Published online Jul 26, 2021. doi: 10.4252/wjsc.v13.i7.861
Peer-review started: March 8, 2021
First decision: March 29, 2021
Revised: April 19, 2021
Accepted: July 5, 2021
Article in press: July 5, 2021
Published online: July 26, 2021
Processing time: 136 Days and 16.6 Hours
Cancer stem cells (CSCs) are tumor cells that share functional characteristics with normal and embryonic stem cells. CSCs have increased tumor-initiating capacity and metastatic potential and lower sensitivity to chemo- and radiotherapy, with important roles in tumor progression and the response to therapy. Thus, a current goal of cancer research is to eliminate CSCs, necessitating an adequate phenotypic and functional characterization of CSCs. Strategies have been developed to identify, enrich, and track CSCs, many of which distinguish CSCs by evaluating the expression of surface markers, the initiation of specific signaling pathways, and the activation of master transcription factors that control stemness in normal cells. We review and discuss the use of reporter gene systems for identifying CSCs. Reporters that are under the control of aldehyde dehydrogenase 1A1, CD133, Notch, Nanog homeobox, Sex-determining region Y-box 2, and POU class 5 homeobox can be used to identify CSCs in many tumor types, track cells in real time, and screen for drugs. Thus, reporter gene systems, in combination with in vitro and in vivo functional assays, can assess changes in the CSCs pool. We present relevant examples of these systems in the evaluation of experimental CSCs-targeting therapeutics, demonstrating their value in CSCs research.
Core Tip: Controversial cancer stem cells (CSCs) research has caused confusion in this discipline. CSCs should be analyzed based on their function with regard to their ability to generate serially transplantable tumors. However, such evaluations are expensive and time-consuming and are fraught with ethical issues. Gene reporter assays can be used as a surrogate measure of the presence of CSCs in a sample. When combined with immunophenotyping and functional assays, reporter systems improve the quality of the evidence. However, there is no standard system; thus, the selection of an appropriate system must carefully consider its utility in previous works for the tumor type that is to be analyzed.
- Citation: Salinas-Jazmín N, Rosas-Cruz A, Velasco-Velázquez M. Reporter gene systems for the identification and characterization of cancer stem cells. World J Stem Cells 2021; 13(7): 861-876
- URL: https://www.wjgnet.com/1948-0210/full/v13/i7/861.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i7.861
Cancer stem cells (CSCs) constitute a small population in the heterogeneous tumor mass and have characteristics and functions of cancer cells and stem cells. In addition to the hallmark alterations of cancer cells, CSCs have the capacity to self-renew and generate a pool of transit-amplifying cells that produce tumor-bulk cells. Accordingly, CSCs can seed tumors when transplanted into immunocompromised or syngeneic animals. Also, CSCs mediate metastasis and the resistance to cytotoxic treatments, including radio- and chemotherapy, leading to minimal residual disease and cancer relapse[1-4].
CSCs differ from tumor-bulk cells with regard to phenotype and function. CSCs have a different gene expression profile and, thus, differentially expressed proteins that can be used as markers. CSCs are quiescent, and when they proliferate, they frequently undergo asymmetric cell division. The gene expression and consequent functional characteristics of CSCs are regulated in part by several key transcription factors that control stemness in embryonic and adult stem cells, including POU class 5 homeobox 1 (POU5F1; OCT4), Nanog homeobox (NANOG), Sex-determining region Y-box 2 (SOX2), Kruppel-like factor 4 (KLF4), and MYC proto-oncogene [5,6].
CSCs have been proposed to be the "seeds" for tumor initiation and development, metastasis, and recurrence in many tumors, based on their ability to repopulate tumor heterogeneity[2,7]. Given their critical function in tumor progression and clinical importance, many strategies for identifying CSCs have been described, including the quantification of the fraction of cancer cells that express markers that are associated with the CSCs phenotype; evaluation of the ability of cancer cells to form colonies in vitro; and assessment of their tumor-initiating potential in xenograft models, the gold standard approach for examining CSCs[8-10].
CSCs are identified by immunophenotyping by analyzing the expression of cell-surface markers. Although this approach is used extensively, it has limited specificity, because dissimilar markers might be expressed in CSCs from disparate tumor subtypes and, in some cases, even between samples of the same subtype[7,11]. Conversely, analyzing tumor-initiating capacity by limiting-dilution xenotransplantation (LDX) is expensive and time-consuming and requires many animals, posing an ethical dilemma for researchers. Further, LDX is unsuitable for high-throughput drug screening[9,12,13].
To supplement existing tools for identifying, isolating, and characterizing CSCs, several reporter gene systems have been developed, having proven to be useful in substituting or complementing the identification of CSCs by immunophenotyping[14,15]. Reporter gene systems have become essential tools in analyzing the contribution of CSCs to cancer progression and developing CSCs-selective therapies. In this report, we provide integral information on the advantages and drawback of reporter gene systems for analyzing and studying CSCs. In addition, we review and discuss their use in the development of CSCs-targeting drugs, providing specific examples.
The classical strategy for identifying, analyzing, isolating, and enriching CSCs entails the analysis of cell surface markers that are differential expressed between CSCs and non-CSCs[16-19]. The first CSCs markers were identified in acute myelogenous leukemia (CD34+CD38-)[20]. Subsequently, markers that are shared between CSCs and normal human stem cells have been used to identify self-renewing CSCs in solid tumors (see[7,11] for reviews).
