existence of disease-associated plasma factors to be sensed by iPSCs.
A PRISMA (Preferred Reported Items for Systematic Reviews and Meta-analysis)-based systematic review of the literature was used to select original studies evaluating the metabolite profiles of FM and ME/CFS body fluids. The MeSH terms “metabolomic” or “metabolites” in combination with FM and ME/CFS disease terms were screened against the PubMed database. Only original studies applying omics technologies, published in English, were included. The data obtained were tabulated according to the disease and type of body fluid analyzed. Coincidences across studies were searched and P-values reported by the original studies were gathered to document significant differences found in the disease groups.
Eighteen previous studies show that some metabolites are commonly altered in ME/CFS and FM body fluids. In vitro cell-based assays have the potential to be developed as screening platforms, providing evidence for the existence of factors in patient body fluids capable of altering morphology, differentiation state and/or growth patterns. Moreover, they can be further developed using approaches aimed at blocking or reversing the effects of specific plasma/serum factors seen in patients. The documented high sensitivity and effective responses of iPSCs to environmental cues suggests that these pluripotent cells could form robust, reproducible reporter systems of metabolic diseases, including ME/CFS and FM. Furthermore, culturing iPSCs, or their mesenchymal stem cell counterparts, in patient-conditioned medium may provide valuable information to predict individual outcomes to stem-cell therapy in the context of precision medicine studies.
This opinion review explains our hypothesis that iPSCs could be developed as a screening platform to provide evidence of a metabolic imbalance in FM and ME/CFS.
Core Tip: Because of the special ability to sense environmental cues we propose that induced pluripotent stem cells (iPSCs) are suitable for use as a sensor system for metabolic disease. As fibromyalgia and myalgic encephalomyelitis/chronic fatigue syndrome body-fluids have unique metabolic profiles, the applicability of iPSC-based bioassays for those conditions are worth investigating. We envisage the development of iPSC platforms that allow differential diagnosis and disease-specific high-throughput drug-screening platforms. Using healthy iPSCs and patient body fluids has significant advantages over using cell lines, primary culture of patient cells or iPSCs derived from patients. A consistent iPSC control-cell line platform will provide a robust metabolic/phenotypic model allowing faster, cost-effective, large cohort screenings.
- Citation: Monzón-Nomdedeu MB, Morten KJ, Oltra E. Induced pluripotent stem cells as suitable sensors for fibromyalgia and myalgic encephalomyelitis/chronic fatigue syndrome. World J Stem Cells 2021; 13(8): 1134-1150
- URL: https://www.wjgnet.com/1948-0210/full/v13/i8/1134.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i8.1134
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a devastating disease of increasing prevalence, affecting millions all over the world, with a higher frequency in women[1,2]. It is a serious medical and economical problem in most countries. Because there are many symptoms that can vary significantly over time in an individual patient, diagnosis is challenging, and extensive delays can increase the burden on patients and their families. The lack of specific biomarkers for the disease[3] impacts not only on our ability to diagnose patients but also limits our capacity to ensure that just ME/CFS patients are recruited into research studies.
Fibromyalgia (FM) is a chronic disease that shares similar characteristics with ME/CFS. Classified as independent diseases, both affect millions of people, have a high prevalence in women, and a lack specific biomarkers, leading to diagnosis based on clinical symptoms and palliative treatments[4]. In FM, the key symptoms are fatigue and pain that often result in a high level of disability. The similarity between the symptoms of these two diseases is quite striking. A study investigating the overlap between ME/CFs and FM concluded that 30% to 70% of the patients diagnosed with FM also met the diagnosis criteria for ME/CFS, and vice versa[5], revealing substantial comorbid appearance in the affected population[6].
Over the last 10 years, researchers have identified a wide variety of potential biomarkers in different cohorts of patients diagnosed with ME/CFS and FM. Investigations have been primarily focused on microRNA profiles, DNA methylation patterns, metabolite differences and cytokine profiles. Unfortunately, no single study has consistently validated geographically distant cohorts, or shown the differences to be specific for ME/CFS of FM. Despite studies not reaching the sizes required to fully validate individual markers, the data provides a compelling case across all studies for the existence of molecular irregularities being present in patient blood samples. With many biomarker studies including only 10-20 patients. Given the associated heterogeneity/complexity of the disease more robust screening platforms are required to test the molecular anomalies.
Preliminary results from peripheral blood mononuclear cells (PBMCs) cultivated with plasma from ME/CFS patients or healthy controls showed differences in behavior of the PBMCs after culture[7], indicating that at least some of the anomalous components of the disease appear to be contained in plasma, either by the presence or by the absence of certain unknown factors. In support of the “ME/CFS-specific plasma factor” reported by Ron Davis and his team at Stanford University, CA, United States, Elisa Oltra´s team at the Catholic University of Valencia, Valencia, Spain, in collaboration with Karl Morten´s group at Oxford University, Oxford, United Kingdom, found that cellular oxygen consumption was altered upon addition of ME/CFS human plasma (unpublished, preliminary data). The impact of ME/CFS plasma on the respiration of the muscle cell line used as reporter in the assays mentioned above is believed to reflect the altered metabolic status of the patients.
Studies of the presence of metabolites associated with FM or ME/CFS, and additional central sensitivity syndromes, have been reviewed recently by Miller et al[4]. By reviewing only studies that used metabolomic approaches, and carrying out a systematic review, we examined the metabolic differences in detail, with a focus on ME/CFS and FM. In addition, we review the characteristics of iPSCs that endow them with the ability to be environmental sensors, and the successful in vitro use of conditioned medium (CM) cultures to evaluate information relevant to iPSC-based bioassay designs. All this provides evidence to support the premise that ME/CFS and FM, and possibly other diseases with a metabolic component, could be diagnosed, or at least initially triaged, using an assay that evaluates the effects of patient plasma on iPSC morphology, growth and/or differentiation capacity.
It is expected that the information gathered in this review will guide future empirical studies towards our hypothesis that iPSCs constitute an ideal cell sensor system for the assay of metabolic disease. In addition to the diagnostic value as a disease sensor system, the information obtained on the effects of plasma CM on stem cells will be useful in future individualized stem-cell therapy programs, as the health status of patients may determine treatment outcomes.