Immunophenotyping is widely used because it is easy and fast, and can be performed without special training. Fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS) with surface markers are the primary strategies for isolating CSCs[21]. FACS can sort by multiple biomarkers simultaneously, has robust specificity, and can be combined with other strategies to analyze the functional characteristics of CSCs, such as fluorescence screening of Hoechst 33342 exclusion[22]. However, FACS requires sterile conditions, and cell sorting is stressful to cells, which can impact their behavior. Further, given that CSCs are a rare population, their sorting requires an excessive number of cells, leading to high experimental costs, and treating cell cultures with trypsin can affect their expression of surface markers[8,23,24].
MACS is a simple antibody-based separation technique that does not requires specialized equipment; however, the number of biomarkers that can be used is limited, and thus, it might be unsuitable for complex CSCs immunophenotypes. The resulting purity is typically higher with FACS, but cell survival rates are better with MACS[14]. Both methods are invaluable in CSCs immunophenotyping. For example, leukemia stem cells have been able to be isolated and characterized by FACS[20,25].
However, the expression of CSCs surface markers depends on the type of tumor and the cell of tumor origin, showing heterogeneity between samples[26]. Thus, the immunophenotype of CSCs from a particular tumor can not be applied to all samples. Moreover, the expression of CSC surface markers can change over time or become susceptible to culture conditions[23,24]. For example, enzymatic dissociation of glioblastoma cells modifies the retention of CD133 at their surface[23], and in “stem-like” pancreatic cancer cells, CD133 is upregulated under hypoxic culture conditions[24]. In addition, the use of different commercial monoclonal antibodies (each with a different specificity) complicates the reproducibility of results[15,27]. Given these caveats, surface marker profiles of CSCs are frequently inconsistent between cancer types. Thus, immunophenotyping alone is considered to be insufficient to demonstrate changes in the CSCs pool and has limited use in developing new prognostic and therapeutic options for cancer[11,28,29].
To overcome these issues, non-membrane CSCs biomarkers have been identified, the most prominent of which is aldehyde dehydrogenase (ALDH). ALDH1 catalyzes the oxidation of aldehydes to carboxylic acids and retinol to retinoic acid, allowing detoxification from drugs and reactive oxygen species[30,31]. ALDH is expressed by normal stem cells, and high levels of ALDH1 activity are observed in CSCs, representing a reliable biomarker for identifying this subset of cells in tumors from many tissues, including breast, bladder, embryonal rhabdomyosarcoma, head and neck squamous cell carcinoma, and lung cancer[30]. Higher ALDH1 expression confers resistance to several chemotherapeutic agents, such as cisplatin, etoposide, fluorouracil, and gefitinib[32]. The selection of a population of interest must be based on the expression levels of the enzyme in the tumor cells, given the heterogeneity in CSCs phenotype between tumors[11]. For example, in breast tumors, 2 subpopulations of CSCs have been identified, but only one is ALDH+[33]. Thus, it is possible that different methods enrich distinct subpopulations of CSCs.
ALDH-based staining is also transient and depends on the presence of its substrate, rendering the system suitable only for a limited period[34,35]. To mitigate these disadvantages, Anorma et al[34] developed and tested a turn-on fluorescent probe (AlDeSense) in vitro and ex vivo. The methyl acetate (MA) group of AlDeSense MA is hydrolyzed by an intracellular esterase to form AlDeSense, and its aldehyde group is then oxidized to carboxylic acid by ALDH1A1 in CSCs, emitting fluorescence. The authors observed a 3-fold increase in fluorescence in spheres that were formed by purified CSCs. For the ex vivo evaluation, they analyzed the lungs of mice that had been injected intravenously with CSCs or non-CSCs through the tail vein to generate metastases. When the lungs were perfused with AlDeSense solution, the signal in the lungs from CSCs-injected mice was higher than in non-CSCs-injected mice. When AlDeSense was injected intratumorally, intratumoral CSCs could be observed in vivo using a whole-body fluorescence imager-but only for 2 weeks postimplantation.
These limitations and the need to track CSCs in vivo during metastasis, angiogenesis, and CSC-stroma interactions, have prompted the development of new tools, including reporter gene systems.
A reporter gene system comprises an easily detectable reporter gene and a regulatory complex of transcriptional control (promoters or enhancers that are constitutive or inducible). The expression of the reporter gene reflects the direct activation of the latter in response to the binding of transcription factors to response elements. Reporters that are under constitutive promoters are used primarily to track cells that have been transduced with the construct[36]. Conversely, inducible reporters are used to monitor biological processes. When a reporter gene construct includes transcriptional control components, it functions as a molecular-genetic sensor that responds to endogenous transcription factors and transcription-regulating complexes that initiate and control reporter gene expression[36,37].
The design and development of reporting systems to analyze such properties as phenotypic plasticity and response to therapy require expertise in genetic engineering[36,38,39]. Moreover, because reporter systems are usually designed to trigger the expression of fluorescent proteins, the incorporation of additional fluorescent dyes into the experiment should be planned carefully to prevent cross-contamination between the signals. Fortunately, there are various fluorescent proteins with a range of excitation and emission spectra (from blue to far red) and distinct structural properties and stability. The selection of the fluorescent protein must also consider its maturation time and half-life in the cell to match the desired application[36,37,40,41]. Alternatively, bioluminescent reporter genes with increased sensitivity can be used for in vivo applications[36]. The combination of luciferase genes with fluorescent protein-coding genes into a single sequence has provided an additional tool for analyzing cell populations in vivo and ex vivo, because this strategy allows 2 signals to be monitored independently[36,37].
In CSCs research, reporter gene systems have many advantages, because they allow live detection and isolation of CSCs from several tumor types. Further, these systems can be combined (simultaneously or sequentially) with other methods that analyze cell viability or the expression of other biomarkers, strengthening the distinction of CSCs vs non-CSCs and increasing the reliability of the evaluation of effects of stimuli on either population.
However, the value of a particular reporter gene systems in tracking a particular type of CSCs is directly proportional to its validation using in vitro and in vivo functional assays. Several reporting systems have been used to identify CSCs from various tumors (Table 1) and have thus become important tools for the study of CSCs biology[12,42-45].
| Promoter gene/element response | Reporter gene | Tumor type | Functional assays performed for validation |
| ALDH | Far red fluorescent protein (mNeptune) | Breast cancer[31] | Sphere formation |
| Limiting-dilution xenotransplantation | |||
| Reprogramming of non-CSCs to CSCs after cytotoxic treatments | |||
| Extravasation potential | |||
| Drug sensitivity in vivo | |||
| tdTomato fluorescent protein | Breast and colon cancer[35] | Sphere formation | |
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vivo | |||
| DsRed2 fluorescent protein | Oral squamous cell carcinoma[29] | Sphere formation | |
| Xenotransplantation assays | |||
| Drug sensitivity in vitro | |||
| CD133 | Luciferase/RFP | Glioma[16] | Sphere formation |
| Transactivation assay | |||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vivo | |||
| Drug sensitivity in vitro | |||
| AFP | GFP | Liver (cholangiocarcinoma) [94] | Sphere formationLimiting-dilution |
| xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Pancreatic cancer[95] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Notch | GFP | Breast cancer[51] | Sphere formation |
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vivo | |||
| Luminescent protein | Lung cancer[52] | Sphere formation | |
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vivo | |||
| Drug sensitivity in vitro | |||
| TERT | GFP | Osteosarcoma[53] | Sphere formation |
| Limiting-dilution xenotransplantation | |||
| Extravasation potential | |||
| Drug sensitivity in vitro | |||
| s-SHIP | GFP | Prostate cancer[96] | Sphere formation |
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| DACH1 | GFP | Breast cancer[97,98] | Sphere formation |
| Xenotransplantation assay | |||
| Transactivation assay | |||
| LGR5 | GFP | Colorectal cancer[99] | Sphere formation |
| Gene expression profiling of flow-sorted cells |
As discussed, CSCs can be characterized by their expression of various differential markers, including: (1) Cell surface proteins, such as CD133; (2) Enzymes, such as ALDH1; and (3) Transcription factors[7,11]. Many reports support that self-renewal markers, including ALDH1A1, POU5F1, and SOX2, reliably distinguish CSCs several cancers[29,38,46,47]. However, because these molecules are intracellular, antibody-based screens are not compatible with functional assays.
To overcome this limitation, reporter systems that are based on ALDH1A1 expression have been developed by cloning fluorescent proteins under the control of the ALDH1A1 promoter[29,31,35]. These systems have been used to identify CSCs in breast cancer[31,35], colon cancer[35], and oral squamous cell carcinoma[29]. They can live track CSCs, allowing one to study CSCs dynamics in their microenvironment, increasing our understanding of CSCs involvement in the formation of metastases, resistance to therapy, and cancer recurrence. For example, a fluorescent reporter system that is based on control of the fusion protein mNeptune-TK by the ALDH1A1 promoter was used to identify a population (mNeptunehigh) in breast cancer cells that were pluripotent, had high sphere-forming capacity, and were more resistant to chemotherapy and radiotherapy[31]. These mNeptunehigh cells were more tumorigenic in immunodeficient mice and generated highly resistant tumors. The reporter system efficiently identified and tracked CSCs in several luminal and mesenchymal breast cancer cell lines[31] and thus might be useful in studying CSCs dynamics in tumors.
A different approach exploits the finding that the glycoprotein CD133 is a CSCs surface marker in various cancers, including breast[11,48], colon[11,49], lung[11], and brain[11,16]. A reporter system that is based on CD133 expression has been developed and used to detect CSCs. Guerra-Rebollo et al[16] transduced human glioblastoma U87 tumor cells with a trifunctional chimeric reporter that expresses Renilla reniformis luciferase, red fluorescent protein, and a truncated version of the herpes simplex virus thymidine kinase sr39tk (tTK), driven by the CD133 promoter. This strategy allowed them to independently monitor the entire tumor population or tumor cell subpopulations with an active CD133 promoter by bioluminescence imaging or confocal microscopy[16]. When culturing U87 cells that were transduced with this reporter, an increase in the formation of tumorspheres was observed. The expression of the tTK gene from the construct selectively killed replicating cells with an active CD133 promoter on treatment with ganciclovir[16].
Other reporter systems center on the activation of specific signaling pathways in CSCs. The Notch pathway, which maintains pluripotent hematopoietic stem cells by inhibiting their differentiation, is specifically involved in preserving self-renewal and amplification in CSCs, supporting tumor formation and mediating resistance to chemotherapeutic agents and recurrence in various tumor types[50]. However, the function and activity of Notch signaling is context-dependent in many tumors[49] and thus can not be considered a universal marker for CSCs. The use of reporter genes that respond to Notch signaling has facilitated the identification of a subset of cells with stem cell activity in breast[51] and lung cancer[52], in which the function of Notch signaling has been examined extensively. These reporter systems detect and monitor CSCs in vitro and in vivo, allowing the study of drug resistance in diverse experimental models. Hassan et al[52] used a Notch-green fluorescent protein (GFP) reporter construct that is activated when Notch intracellular domain translocate to the nucleus. They identified a subset of lung cancer cells with high Notch activity (GFPbright) with increased ability to form tumorspheres and generate GFPbright and GFPdim cell populations. Similarly, GFPbright cells were resistant to chemotherapy and tumorigenic in serial xenotransplantation assays, demonstrating that only cells with active Notch signaling could self-renew[52].