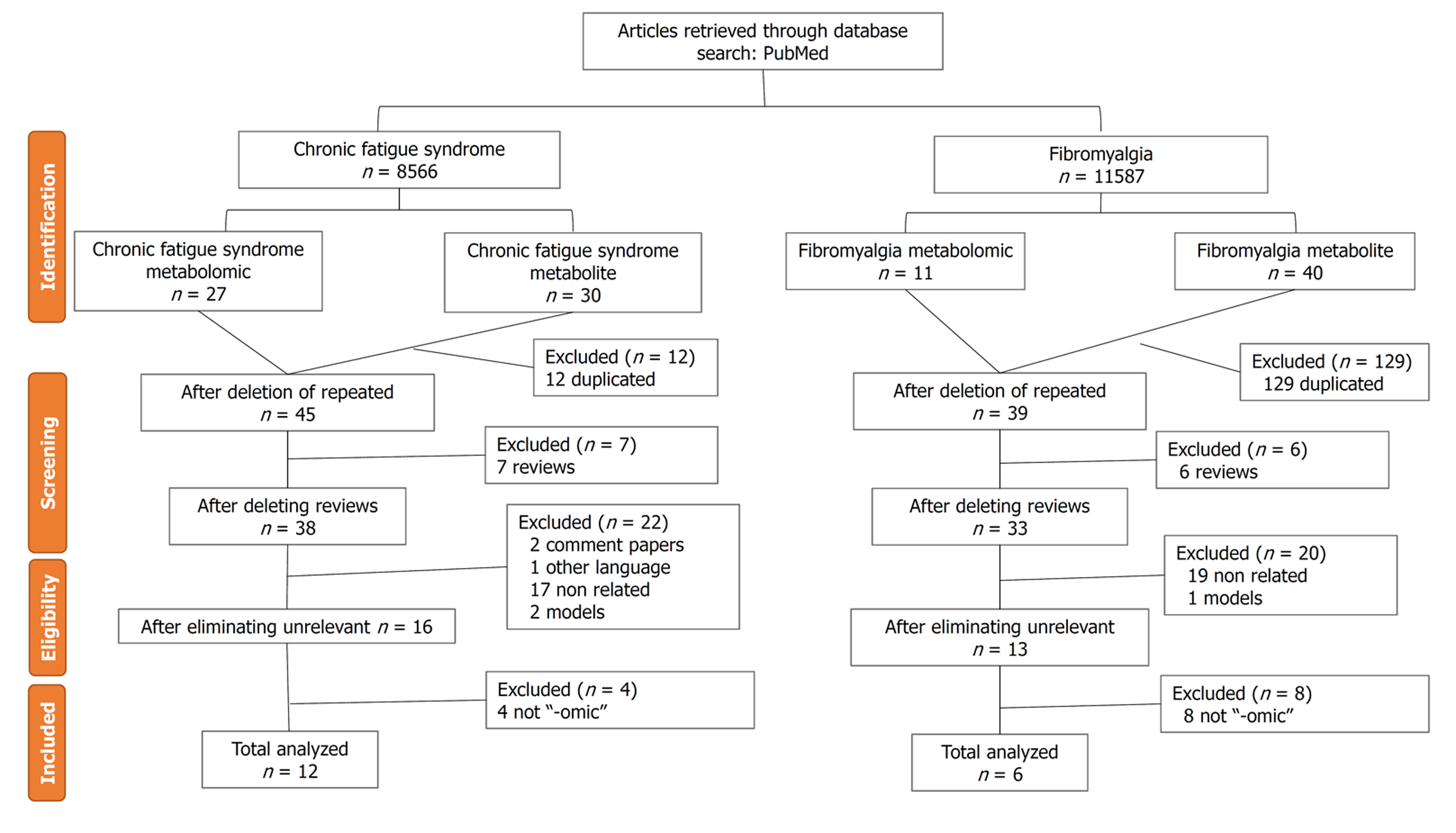
By applying a PRISMA (Preferred Reported Items for Systematic Reviews and Meta-analysis)[8] using the MeSH terms “metabolomic” or “metabolites” in combination with the disease term in the PubMed database, a total of 30 publications of studies of metabolic disorders in ME/CFS, and 40 of FM, were found (Figure 1). After removing duplicate publications and applying filters for reviews, languages other than English and Spanish, animal models, not “-omic” studies, or unrelated topics, a total of 12 original research articles for ME/CFS and six for FM were selected (Figure 1). The searches were performed between February and April 2020 by a single investigator. The protocol was not registered.
An examination of the literature found that the study results were reported in two different body fluid types, blood plasma and serum (Supplementary Table 1), and urine (Table 1). Other sample types like feces were not included in this analysis because they tended to be reported in studies of microbiome composition rather than altered metabolites. Supplementary Table 1 summarizes the results of nine different studies performed by six research groups that detected a wide variety of metabolites relative to controls in the plasma of ME/CFS patients. A total of 391 ME/CFS individuals, 529 healthy participants, and 97 depressed patients, were evaluated in the studies. ME/CFS patients were diagnosed using the Fukuda, CDC or Canadian criteria, except for the participants in the study published by Zhang et al[10] in 2019, which did not report the diagnosis criteria. The most frequent method of analysis was liquid chromatography/mass spectroscopy (LC/MS), used in 4/13 (31%) of the studies. Whenever possible, metabolites were classified as increased or decreased, and bolded to highlight higher potential significance for the disease when reported by more than one study. Registered metabolite coincidences include choline (P < 0.03)[9], (P = 0.017)[13], (P < 0.014), and (P = 0.008)[15]; ceramides, (P < 0.01)[9], (P = 0.1)[13], and (P = 0.03)[17]; carnitines (P = 0.057)[9], and (P = 0.017)[13]; glucose (P < 0.05)[10] and (P = 0.009)[15]; ATP (P = 0.002)[15] and (P = NA)[16]; and two amino acids: lysine and tyrosine (P < 0.05)[10] and (P < 0.05)[13].
| Ref. | Population studied/study design | Diagnosis criteria | Technique | Metabolites with increased levels | Metabolites with decreased levels | |||||
| Erasmus et al[19], 2019 | 31 high CFS (patients) (31 F, 0 M). 39 low CFS (control) (39 F, 0 M). Age range, median and mean age NA | NA | H-NMR | Salicylic acid | P = 0.06 | |||||
| Glucose | ||||||||||
| Acetaminophen | P = 0.08 | |||||||||
| Acetaminophen-glucuronide | ||||||||||
| Zhang et al[10], 2019 | 105 CFS (65 F, 40 M). 97 DD (56 F, 41 M). 190 HC (69 F, 121 M). Age range 20-49. Median and mean age NA | NA | H-NMR | M: Taurine | P < 0.01 | |||||
| M: Tyrosine | ||||||||||
| M: Leucine | ||||||||||
| M: Valine | ||||||||||
| M: Isoleucine | ||||||||||
| M: Aspartate | ||||||||||
| M: Ethylmalonate | ||||||||||
| M: Lactate | ||||||||||
| M: Threonine | ||||||||||
| M: Trans-Aconitate | ||||||||||
| M: 2-Aminohippurate | ||||||||||
| M: N-Phenylacetylglycine | ||||||||||
| M: N-Acetylaspartate | ||||||||||
| M: Thymol | ||||||||||
| B: Dimethylamine | ||||||||||
| B: Histidine | ||||||||||
| B: 4-Aminohippurate | ||||||||||
| B: Hippurate | ||||||||||
| B: Glutamine | ||||||||||
| B: Isocitrate | P > 0.05 | |||||||||
| B: 1-Methylnicotinamide | ||||||||||
| B: N-Acetylglutamine | ||||||||||
| B: Creatinine | ||||||||||
| F: Isobutyrate | ||||||||||
| F: Lysine | ||||||||||
| F: Alanine | ||||||||||
| F: Hypoxanthine | ||||||||||
| F: Fucose | ||||||||||
| F: N,N-dimethylglycine | ||||||||||
| F: Urea | ||||||||||
| F: Carnosine | ||||||||||
| F: 3-Hydroxymandelate | ||||||||||
| F: Indole-3-lactate | ||||||||||
| McGregor et al[20], 2019 | 11 ME/CFS (11F, 0 M) Mean age 30.9. 25 HC (24 F, 1 M). Mean age 33. 6 | Canadian criteria | H-NMR | Mannitol | P < 0.001 | Acetate | P < 0.001 | |||
| Urea | ||||||||||
| Methylhistidine | P < 0.01 | Pyruvate | ||||||||
| Chi et al[21], 2018 | 98 CFS (77 F, 21 M). Age range 21-69. Median age 43. 99 HC (76 F, 23 M). Age range 19-65 yr. Median age 39 yr. Mean age: NA | United States centers | LC-MS | AE: Methyladenosine | P < 0.05 | BE: Indoleacetaldehyde | P < 0.05 or P < 0.01 | |||
| BE: Phytosphingosine | ||||||||||
| BE: Pyroglutamic acid | ||||||||||
| BE: Creatinine | ||||||||||
| AE: Acetylcarnitine | ||||||||||
| AE Capric acid | ||||||||||
| AE: Corticosterone | ||||||||||
| AE: Creatinine | AE: Levonorgestrel | |||||||||
| AE: Pantothenic acid | ||||||||||
| AE: Pyroglutamic acid | ||||||||||
| AE: Xanthosine | ||||||||||
| AE: Xanthurenic acid | ||||||||||
| AE: Nonanedioic acid | ||||||||||
| Ruiz-Núñez et al[14], 2018 | 98 CFS (77 F, 21 M). Age range 21-69 yr. Median age 43 yr. 99 HC (76 F, 23 M). Age range 19-65 yr. Median age 39 yr. Mean age: NA | Fukuda criteria | LC-MS | NA | Nutritional factor | Urinary Iodine | P < 0.001 | |||
| McGregor et al[11], 1996 | 20 CFS (16 F, 4 M). Age range 17-58 yr. Mean age 39.4 yr. 45 HC (32 F, 13 M). Age range 12-74 yr. Mean age 37.1 yr. Median age NA | CDC criteria | GC-MS | Aminohydroxy-N-methylpyrrolidine CFSUM1 | P < 0.00003 | CFSUM2 | P < 0.0007 | |||
| Tyrosine | P < 0.02 | Alanine | P < 0.005 | |||||||
| β-alanine | ||||||||||
| Aconitic acid | P < 0.05 | Glutamic acid | P < 0.02 | |||||||
| Succinic acid | ||||||||||
It is interesting that the levels of some cholines, ATP and glucose were found repeatedly decreased in more than one study. A decrease in ATP fits with the constant lack of energy associated with ME/CFS, although possibly unspecific. Acyl cholines appeared as a relevant group of metabolites presenting with a clear reduction in the study by Germain et al[9], including palmitoylcholine, linoleoylcholine and arachidonoylcholine. Their decrease has been related to low blood pressure which is one of the many symptoms associating with ME/CFS[18]. By contrast, the amino acids lysine and tyrosine were found to be increased, and interestingly, carnitine and ceramides were detected in both, increased and decreased levels, depending on the study. Methodological differences including time between sample collection and processing, and whether patients were fasted or in the post-prandial state, might explain some of the current contradictions between studies. Alternatively, perhaps the differences point to subgroups of patients with different metabolic profiles.