Another reporter gene system has been developed to detect CSCs, in which GFP is driven by the telomerase reverse-transcriptase (TERT) promoter, successfully enriching human osteosarcoma stem cells[53]. These GFP+ cells had greater sphere-forming ability and enhanced stem cell-like properties, such as invasiveness, metastatic activity, and resistance to chemotherapeutic agents in vitro and in vivo[53]. Further, the subpopulation in which the hTERT promoter was activated had significantly higher tumorigenic activity in vivo. In orthotopic and ectopic transplantations, the GFP+ cells consistently formed tumors at a lower number of injected cells; these tumors were phenotypically diverse and could initiate new tumors after serial transplantation[53]. However, certain osteosarcoma cell lines and two-thirds of clinical osteosarcoma samples are telomerase-negative, rendering TERT-dependent labeling unsuitable for some patients.
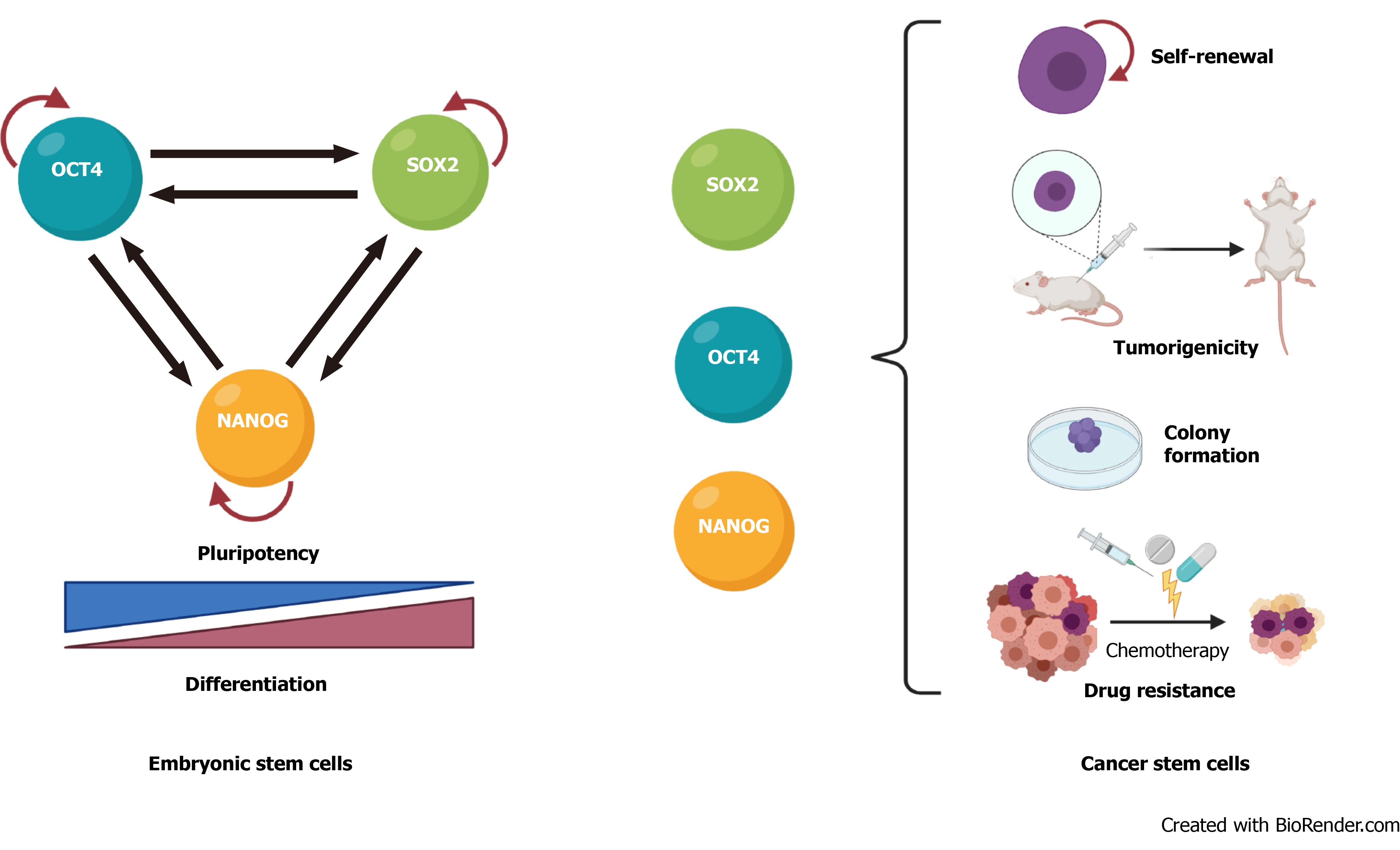
The transcription factors that regulate stemness in normal stem cells are also involved in cancer progression and CSC biology. In mouse embryonic stem cells, these factors form interconnected feed-forward transcriptional loops to establish and reinforce cell type-specific gene expression programs[54,55] (Figure 1A). The ensemble of core transcription factors and their regulatory loops constitutes core transcriptional regulatory circuitry in many signaling pathways that regulate CSCs functions[56] (Figure 1B).