Another body fluid commonly studied for the detection of disease-associated metabolites is urine. Only six studies performed by five independent research groups were found. Summary Table 1 displays main findings reported by the five studies. A total of 312 ME/CFS patients were examined in those investigations and compared with 367 healthy subjects. However, the study by the Erasmus et al[19] compared severely affected CFS patients with mild cases.
The diagnostic criteria applied towards patient recruitment was as previously mentioned with the exception of the study by Chi et al[21] that reported the diagnosis criteria from United States centers without any further specification. Another difference found between plasma and urine analysis was in the use of analytic techniques, with H-MRI being the most commonly used (in 3/6 or 50% of the studies, Table 1).
Following the same format as in Supplementary Table 1. Metabolites showing significant different levels were categorized as increased or decreased, and the significance was set at P < 0.1, as indicated. This time, creatinine (P < 0.05)[10] and (P < 0.05)[21]; alanine (P < 0.05)[10] and (P < 0.005)[11]; and urea (P < 0.05)[10] and (P < 0.001)[20] were dysbalanced in more than one independent study and are bolded in Table 1. Urea was the only metabolite that was persistently decreased, and the most significant, with a P value < 0.001. By contrast, creatinine and alanine were found to be both increased and decreased across different studies. Once more the contradictory results may suggest differences in methods or perhaps be indicative of patient subgroups.
The populations included in all six studies were consistently diagnosed with the American College of Rheumatology criteria[22]. The body fluids analyzed were plasma (two studies), serum (one study), whole blood (one study) and urine (two studies). A total of 192 FM patients and 189 healthy controls, seven osteoarthritis (OA) patients, and nine patients with rheumatoid arthritis (RA)[26] were evaluated in the six studies (Table 2). The objective of the last study was to find specific molecular differences across diseases presenting some overlapping symptoms with FM, such as OA and RA, and set up a tool to easily distinguish them in the clinic. Once more, metabolites showing significantly (P < 0.05) increased or decreased levels with respect to control groups are indicated, and bolded when reported by more than one study. As shown on Table 2, only three bolded metabolites stand out from the rest: phosphocholines (P < 0.007)[23], and (P = NA)[24], in particular phosphocholine 16:0, (P < 0.001); glutaric acid (P = NA)[25], and (P = 0.00011)[27]; and taurine (P < 0.001)[25], and (P = 0.0007)[28]. Furthermore, a different type of phosphocholine, lysoPCs, were increased in both plasma studies[23,24]. This class of metabolites is linked to the metabolism of lipids and was also altered in plasma from ME/CFS patients, as indicated by the uncompensated levels of ceramides (Supplementary Table 1). An increase in amino acid metabolism, as shown by significant changes in taurine in both plasma and urine samples from FM patients, was also reported. In contrast, glutaric acid levels associated with energy metabolism pathways were found to be both increased and decreased in the two different sample types.
| Ref. | Population studied/study design | Diagnosis criteria | Sample | Technique | Metabolite with increased levels | Metabolite with decreased levels | |||||||
| Menzies et al[23], 2020 | 20 FMS (20 F, 0 M). 20 HC (20 F, 0 M). Mean age in both groups 41.9 yr. Age range and medium age NA | ACR 1990 criteria | Plasma | LC-MS | Amino acid metabolism | Taurine | P < 0.001 | Energy metabolism | 2-hydroxyglutarate | P < 0.001 | |||
| Methionine sulfoxide | |||||||||||||
| Anthranilate | |||||||||||||
| Lipid metabolism | Lysophosphocholines (16:0) | ||||||||||||
| Lysophosphocholines (18:0) | |||||||||||||
| Lysophosphocholines (20:4) | P = 0.001 | Alpha-ketoglutaric acid | NA | ||||||||||
| Lysophosphocholines (22:6) | |||||||||||||
| Lysophosphocholines (18:1) | P = 0.003 | ||||||||||||
| Lysophosphocholines (16:1) | P = 0.007 | ||||||||||||
| Caboni et al[24], 2014 | 22 FMS (22 F, 0 M). Age range 27-72 yr. Mean age 52 yr. 21 HC (21 F, 0 M). Age range 27-67 yr. Mean age 50 yr. Median age NA | ACR criteria | Plasma | LC-Q-TOF/MS | 1-hexadecanoyl-sn-glycero-3-phosphocholine [PC (16:0)] | NA | NA | ||||||
| 1-tetradecanoyl-sn-glycero-3-phosphocholine [PC (14:0)] | |||||||||||||
| Clos-Garcia et al[25], 2019 | 105 FMS (73 F, 32 M). 54 HC (26 F, 28 M). Age range and mean and medium age NA | ACR 2010 criteria | Serum | UPLC-MS | Ornithine | NA | Platelet activating factor (PAF-16) | NA | |||||
| L-arginine | |||||||||||||
| Nε-Methyl-L-lysine | |||||||||||||
| L-glutamate | |||||||||||||
| Asymmetric dimethylarginine (ADMA) | |||||||||||||
| l-threonine/DL-homoserin | |||||||||||||
| L-glutamine | |||||||||||||
| Hackshaw et al[26], 2013 | 10 FMS (9 F, 1 M). Mean age 50 yr. 10 OA (7 F, 3 M). Mean age 65 yr. 10 RA (9 F, 1 M). Mean age 60 yr. Age range and median age NA | ARC criteria | Blood | UHPLC/MS/MS | Trans-urocanate | NA | NA | NA | |||||
| Asparagine | |||||||||||||
| Heme | |||||||||||||
| Cysteine–glutathione disulfide | |||||||||||||
| Glucose | |||||||||||||
| Pyruvate | |||||||||||||
| Nicotinamide adenine dinucleotide (NAD+) | |||||||||||||
| Malatji et al[27], 2019 | 17 FMS (17 F, 0 M). 62 HC (62 F, 0 M), 11 CF, 10 CO, 41 CN (age range 18-22 yr). Median and mean age: NA | ACR 1990 criteria | Urine | GC-MS | 3-D-Ribohexonic acid | P = 0.0000089 | |||||||
| Sorbose | P < 0.00001 | ||||||||||||
| Threonic acid | |||||||||||||
| Erythropentonic acid | |||||||||||||
| 2-Keto-1-gluconic acid | |||||||||||||
| 2-hydroxyglutaric acid | P = 0.000013 | ||||||||||||
| Arabinose | P = 0.000017 | ||||||||||||
| 2-D-3,5-DHPL | P = 0.000019 | ||||||||||||
| 2,3,4-Trihydroxybutyl-L | P = 0.000021 | ||||||||||||
| Phosphoric acid | P = 0.00011 | ||||||||||||
| Glutaric acid | |||||||||||||
| Oxalic acid | |||||||||||||
| Rhamnose | |||||||||||||
| Tagatose | P = 0.00024 | ||||||||||||
| 4-HBA | P = 0.00109 | ||||||||||||
| Malic acid | P = 0.024 | ||||||||||||
| Malatji et al[28], 2017 | 18 FMS (18 F, 0 M). 41 HC (41 F, 0 M), 11 FR, 10 AM, 20 YHC (age range 18-22 yr). Median and mean age NA | ARC 1990 criteria | Urine | QE-MS | Gut microbiome | Succinic acid | P = 0.0001 | NA | |||||
| Lactic acid | P = 0.0044 | ||||||||||||
| Hippuric | P = 0.09 | ||||||||||||
| Osmolytes | Taurine | P = 0.0007 | |||||||||||
| TMAO | P = 0.006 | ||||||||||||
CM, also known as the cell secretome, corresponds to the media harvested from cultured cells, where the cells are excluded or removed. It is composed of all classes of soluble mediator molecules secreted by the cell, often originating from the endoplasmic reticulum, the Golgi apparatus, or from less common vesicle secretory pathways, including exosome release. The excreted molecules include small metabolites, growth factors, hormones, cytokines, and other proteins, in addition to molecules of other types. All are essential components of different biological processes including cell growth[29]. It is known that cell proliferation is dependent on CM composition, mainly in response to the levels of nutrients and growth factors that are present. By analogy, blood plasma and other body fluids can be considered the individual’s CM, containing all secreted molecules from the different cell types and tissues in the body. In the last few years, researchers have used CM to address several medical and health associated questions. Because of the vast array of procedures used on a wide range of targeted cultured cells with different experimental questions, a few examples were selected to illustrate the reference framework of this opinion review. Studies that evaluated adult stem cells, primary cells isolated from patients, and the more commonly used cultured cell lines as distinct sources of CM (Table 3), highlight the medical relevance of these fluids (Table 3).
| Ref. | Disease/condition | Conditioned medium/culture supernatant | Target cultured cells | Results/effects |
| Raimondo et al[30], 2019 | Multiple myeloma (MM) | Exosomes from MM.1S cell line. Exosomes from bone marrow | hTERT-MSC (human telomerase reverse transcriptase stromal cell line) | Osteoclastogenesis (Osteoclast formation). Blockade of osteogenic differentiation of MSC with increased IL-8 expression |
| Bougaret et al[31], 2017 | Obesity-associated increased risk of metastasis in breast cancer post-menopausal patients | Supernatant of MCF-7 cells cocultured with mature adipocytes | Human umbilical vein endothelial cells (HUVECs) | Increased HUVEC cell proliferation, endothelial tube formation, migration, IL-6 levels and decrease in Leptin and VEGFR HUVEC receptors |
| Bai et al[32], 2016 | Trichinella spiralis skeletal muscle infection | Supernatant of murine macrophage cell line J774A.1 infected with ML-ESP | Murine myoblast cell line C2C12 | Avoid formation of myotubes by decreased MyoD, myogenin and MyHC protein expression affecting the differentiation of myoblasts. Suppresses the cell cycle influencing p21 and D1 protein expression |
| Kozakowska et al[33], 2015 | Diabetes-associated angiogenesis deficiency | Supernatant of C2C12 myoblasts overexpressing HO-1 or a luciferase gene (control) | Intramuscular injection in mice | Improved skeletal muscle regeneration after ischemia by increased neovascularization with lower number of granulocytes and higher proportion of lymphocytes |
| Wobus et al[34], 2015 | Breast cancer | Supernatant of MFC-7, MDA-MB231 breast carcinomas with respect to MCF-10 (control) | Primary Mesenchymal Stromal Cells (MSC) and MSC cell line SCP-1 | Modulate genes belonging to proliferation and differentiation. Downregulation of chemokine receptor CXCL12 expression |
| Cabbage et al[35], 2014 | Atherosclerosis | Supernatant of the macrophage cell line (RAW 264.7) inoculated with Chlamydia pneumoniae | BSMC (bovine aortic smooth muscle cells) and perivascular Sca-1+, CD31-, CD45- cells from apoE-/- mouse aortas | Accelerate the conversion of smooth muscle cells to calcifying cells. Increase the production of collagen II causing osteochondrocytic phenotypes |
| Fasslrinner et al[36], 2012 | Graft vs host disease/Inflammation | MLR supernatant (Mixed Lymphocyte Reaction) from PBMCs | Mesenchymal stromal cells (MSC) from the bone marrow | Decrease in the population doubling compared to control. Increased IL-8, IL-6 and Ang-1 levels. Induced osteogenic differentiation and suppress adipogenic differentiation |
| Zhao et al[37], 2007 | Amyotrophic lateral sclerosis (ALS) | SOD1-G93A ALS mice CNS extracts | hMSC (human mesenchymal stem cell) | Morphological change from fibroblast to neuron phenotype without detectable apoptosis and death. Delay in proliferation of hMSC due to increased differentiation rate |
| Vaisman et al[38], 1996 | Granulopoiesis in Anorexia Nervosa | Plasma and supernatant of cultured PBMCs | Bone marrow GM-CSF (granulocyte macrophage colony forming cells) | Decreased numbers of GM-CFS |
| Roodman et al[39], 1992 | Paget’s disease | Supernatant of cultured Paget’s nonadherent bone marrow mononuclear cells | Bone marrow cells from healthy donors | Increased MNC (multinucleated cell) formation and IL-6 levels |
To the best of our knowledge, the first report of the use of CM from patients was almost 30 years ago[39]. In that pioneering study, the authors tried to understand more about the pathogenicity of Paget disease, a health problem that increases the formation of osteoclasts directly affecting the generation and regeneration of bones. The research focused on obtaining the supernatant from nonadherent mononuclear cells from the bone marrow of patients suffering from the disease. Once the CM was obtained, it was applied to selected target cells (bone marrow cells from healthy participants). The results indicated an increase in monoclonal cell formation that was linked to an increased release of interleukin 6 (IL-6), which is an important regulator of bone mass and is linked to the processes of aging and inflammation[40].