High expression of OCT4 correlates with self-renewal, chemoresistance, and tumorigenic potential of bladder, breast, and glial cells[57-59] and an unfavorable prognosis in cervical, breast, and esophageal squamous cancers[58,60,61]. SOX2 is important in maintaining self-renewal and tumorigenesis and inhibiting differentiation in CSCs from melanoma, lung adenocarcinoma, and lymphoma tissue[62-64], and its elevated expression correlates positively with drug resistance and poor survival in prostate, breast, and glioma cancer patients[65-69]. Overexpression of NANOG in CSCs promotes tumorigenicity by regulating self-renewal and proliferation in prostate, ovarian, and head and neck squamous cells[4,70-72] and is an unfavorable prognostic marker in colorectal, renal, and rectal cancer patients[73-75]. KLF4 is a bifunctional transcription factor that can be an oncogenic or tumor suppressor signal, depending on the type of cancer[76]; lower KLF4 expression contributes to cellular hyperproliferation and malignant transformation in meningioma and prostate cancer[77,78], but upregulation of KLF4 promotes tumor progression in osteosarcoma, breast, and gastrointestinal cancer[79-81]. MYC is usually dysregulated in human cancers, in which it cooperates with other factors during tumorigenesis and promotes invasiveness in CSCs[7,82,83].
Based on their relevance in cancer progression, pluripotent stem cell transcription factors have been used to develop reporter gene systems that are based on their promoters (Table 2). Several promoter-reporter constructs that incorporate portions of the Oct4, Sox2, and Nanog promoters have been used widely to monitor the reprogramming of murine somatic cells into an induced pluripotent state[38]. However, the expression levels of these transcription factors might be lower in CSCs and vary significantly between samples[84-88]. Moreover, the large promoter regions that are used in such constructs invariably contain response elements for other transcription factors, potentially reducing reporter specificity, limiting their application in identifying CSCs. Further, some of these genes, such as Oct4, have alternate transcripts and pseudogenes, complicating their detection[38,39].
| Promoter gene/element response | Reporter gene | Tumor type | Functional assays performed for validation |
| NANOG | GFP; Luminescent protein | Breast cancer[100] | Sphere formation |
| Limiting-dilution xenotransplantation | |||
| Extravasation potential | |||
| In vitro limiting-dilution assays | |||
| Breast cancer[98] | Transactivation assay | ||
| Prostate cancer[70] | Sphere formation | ||
| Extravasation potential | |||
| Drug sensitivity in vitro | |||
| Limiting-dilution xenotransplantation | |||
| Nasopharynx cancer [101] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Extravasation potential | |||
| Drug sensitivity in vivo | |||
| Drug sensitivity in vitro | |||
| Liver (hepatocellular carcinoma)[45] | Sphere formationLimiting-dilution xenotransplantation | ||
| Extravasation potential | |||
| Drug sensitivity in vitro | |||
| Ovary cancer[89] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Ovary cancer[72] | Sphere formation | ||
| Xenotransplantation assay | |||
| Drug sensitivity in vitro | |||
| Extravasation potential | |||
| SOX2 | tdTomato fluorescent protein/ Luminescent protein; GFP; Luminescent protein | Breast cancer[17] | Sphere formation |
| Breast cancer[102] | Sphere formation | ||
| Breast cancer[43] | Sphere formation | ||
| Drug sensitivity in vitro | |||
| Transactivation assay | |||
| Breast cancer[98] | Transactivation assay | ||
| Glioma[46] | Sphere formation | ||
| Xenotransplantation assay | |||
| In vitro limiting-dilution assays | |||
| Skin cancer[103] | Limiting-dilution xenotransplantation | ||
| Drug sensitivity in vivo | |||
| Cervical cancer[104] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Cervical cancer[105] | Sphere formation | ||
| Transactivation assay | |||
| Thyroid cancer[106] | Transactivation assay | ||
| Teratomas from neoplastic hPSCs[12] | Transactivation assay | ||
| Drug sensitivity in vivo | |||
| Progenitor assays (clonogenic and multilineage hematopoietic differentiation) | |||
| OCT4 | GFP; Luminescent protein | Liver (hepatocellular carcinoma)[47] | Sphere formation |
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Melanoma[92] | Sphere formation | ||
| Drug sensitivity in vitro | |||
| Xenotransplantation assay | |||
| Sarcoma[18] | Xenotransplantation assay | ||
| Drug sensitivity in vitro | |||
| Drug sensitivity in vivo | |||
| Breast cancer[43] | Sphere formation | ||
| Drug sensitivity in vitro | |||
| Transactivation assay | |||
| Teratomas from neoplastic hPSCs[12] | Transactivation assay | ||
| Drug sensitivity in vitro | |||
| Progenitor assays (clonogenic and multilineage hematopoietic differentiation) | |||
| SOX2-OCT4 | GFP; mCherry fluorescent protein; Luminescent protein; Luminescent protein/ RFP | Breast cancer[38] | Sphere formation |
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Sarcoma[42] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Prostate cancer[107] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vivo | |||
| Drug sensitivity in vitro | |||
| Gastric cancer[44] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Malignant mesothelioma[108] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Head and neck squamous cancer[109] | Sphere formation | ||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Glioma[16] | Sphere formation | ||
| Transactivation assay | |||
| Limiting-dilution xenotransplantation | |||
| Drug sensitivity in vitro | |||
| Drug sensitivity in vivo |
Thus, systems that use reporter genes under the transcriptional control of promoters that are active specifically in human CSCs have been generated[15,35]. For example, a GFP reporter that is driven by the NANOG promoter was developed to enrich and track ovarian CSCs[72,89]. Wiechert et al[89] introduced this reporter into cisplatin-naïve, high-grade, serous ovarian cancer patient-derived xenografts and ovarian cancer cell lines. GFP+ cells expressed higher levels of stem cell transcription factors (NANOG, SOX2, and POU5F1) and CSC surface markers (CD44, CD133, CD117, CD49f, and CD24) and showed increased tumor-initiating potential. GFP+CD49f+ patient-derived cells were enriched using the reporter system and CD49f staining. Further, the reporter system allowed the group to visualize dynamic changes in stemness in response to cisplatin treatment and to analyze the self-renewing capacity of cisplatin resistant cells[89].