In contrast, among the most recent publications in this area of research, we found a study by Raimondo et al[30] on multiple myeloma (MM), a terrible disease that also affects the formation of bones. Here, the CM corresponded to extracellular vesicles, named exosomes, isolated from bone marrow plasma samples of MM patients, and the target cells were a type of telomerase mesenchymal stromal cell (hTERT-MSC). The effect of culturing both together was on the formation of osteoclasts. Specifically, MM-exosomes induced osteoclastogenesis and increased the levels of expression of IL-8, whose function is linked to bone destruction in this disease. These two examples show that factors contained in the CM or body fluids contain properties that affect the growth and differentiation of target cells (i.e. the presumed reporter cell system in the assay).
The CM bibliography in Table 3 includes a wide variety of sample types. In many studies, the CM constituents were present in the cell culture supernatants of a wide variety of samples, including cancer cell lines[31,34]; primary cultures, such as PBMCs[36,38]; and macrophages infected with parasites[32] or bacteria[35]; or myoblasts[33]. One study also included purified exosomes in the CM. The purified fraction mediated cell-to-cell signaling assumed to promote activity or reflect the cell´s physiological state[30]. In addition to the CM the type of target reporter cell that is used is also an important element of the reporter bioassay. Some studies show different types of MSCs acting as target cells (Table 3)[30,34,36,37]. Cultures of this type of adult stem cell are highly relevant to this review, as MSCs share some similarities with iPSCs and can also be derived from them[41].
MSC differentiation to adipocytes, chondrocytes, or osteocytes, is altered or blocked when the immune system is impaired or activated. An example of this was demonstrated by Fasslrinner et al[36]. The research focused on evaluating the inflammatory response caused by graft vs host disease. In that study, mixed lymphocyte reaction (MLR) supernatants were extracted from a collection of different PBMCs donors and cultured with MSCs obtained from bone marrow collected from healthy donors. The results showed suppression of the potential of MSCs to differentiate to adipose tissue; and in exchange, potentiation of osteogenic differentiation by exposure to patient MLR supernatants.
In murine myoblasts, another type of target cell, the supernatant collected after culturing murine macrophages with products released by Trichinella spiralis larvae affected myotube-formation proteins, inducing inhibition of differentiation[32].
Apart from changes in lineage differentiation, growth rates of stem cells are also affected by modifications in the media composition. Supernatants of cultured anorexia nervosa PBMCs decreased granulopoiesis[38] in bone marrow granulocyte macrophage-colony forming cells, which validated the existence of a “factor or factors” in the supernatant of patient blood cells that affected the growth of cultured cells. Interestingly, inflammatory diseases have been evaluated in MSCs[34,36,37], a cell type that has anti-inflammatory properties. To conclude, it can be deduced from Table 3, that a wide variety of diseases have been studied by CM approaches.
iPSCs are known to be particularly sensitive to environmental cues, and thus may allow us to model what might be happening in vivo when incubated in the presence of abnormal metabolites from patients in the growing medium. The aim of a recent study by Rodimova et al[42] was to evaluate the metabolic status of iPSCs during dermal and epidermal differentiation by optical metabolic imaging. When iPSCs differentiated into dermal fibroblasts and keratinocyte progenitor cells, a metabolic shift towards a more oxidative state, as indicated by FAD/NAD(P)H redox ratios was observed. While the biosynthetic processes occurring in dermal fibroblasts lowered the intracellular pH, no intracellular pH shift was observed in keratinocyte progenitor cells. The pH levels in dermal fibroblasts were consistent with an OXPHOS metabolic pathway switch.
The differences in energy metabolism in undifferentiated and differentiated cells are well known. Undifferentiated cells need larger amounts of energy in a shorter period of time when they are actively proliferating. Although the OXPHOS pathway can produce more molecules of ATP in the final output, the rapid production of ATP by glycolysis seems to be a crucial feature for undifferentiated cells. Energy-producing pathways may also be affected differently in plasma-containing cultures. Interestingly, the plasticity of iPSCs is attributed to the presence of a specific mitochondrial phenotype that reflects the somatic cell type from which the stem cells were derived. Some iPSC cell lines may thus perform better than other when used as sensors of environmental cues.
When metabolism is altered, epigenetic changes may occur that may directly affect its physiology[43]. Glucose-derived metabolites like acetyl-CoA, NAD+, S-adenosyl methionine (SAM), L-proline, alpha-ketoglutarate (αKG), and fatty acids, have been found to be associated with regulation of the epigenome and stem cell pluripotency[44]. Because of the reprogrammed open status of iPSC chromatin[45], iPSCs should be especially sensitive to the nutritional environment.
There are two types of pluripotent states: naïve and primed (i.e. closer to the differentiated phenotype commitment). They differ in the energy metabolic pathway used to produce ATP. In the case of naïve pluripotent stem cells, glycolysis and OXPHOS can both be used as energy metabolic pathways. By contrast, primed pluripotent stem cells obtain energy only from glycolysis even if oxygen is present in the environment[46]. It seems that factors inhibiting the OXPHOS pathway would be of use in the transition from naïve to primed pluripotency. For example, methionine withdrawal, which promotes the OXPHOS pathway, inhibits glycolysis and triggers differentiation towards a fibroblast phenotype.
An additional cue to be sensed by iPSCs includes the glucose uptake process. Increased glucose uptake in combination with glycolysis activation, enhances iPSC colony formation[47], leading to macroscopic, easily observable morphological changes, and self-renewal rates. It has been reported that hyperosmotic stress has an impact on cell shape caused by the AQP1 cytoskeletal remodeling protein[48]. AQP1 water channels in membranes are increased by hyperosmolarity. In stem cells, various cell shapes can affect downstream differentiation options. To examine the effects of a high glucose environment and the effects of hyperosmotic stress on iPSCs, a group of investigators[49] treated cells with either high glucose or high mannitol, as a hyperosmolar agent and a metabolic control. Both compounds increased the number of cellular AQP1 channels, but only high glucose increased pluripotency markers. Knowing the signals that regulate the osmosis pathway allows investigators to direct iPSC fate options, indicating once more their suitability as sensors of the metabolites present in plasma. The effects of changes in oxygen level were also studied by the authors. Hypoxic environments, in which the oxygen requirement exceeds the available supply, increase reprogramming efficiency and help cells maintain their pluripotent state[49]. That is an important observation, and supports iPSCs as a potential sensor for the diagnosis of ME/CFS, because, as previously mentioned, preliminary results obtained by Oltra and Morten showing that the addition of ME/CFS plasma to cell cultures increased oxygen consumption.
Additional environmental factors that affect iPSC cultures are variables to be controlled in the reporter system set, and include cell density associated with acidification of the culture medium[50]; cell-cell contact interactions, the extracellular matrix; physical forces, and the physiochemical environment[51]. It is important to keep in mind that selection of the appropriate growth conditions will influence the robustness and quality of the sensor assay based on iPSCs. The conditions may vary according to the needs, types of metabolites, or soluble factors to be sensed.
Choline is an essential nutrient in humans and is a functional component of every cell membrane[52]. Previous studies have shown that many compounds containing choline are present in ME/CFS and FM patients at levels that vary from those in healthy subjects. As shown on Supplementary Table 1, choline levels are decreased in all ME/CFS studies so far performed. A decrease in choline levels often correlates with disorders of the liver, lymphocytes, and skeletal muscle. Acyl cholines, can be long-chain or short-chain compounds, and both are active in the regulation of blood pressure. Interestingly, only long-chain acyl cholines are related to acetylcholine, a crucial neurotransmitter present in both the central and peripheral nervous systems. Germain et al[9] reported that acyl cholines negatively regulate the expression of acetylcholine, which may explain brain fog and memory loss symptoms in ME/CFS patients. Interestingly, decreased levels of acetylcholine, which is a major neurotransmitter in the brain, may explain the occurrence of patient cognitive problems.
Increased choline levels were found in different parts of the brain in ME/CFS patients compared with controls[53,54]. The finding is not shown in any of the summary tables included in this review because it was reported in only one metabolite analysis and not as an -omics finding. Nevertheless, low-scope studies[55] and/or studies in tissues may help interpret the results of wider scope studies done on body fluids. The only increased choline metabolite reported in -omics studies was phosphocholine in the plasma of FM patients (Table 2). The increased level of a metabolite in a body fluid does not necessarily mean that it was also increased in all tissues or in other body fluids.
Ceramides are related to lipid formation, specifically sphingolipids. Differences have so far been found in only the plasma of ME/CFS patients (Supplementary Table 1). Ceramides are needed for the formation of extracellular vesicles and exosomes[56], which could explain the increased numbers reported by Oltra et al[57] in the plasma of severely affected ME/CFS patients. Variable levels of ceramides have also been found in a number of other pathologies[58], consequently neither ceramides nor compounds containing choline, are considered specific to ME/CFS. Alterations could be a consequence of disease rather than a cause. Nevertheless, the proposal here is to provide a system capable of sensing complex cues, not variations of single metabolites.
Metabolomic screening has limitations in identifying compound variants or unusual molecules in the analyzed fractions because of a lack of reference compounds for comparison-based identification. The vast majority of metabolites isolated by metabolomics are not identified. That is probably why specific ME/CFS or FM biomarker metabolites have not yet been found. Study heterogeneity resulting from the methods and/or the diagnostic criteria used for patient selection constitute additional limitations for meta-analysis.
No single study included in our analysis confirmed our initial hypothesis that iPSCs are the ideal target cells to use as sensor reporters of CM components in ME/CFS and FM plasma. However, the absence of studies supports the novelty of the proposed idea, and the possibility of previous reports evaluating the impact of CM on iPSCs growth and differentiation supportive of our hypothesis. Table 3 includes a series of studies using different types of mesenchymal stem or stromal cells as targets of CM from other sources, leading to changes in their differentiation potential and growth capability. As MSCs and iPSCs share some features, including the capacity to self-renew and the potential to differentiate into other tissue lineages[59], it is assumed that the results obtained with MSCs can be extended to iPSCs. It is also hypothesized that the increased plasticity of iPSCs leads to improved sensitivity and cell homogeneity, leading to more reproducible batch-to-batch results.
Data from several review articles reported that pluripotent stem cells were sensitive to a wide variety of stimuli, metabolic pathways for energy production[47], intracellular pH[60], hyperosmotic stress[48], the presence or absence of oxygen[49], cell density[50], and the composition of the medium[61], all of which contributing to outcomes of iPSC cultures. We failed to find studies of iPSC sensitivity to environmental factors compared with other stem cell types or cultured cells, but the data obtained here supports our proposal to a certain extent. Moreover, it should be of interest to invest future research efforts to provide empirical data for the currently undocumented comparisons. The fact that iPSCs have also been used to test toxicity, mutagenicity, teratogenicity, and carcinogenic potential effects of environmental factors[62], further supports the adequacy of iPSCs, as a sensor system to assay soluble factors present in the plasma of patients with metabolic dysfunction, as proposed in this review.
An important limitation of the proposed bioassay comes from its inherent low specificity, as different metabolites may induce similar gross phenotypic changes in iPSCs. Nevertheless, the method should at least serve as an initial selective process for triage and to screen healthy individuals. The bioassay should increase the success of transcriptomic and proteomic CM-treated iPSCs to uncover pathway modulation by disease-specific factors. The additional relevance of this proposal being the assessment of the impact of the disease on patient endogenous stem cells. Furthermore, the system proposed here might help to estimate the potential response to stem-cell therapy of patients according to their metabolic status, perhaps leading to more robust outcomes of stem-cell therapy in future clinical trials.
Evaluation of additional subcellular changes including mitochondrial function in iPSCs after CM addition, and the mitochondrial characteristics in the CM cell sensors are additional aspects that deserve investigation in future research studies. Similarly, evaluation of the endocytic pathway, exosome release, and paracrine factors are additional aspects of sensor cells that can inform on the pathophysiology under study, helping identify early, specific disease biomarkers and predict individual responses to stem-cell therapy.
To the best of our knowledge, this is the first comprehensive systematic review of ME/CFS and FM metabolic profiles. It is an update of the metabolic differences reported by more than one independent study, and the discrepancies that exist may reflect patient heterogeneity in these two overlapping diseases. Possible associations between dysregulated metabolites and disease symptoms were also found.
The CM bibliography reveals the prevalence of studies that investigated the inflammatory components of disease and a preference for the use of adult stem cells as a target reporter system, perhaps because of the attributed immune-regulatory properties of MSCs. It also highlighted a variety of CM ranging from cell culture supernatants to human body fluids either as complex mixtures or as functional purified fractions such as exosomes.
Finally, even though we did not find any articles reporting the use of iPSCs as a disease sensor, which might reflect the novelty of our hypothesis, we managed to gather information supportive of the high sensitivity and environmental response of iPSCs. The environmental cues included several physical and chemical factors and soluble mediators, which are seen as evidence of exceptional sensor capacity for future assay of the ME/CFS and FM disease-associated “plasma factors” and essentially the main overall goal pursued here.
Fibromyalgia (FM) and myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) are multifactorial immuno-metabolic disorders lacking biomarker-based diagnostic methods. Comorbidity is frequent, and the prevalence is increased in women, affecting as much as 5% of the population globally. Available clinical treatments are symptom-palliative only.
A diagnostic bioassay of FM and ME/CFS would reduce the time to diagnosis, clinical costs, and permit the development of effective, curative, treatments. Methods capable of detecting disease-associated “plasma factors” could serve this purpose even if the nature of the detected factors remain unknown.
Identification of metabolic imbalances associated with FM and ME/CFS provides the background needed to develop a cell-based diagnostic bioassay for FM and ME/CFS.
The methods included a PRISMA (Preferred Reported Items for Systematic Reviews and Meta-analysis)-based systematic review of the literature analyzing FM and ME/CFS metabolic profiles, and the technical evidence supporting induced pluripotent stem cells (iPSCs) as sensors of environmental imbalance.
More than one study found statistically significant changes (P < 0.05) in body-fluid metabolites, particularly cholines, ceramides, and some amino acids in FM and ME/CFS patients. Environmental cues can affect stem cell phenotype.
FM and ME/CFS metabolite profiles support metabolic imbalance. The lack of previous research exploring the hypothesis raised confirms the novelty of our proposal.