Another example is a lentiviral reporter system, called SORE6, that was developed by Tang et al[38]. The system comprises 6 concatenated repeats of the SOX2 and OCT4 response elements from the proximal human NANOG promoter, controlling the expression of reporter genes (GFP or mCherry). This tool was validated in vitro and in vivo in several models of breast cancer, tracking self-renewal, the generation of heterogeneous offspring, tumor- and metastasis-initiating activity, CSCs plasticity, and the response to therapeutics in real time. SORE6+ cells underwent asymmetrical cell division, generated SORE6- cells, and initiate tumors in serial transplantation, demonstrating that they have tumor-initiating ability and long-term self-renewal[38]. However, no CD44+CD24- cells (the subset commonly reported as breast CSCs) were enriched in the SORE6+ fraction, and there was no overlap with the ALDH1-positive population, suggesting that heterogeneity exists even within stem cell populations, as published[90]. Thus, CSCs detection with SORE6 in breast cancer is more robust than with typical biomarkers, rendering the system ideal for the preclinical evaluation of new drugs.
These examples indicate that systems that report the expression and activity of NANOG, SOX2, and OCT4 are valuable tools for studying CSCs, accelerating the development of more efficient and specific reporter systems and transgene delivery strategies. As discussed, the validation of such systems will require extensive and meticulously planned preclinical testing.
The number of CSCs affects tumor progression, disease recurrence, promotion of angiogenesis, evasion of the immune system, and resistance to conventional anticancer therapies[1,2,18,91]. Increased CSC content in a tumor has also been associated with a more aggressive form and metastatic type[10,33,92,93]. Although certain therapeutic agents that target CSCs have been described[1,10,11,28], it is evident that new selective treatments should be developed. Several studies have demonstrated the value of reporter gene systems in identifying drugs that target CSCs and determining their mechanisms of action (Table 2). For example, the combination of reporting systems with cell viability-tracking dyes can distinguish agents that induce differentiation and the loss of self-renewing pluripotency vs those cause direct cytotoxicity.
In sarcomas, stemness is coordinated by the expression of the pluripotency factor SOX2. Accordingly, the SORE6 reporter system has been used to study the response of CSCs to therapeutics agents in patient-derived cell lines from undifferentiated pleomorphic sarcoma[42]. The simultaneous analysis of SORE6 and caspase-3 activation identified the differential mechanism that was associated with the ability of trabectedin and EC-8042 to reduce the CSC fraction. Trabectedin is an efficient inducer of apoptosis in SORE6+ and SORE6- cells, but EC-8042 reduces the percentage of SORE6+ cells before the apoptotic effect becomes evident, suggesting that EC-8042 switches off SORE6-related transcriptional activity and CSC-associated properties[42].
Similarly, Pádua et al[44] used the SORE6 reporter system to characterize CSCs from gastric cancer and evaluate small molecules in a high-throughput screen[44]. SORE6+ gastric cancer cells from the AGS and Kato III cell lines underwent increased sphere formation and tumorigenicity. Kato III SORE6+ cells had higher levels of ALDH1 compared with SORE6− cells, but AGS cells did not express ALDH1. No other CSC-marker was enriched in SORE6+ cells from either cell line, consistent with several reports that have demonstrated that stemness transcription factors are better markers of CSCs. In the same work, the authors screened 1200 compounds from the Prestwick chemical library in SORE6+ or SORE- cells and observed that monensin induces a reduction in cell number selective towards the SORE6+ population[44]. Given that SORE+ cells are resistant to 5-FU, the identification of monensin as a gastric CSC-targeting drug might guide the development of future adjuvant therapies.
These examples, with those in Table 2, demonstrate that the transcriptional activity of pluripotency transcription factors can be used as a marker of CSCs in various tumor types. Thus, reporter systems can be implemented as a core component of analyses that identify compounds and molecules that target CSCs.
In summary, the appropriate selection of a gene reporter system eliminates common obstacles in the CSCs research, based on their ability to:
Allow direct quantification and isolation of cells with CSCs properties in preclinical tumor models and freshly excised tumors.
Track cells in time and space (in vitro and in vivo) in the analysis of CSCs niches, interactions between CSCs and their microenvironment, and interactions with neighboring cells.
Circumvent direct cell staining procedures and avoid issues with label dilution phenomena.
Track functional properties of CSCs, such as their phenotypic plasticity.
Identify selective agents that target CSCs and could be useful for preclinical testing of anticancer drugs with high sensitivity.
Lastly, it is expected that new reporter gene systems will be generated in the coming years, after the identification of additional CSCs-specific promoters and response elements. Those reporter gene systems could be combined with genetic-editing strategies, such as the CRISPR/Cas9 system, to improve their specificity and reliability by reducing genomic instability due by the integration of indirect genetic markers through viral vectors.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
| 1. | Zeng X, Liu C, Yao J, Wan H, Wan G, Li Y, Chen N. Breast cancer stem cells, heterogeneity, targeting therapies and therapeutic implications. Pharmacol Res. 2021;163:105320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 92] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 2. | Huang T, Song X, Xu D, Tiek D, Goenka A, Wu B, Sastry N, Hu B, Cheng SY. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10:8721-8743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 288] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 3. | Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 937] [Cited by in RCA: 935] [Article Influence: 155.8] [Reference Citation Analysis (35)] |
| 4. | Pedregal-Mallo D, Hermida-Prado F, Granda-Díaz R, Montoro-Jiménez I, Allonca E, Pozo-Agundo E, Álvarez-Fernández M, Álvarez-Marcos C, García-Pedrero JM, Rodrigo JP. Prognostic Significance of the Pluripotency Factors NANOG, SOX2, and OCT4 in Head and Neck Squamous Cell Carcinomas. Cancers (Basel). 2020;12:1794. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 338] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 6. | Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947-956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3597] [Cited by in RCA: 3369] [Article Influence: 168.5] [Reference Citation Analysis (0)] |
| 7. | Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 896] [Cited by in RCA: 1195] [Article Influence: 239.0] [Reference Citation Analysis (0)] |
| 8. | Liu L, Borlak J. Advances in Liver Cancer Stem Cell Isolation and their Characterization. Stem Cell Rev Rep. 2021;epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | Velasco-Velázquez MA, Velázquez-Quesada I, Vásquez-Bochm LX, Pérez-Tapia SM. Targeting Breast Cancer Stem Cells: A Methodological Perspective. Curr Stem Cell Res Ther. 2019;14:389-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Agliano A, Calvo A, Box C. The challenge of targeting cancer stem cells to halt metastasis. Semin Cancer Biol. 2017;44:25-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 155] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 11. | Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauß A, Blaudszun AR, Yevsa T, Fricke S, Kossatz-Boehlert U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front Immunol. 2020;11:1280. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 357] [Cited by in RCA: 581] [Article Influence: 116.2] [Reference Citation Analysis (0)] |
| 12. | Sachlos E, Risueño RM, Laronde S, Shapovalova Z, Lee JH, Russell J, Malig M, McNicol JD, Fiebig-Comyn A, Graham M, Levadoux-Martin M, Lee JB, Giacomelli AO, Hassell JA, Fischer-Russell D, Trus MR, Foley R, Leber B, Xenocostas A, Brown ED, Collins TJ, Bhatia M. Identification of drugs including a dopamine receptor antagonist that selectively target cancer stem cells. Cell. 2012;149:1284-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 336] [Cited by in RCA: 378] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 13. | Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1786] [Cited by in RCA: 1905] [Article Influence: 119.1] [Reference Citation Analysis (0)] |
| 14. | Torre-Healy LA, Berezovsky A, Lathia JD. Isolation, characterization, and expansion of cancer stem cells. In: Methods in Molecular Biology. Humana Press Inc., 2017: 133–143. |
| 15. | Saygin C, Samour M, Chumakova A, Jarrar A, Lathia JD, Reizes O. Reporter Systems to Study Cancer Stem Cells. Methods Mol Biol. 2016;1516:319-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Guerra-Rebollo M, Garrido C, Sánchez-Cid L, Soler-Botija C, Meca-Cortés O, Rubio N, Blanco J. Targeting of replicating CD133 and OCT4/SOX2 expressing glioma stem cells selects a cell population that reinitiates tumors upon release of therapeutic pressure. Sci Rep. 2019;9:9549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 17. | Liang S, Furuhashi M, Nakane R, Nakazawa S, Goudarzi H, Hamada J, Iizasa H. Isolation and characterization of human breast cancer cells with SOX2 promoter activity. Biochem Biophys Res Commun. 2013;437:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Sangiolo D, Mesiano G, Gammaitoni L, Leuci V, Todorovic M, Giraudo L, Cammarata C, Dell'Aglio C, D'Ambrosio L, Pisacane A, Sarotto I, Miano S, Ferrero I, Carnevale-Schianca F, Pignochino Y, Sassi F, Bertotti A, Piacibello W, Fagioli F, Aglietta M, Grignani G. Cytokine-induced killer cells eradicate bone and soft-tissue sarcomas. Cancer Res. 2014;74:119-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 19. | Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21:1119-1131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 266] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 20. | Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4851] [Cited by in RCA: 4850] [Article Influence: 173.2] [Reference Citation Analysis (1)] |
| 21. | de Wynter EA, Coutinho LH, Pei X, Marsh JC, Hows J, Luft T, Testa NG. Comparison of purity and enrichment of CD34+ cells from bone marrow, umbilical cord and peripheral blood (primed for apheresis) using five separation systems. Stem Cells. 1995;13:524-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 86] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Montanaro F, Liadaki K, Schienda J, Flint A, Gussoni E, Kunkel LM. Demystifying SP cell purification: viability, yield, and phenotype are defined by isolation parameters. Exp Cell Res. 2004;298:144-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 117] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 23. | Lv D, Ma QH, Duan JJ, Wu HB, Zhao XL, Yu SC, Bian XW. Optimized dissociation protocol for isolating human glioma stem cells from tumorspheres via fluorescence-activated cell sorting. Cancer Lett. 2016;377:105-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Hashimoto O, Shimizu K, Semba S, Chiba S, Ku Y, Yokozaki H, Hori Y. Hypoxia induces tumor aggressiveness and the expansion of CD133-positive cells in a hypoxia-inducible factor-1α-dependent manner in pancreatic cancer cells. Pathobiology. 2011;78:181-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 25. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7708] [Article Influence: 350.4] [Reference Citation Analysis (0)] |