Empirical testing of the influence of FM and ME/CFS “plasma factors” on iPSCs growth and behavior is warranted.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tomizawa M S-Editor: Gao CC L-Editor: Filipodia P-Editor: Zhang YL
| 1. | Reyes M, Nisenbaum R, Hoaglin DC, Unger ER, Emmons C, Randall B, Stewart JA, Abbey S, Jones JF, Gantz N, Minden S, Reeves WC. Prevalence and incidence of chronic fatigue syndrome in Wichita, Kansas. Arch Intern Med. 2003;163:1530-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 296] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 2. | Estévez-López F, Mudie K, Wang-Steverding X, Bakken IJ, Ivanovs A, Castro-Marrero J, Nacul L, Alegre J, Zalewski P, Słomko J, Strand EB, Pheby D, Shikova E, Lorusso L, Capelli E, Sekulic S, Scheibenbogen C, Sepúlveda N, Murovska M, Lacerda E. Systematic Review of the Epidemiological Burden of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Across Europe: Current Evidence and EUROMENE Research Recommendations for Epidemiology. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 3. | Missailidis D, Annesley SJ, Fisher PR. Pathological Mechanisms Underlying Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Diagnostics (Basel). 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 4. | Miller JS, Rodriguez-Saona L, Hackshaw KV. Metabolomics in Central Sensitivity Syndromes. Metabolites. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 5. | Aaron LA, Burke MM, Buchwald D. Overlapping conditions among patients with chronic fatigue syndrome, fibromyalgia, and temporomandibular disorder. Arch Intern Med. 2000;160:221-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 393] [Cited by in RCA: 388] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 6. | Blomberg J, Gottfries CG, Elfaitouri A, Rizwan M, Rosén A. Infection Elicited Autoimmunity and Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: An Explanatory Model. Front Immunol. 2018;9:229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 7. | Esfandyarpour R, Kashi A, Nemat-Gorgani M, Wilhelmy J, Davis RW. A nanoelectronics-blood-based diagnostic biomarker for myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS). Proc Natl Acad Sci U S A. 2019;116:10250-10257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 8. | Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009;6:e1000100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11206] [Cited by in RCA: 11041] [Article Influence: 690.1] [Reference Citation Analysis (0)] |
| 9. | Germain A, Barupal DK, Levine SM, Hanson MR. Comprehensive Circulatory Metabolomics in ME/CFS Reveals Disrupted Metabolism of Acyl Lipids and Steroids. Metabolites. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 10. | Zhang F, Wu C, Jia C, Gao K, Wang J, Zhao H, Wang W, Chen J. Artificial intelligence based discovery of the association between depression and chronic fatigue syndrome. J Affect Disord. 2019;250:380-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | McGregor NR, Dunstan RH, Zerbes M, Butt HL, Roberts TK, Klineberg IJ. Preliminary determination of the association between symptom expression and urinary metabolites in subjects with chronic fatigue syndrome. Biochem Mol Med. 1996;58:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Germain A, Ruppert D, Levine SM, Hanson MR. Prospective Biomarkers from Plasma Metabolomics of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Implicate Redox Imbalance in Disease Symptomatology. Metabolites. 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 13. | Nagy-Szakal D, Barupal DK, Lee B, Che X, Williams BL, Kahn EJR, Ukaigwe JE, Bateman L, Klimas NG, Komaroff AL, Levine S, Montoya JG, Peterson DL, Levin B, Hornig M, Fiehn O, Lipkin WI. Insights into myalgic encephalomyelitis/chronic fatigue syndrome phenotypes through comprehensive metabolomics. Sci Rep. 2018;8:10056. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 14. | Ruiz-Núñez B, Tarasse R, Vogelaar EF, Janneke Dijck-Brouwer DA, Muskiet FAJ. Higher Prevalence of "Low T3 Syndrome" in Patients With Chronic Fatigue Syndrome: A Case-Control Study. Front Endocrinol (Lausanne). 2018;9:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 15. | Germain A, Ruppert D, Levine SM, Hanson MR. Metabolic profiling of a myalgic encephalomyelitis/chronic fatigue syndrome discovery cohort reveals disturbances in fatty acid and lipid metabolism. Mol Biosyst. 2017;13:371-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 110] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 16. | Yamano E, Sugimoto M, Hirayama A, Kume S, Yamato M, Jin G, Tajima S, Goda N, Iwai K, Fukuda S, Yamaguti K, Kuratsune H, Soga T, Watanabe Y, Kataoka Y. Index markers of chronic fatigue syndrome with dysfunction of TCA and urea cycles. Sci Rep. 2016;6:34990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 17. | Naviaux RK, Naviaux JC, Li K, Bright AT, Alaynick WA, Wang L, Baxter A, Nathan N, Anderson W, Gordon E. Metabolic features of chronic fatigue syndrome. Proc Natl Acad Sci USA. 2016;113:E5472-E5480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 254] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 18. | Committee on the Diagnostic Criteria for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome; Board on the Health of Select Populations; Institute of Medicine. Beyond Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Redefining an Illness. Washington (DC): National Academies Press (US); 2015. [PubMed] [DOI] [Full Text] |
| 19. | Erasmus E, Mason S, van Reenen M, Steffens FE, Vorster BC, Reinecke CJ. A laboratory approach for characterizing chronic fatigue: what does metabolomics tell us? Metabolomics. 2019;15:158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | McGregor NR, Armstrong CW, Lewis DP, Gooley PR. Post-Exertional Malaise Is Associated with Hypermetabolism, Hypoacetylation and Purine Metabolism Deregulation in ME/CFS Cases. Diagnostics (Basel). 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 21. | Chi AP, Wang ZN, Shi B, Yang XF, Min RX, Song J. [Comparison of differential metabolites in urine of the middle school students with chronic fatigue syndrome before and after exercise]. Zhongguo Yingyong Shengli Xue Zazhi. 2018;34:340-344 349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 22. | Wolfe F, Smythe HA, Yunus MB, Bennett RM, Bombardier C, Goldenberg DL, Tugwell P, Campbell SM, Abeles M, Clark P. The American College of Rheumatology 1990 Criteria for the Classification of Fibromyalgia. Report of the Multicenter Criteria Committee. Arthritis Rheum. 1990;33:160-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5950] [Cited by in RCA: 5745] [Article Influence: 164.1] [Reference Citation Analysis (0)] |
| 23. | Menzies V, Starkweather A, Yao Y, Thacker LR 2nd, Garrett TJ, Swift-Scanlan T, Kelly DL, Patel P, Lyon DE. Metabolomic Differentials in Women With and Without Fibromyalgia. Clin Transl Sci. 2020;13:67-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 24. | Caboni P, Liori B, Kumar A, Santoru ML, Asthana S, Pieroni E, Fais A, Era B, Cacace E, Ruggiero V, Atzori L. Metabolomics analysis and modeling suggest a lysophosphocholines-PAF receptor interaction in fibromyalgia. PLoS One. 2014;9:e107626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 25. | Clos-Garcia M, Andrés-Marin N, Fernández-Eulate G, Abecia L, Lavín JL, van Liempd S, Cabrera D, Royo F, Valero A, Errazquin N, Vega MCG, Govillard L, Tackett MR, Tejada G, Gónzalez E, Anguita J, Bujanda L, Orcasitas AMC, Aransay AM, Maíz O, López de Munain A, Falcón-Pérez JM. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine. 2019;46:499-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 147] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 26. | Hackshaw KV, Rodriguez-Saona L, Plans M, Bell LN, Buffington CA. A bloodspot-based diagnostic test for fibromyalgia syndrome and related disorders. Analyst. 2013;138:4453-4462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | Malatji BG, Mason S, Mienie LJ, Wevers RA, Meyer H, van Reenen M, Reinecke CJ. The GC-MS metabolomics signature in patients with fibromyalgia syndrome directs to dysbiosis as an aspect contributing factor of FMS pathophysiology. Metabolomics. 2019;15:54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 28. | Malatji BG, Meyer H, Mason S, Engelke UFH, Wevers RA, van Reenen M, Reinecke CJ. A diagnostic biomarker profile for fibromyalgia syndrome based on an NMR metabolomics study of selected patients and controls. BMC Neurol. 2017;17:88. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 29. | Dowling P, Clynes M. Conditioned media from cell lines: a complementary model to clinical specimens for the discovery of disease-specific biomarkers. Proteomics. 