| 26. | Immervoll H, Hoem D, Sakariassen PØ, Steffensen OJ, Molven A. Expression of the "stem cell marker" CD133 in pancreas and pancreatic ductal adenocarcinomas. BMC Cancer. 2008;8:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 165] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 27. | Osmond TL, Broadley KW, McConnell MJ. Glioblastoma cells negative for the anti-CD133 antibody AC133 express a truncated variant of the CD133 protein. Int J Mol Med. 2010;25:883-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 28. | Duan H, Liu Y, Gao Z, Huang W. Recent advances in drug delivery systems for targeting cancer stem cells. Acta Pharm Sin B. 2021;11:55-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 156] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 29. | Shanmugam G, Mohan A, Kumari K, Louis JM, Soumya Krishnan U, Balagopal PG, George NA, Sebastian P, Maliekal TT. A novel reporter construct for screening small molecule inhibitors that specifically target self-renewing cancer cells. Exp Cell Res. 2019;383:111551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 30. | Tomita H, Tanaka K, Tanaka T, Hara A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget. 2016;7:11018-11032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 321] [Cited by in RCA: 435] [Article Influence: 54.4] [Reference Citation Analysis (0)] |
| 31. | Bidan N, Bailleul-Dubois J, Duval J, Winter M, Denoulet M, Hannebicque K, El-Sayed IY, Ginestier C, Forissier V, Charafe-Jauffret E, Macario M, Matsunaga YT, Meignan S, Anquez F, Julien S, Bonnefond A, Derhourhi M, Le Bourhis X, Lagadec C. Transcriptomic Analysis of Breast Cancer Stem Cells and Development of a pALDH1A1:mNeptune Reporter System for Live Tracking. Proteomics. 2019;19:e1800454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 32. | Huang CP, Tsai MF, Chang TH, Tang WC, Chen SY, Lai HH, Lin TY, Yang JC, Yang PC, Shih JY, Lin SB. ALDH-positive lung cancer stem cells confer resistance to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Lett. 2013;328:144-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 33. | Luo M, Brooks M, Wicha MS. Epithelial-mesenchymal plasticity of breast cancer stem cells: implications for metastasis and therapeutic resistance. Curr Pharm Des. 2015;21:1301-1310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 164] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 34. | Anorma C, Hedhli J, Bearrood TE, Pino NW, Gardner SH, Inaba H, Zhang P, Li Y, Feng D, Dibrell SE, Kilian KA, Dobrucki LW, Fan TM, Chan J. Surveillance of Cancer Stem Cell Plasticity Using an Isoform-Selective Fluorescent Probe for Aldehyde Dehydrogenase 1A1. ACS Cent Sci. 2018;4:1045-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 35. | Gener P, Gouveia LP, Sabat GR, de Sousa Rafael DF, Fort NB, Arranja A, Fernández Y, Prieto RM, Ortega JS, Arango D, Abasolo I, Videira M, Schwartz S Jr. Fluorescent CSC models evidence that targeted nanomedicines improve treatment sensitivity of breast and colon cancer stem cells. Nanomedicine. 2015;11:1883-1892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 36. | Serganova I, Blasberg RG. Molecular Imaging with Reporter Genes: Has Its Promise Been Delivered? J Nucl Med. 2019;60:1665-1681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 37. | Li M, Wang Y, Liu M, Lan X. Multimodality reporter gene imaging: Construction strategies and application. Theranostics. 2018;8:2954-2973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 38. | Tang B, Raviv A, Esposito D, Flanders KC, Daniel C, Nghiem BT, Garfield S, Lim L, Mannan P, Robles AI, Smith WI Jr, Zimmerberg J, Ravin R, Wakefield LM. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports. 2015;4:155-169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 39. | Jarrar A, Chumakova A, Hitomi M, Lathia JD. Enrichment and Interrogation of Cancer Stem Cells. In: Cancer Stem Cells. Elsevier, 2016: 59–98. |
| 40. | Mohammadi Z, Karamzadeh A, Tabatabaiefar MA, Khanahmad H, Shariati L. Evidence for expression of promoterless GFP cassette: Is GFP an ideal reporter gene in biotechnology science? Res Pharm Sci. 2019;14:351-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 41. | Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2252] [Cited by in RCA: 2123] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 42. | Menendez ST, Rey V, Martinez-Cruzado L, Gonzalez MV, Morales-Molina A, Santos L, Blanco V, Alvarez C, Estupiñan O, Allonca E, Rodrigo JP, García-Castro J, Garcia-Pedrero JM, Rodriguez R. SOX2 Expression and Transcriptional Activity Identifies a Subpopulation of Cancer Stem Cells in Sarcoma with Prognostic Implications. Cancers (Basel). 2020;12:964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 43. | Vásquez-Bochm LX, Velázquez-Paniagua M, Castro-Vázquez SS, Guerrero-Rodríguez SL, Mondragon-Peralta A, De La Fuente-Granada M, Pérez-Tapia SM, González-Arenas A, Velasco-Velázquez MA. Transcriptome-based identification of lovastatin as a breast cancer stem cell-targeting drug. Pharmacol Rep. 2019;71:535-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 44. | Pádua D, Barros R, Amaral AL, Mesquita P, Freire AF, Sousa M, Maia AF, Caiado I, Fernandes H, Pombinho A, Pereira CF, Almeida R. A SOX2 Reporter System Identifies Gastric Cancer Stem-Like Cells Sensitive to Monensin. Cancers (Basel). 2020;12:495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 45. | Shan J, Shen J, Liu L, Xia F, Xu C, Duan G, Xu Y, Ma Q, Yang Z, Zhang Q, Ma L, Liu J, Xu S, Yan X, Bie P, Cui Y, Bian XW, Qian C. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology. 2012;56:1004