2011;11:794-804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 30. | Raimondo S, Saieva L, Vicario E, Pucci M, Toscani D, Manno M, Raccosta S, Giuliani N, Alessandro R. Multiple myeloma-derived exosomes are enriched of amphiregulin (AREG) and activate the epidermal growth factor pathway in the bone microenvironment leading to osteoclastogenesis. J Hematol Oncol. 2019;12:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 31. | Bougaret L, Delort L, Billard H, Lequeux C, Goncalves-Mendes N, Mojallal A, Damour O, Vasson MP, Caldefie-Chezet F. Supernatants of Adipocytes From Obese Versus Normal Weight Women and Breast Cancer Cells: In Vitro Impact on Angiogenesis. J Cell Physiol. 2017;232:1808-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 32. | Bai X, Wang XL, Tang B, Shi HN, Boireau P, Rosenthal B, Wu XP, Liu MY, Liu XL. The roles of supernatant of macrophage treated by excretory-secretory products from muscle larvae of Trichinella spiralis on the differentiation of C2C12 myoblasts. Vet Parasitol. 2016;231:83-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 33. | Kozakowska M, Kotlinowski J, Grochot-Przeczek A, Ciesla M, Pilecki B, Derlacz R, Dulak J, Jozkowicz A. Myoblast-conditioned media improve regeneration and revascularization of ischemic muscles in diabetic mice. Stem Cell Res Ther. 2015;6:61. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 34. | Wobus M, List C, Dittrich T, Dhawan A, Duryagina R, Arabanian LS, Kast K, Wimberger P, Stiehler M, Hofbauer LC, Jakob F, Ehninger G, Anastassiadis K, Bornhäuser M. Breast carcinoma cells modulate the chemoattractive activity of human bone marrow-derived mesenchymal stromal cells by interfering with CXCL12. Int J Cancer. 2015;136:44-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Cabbage S, Ieronimakis N, Preusch M, Lee A, Ricks J, Janebodin K, Hays A, Wijelath ES, Reyes M, Campbell LA, Rosenfeld ME. Chlamydia pneumoniae infection of lungs and macrophages indirectly stimulates the phenotypic conversion of smooth muscle cells and mesenchymal stem cells: potential roles in vascular calcification and fibrosis. Pathog Dis. 2014;72:61-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 36. | Fasslrinner F, Wobus M, Duryagina R, Müller K, Stopp S, Wehner R, Rauner M, Hofbauer LC, Schmitz M, Bornhäuser M. Differential effects of mixed lymphocyte reaction supernatant on human mesenchymal stromal cells. Exp Hematol. 2012;40:934-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Zhao CP, Zhang C, Wang YH, Zhou SN, Zhou C, Li WY, Yu MJ. Signals in pathological CNS extracts of ALS mice promote hMSCs neurogenic differentiation in vitro. Cell Biol Int. 2007;31:1428-1435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 38. | Vaisman N, Barak Y, Hahn T, Karov Y, Malach L, Barak V. Defective in vitro granulopoiesis in patients with anorexia nervosa. Pediatr Res. 1996;40:108-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 39. | Roodman GD, Kurihara N, Ohsaki Y, Kukita A, Hosking D, Demulder A, Smith JF, Singer FR. Interleukin 6. A potential autocrine/paracrine factor in Paget's disease of bone. J Clin Invest. 1992;89:46-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 257] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 40. | Harmer D, Falank C, Reagan MR. Interleukin-6 Interweaves the Bone Marrow Microenvironment, Bone Loss, and Multiple Myeloma. Front Endocrinol (Lausanne). 2018;9:788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 204] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 41. | Kim S, Kim TM. Generation of mesenchymal stem-like cells for producing extracellular vesicles. World J Stem Cells. 2019;11:270-280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 42. | Rodimova SA, Meleshina AV, Kalabusheva EP, Dashinimaev EB, Reunov DG, Torgomyan HG, Vorotelyak EA, Zagaynova EV. Metabolic activity and intracellular pH in induced pluripotent stem cells differentiating in dermal and epidermal directions. Methods Appl Fluoresc. 2019;7:044002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Spyrou J, Gardner DK, Harvey AJ. Metabolism Is a Key Regulator of Induced Pluripotent Stem Cell Reprogramming. Stem Cells Int. 2019;2019:7360121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 44. | Lees JG, Gardner DK, Harvey AJ. Pluripotent Stem Cell Metabolism and Mitochondria: Beyond ATP. Stem Cells Int. 2017;2017:2874283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 45. | Zhang S, Moy W, Zhang H, Leites C, McGowan H, Shi J, Sanders AR, Pang ZP, Gejman PV, Duan J. Open chromatin dynamics reveals stage-specific transcriptional networks in hiPSC-based neurodevelopmental model. Stem Cell Res. 2018;29:88-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 46. | Teslaa T, Teitell MA. Pluripotent stem cell energy metabolism: an update. EMBO J. 2015;34:138-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 173] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 47. | Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerías A, Batchelder EM, Plongthongkum N, Lutz M, Berggren WT, Zhang K, Evans RM, Siuzdak G, Izpisua Belmonte JC. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 402] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 48. | Madonna R, Shelat H, De Caterina R, Geng YJ. Aquaporin-1 is Required for Vascular Differentiation of Human Induced Pluripotent Stem Cells Following Exposure to Glucose-Induced Hyperosmolarity. Circulat. 2011;124:A17216. |
| 49. | Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 567] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 50. | Liu W, Ren Z, Lu K, Song C, Cheung ECW, Zhou Z, Chen G. The Suppression of Medium Acidosis Improves the Maintenance and Differentiation of Human Pluripotent Stem Cells at High Density in Defined Cell Culture Medium. Int J Biol Sci. 2018;14:485-496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 51. | Serra M, Brito C, Correia C, Alves PM. Process engineering of human pluripotent stem cells for clinical application. Trends Biotechnol. 2012;30:350-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 222] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 52. | Sanders LM, Zeisel SH. Choline: Dietary Requirements and Role in Brain Development. Nutr Today. 2007;42:181-186. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 71] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 53. | Puri BK, Counsell SJ, Zaman R, Main J, Collins AG, Hajnal JV, Davey NJ. Relative increase in choline in the occipital cortex in chronic fatigue syndrome. Acta Psychiatr Scand. 2002;106:224-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 49] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 54. | Mueller C, Lin JC, Sheriff S, Maudsley AA, Younger JW. Evidence of widespread metabolite abnormalities in Myalgic encephalomyelitis/chronic fatigue syndrome: assessment with whole-brain magnetic resonance spectroscopy. Brain Imaging Behav. 2020;14:562-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 55. | Jerjes WK, Taylor NF, Peters TJ, Wessely S, Cleare AJ. Urinary cortisol and cortisol metabolite excretion in chronic fatigue syndrome. Psychosom Med. 2006;68:578-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 56. | Elsherbini A, Bieberich E. Ceramide and Exosomes: A Novel Target in Cancer Biology and Therapy. Adv Cancer Res. 2018;140:121-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 57. | Almenar-Pérez E, Ovejero T, Sánchez-Fito T, Espejo JA, Nathanson L, Oltra E. Epigenetic Components of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome Uncover Potential Transposable Element Activation. Clin Ther. 2019;41:675-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 58. | Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 2018;19:175-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1156] [Cited by in RCA: 1384] [Article Influence: 197.7] [Reference Citation Analysis (0)] |
| 59. | Zomer HD, Vidane AS, Gonçalves NN, Ambrósio CE. Mesenchymal and induced pluripotent stem cells: general insights and clinical perspectives. Stem Cells Cloning. 2015;8:125-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 60. | Gao W, Zhang H, Chang G, Xie Z, Wang H, Ma L, Han Z, Li Q, Pang T. Decreased intracellular pH induced by cariporide differentially contributes to human umbilical cord-derived mesenchymal stem cells differentiation. Cell Physiol Biochem. 2014;33:185-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Chen KG, Mallon BS, McKay RD, Robey PG. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14:13-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 262] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 62. | Sokolov MV, Neumann RD. Changes in human pluripotent stem cell gene expression after genotoxic stress exposures. World J Stem Cells. 2014;6:598-605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |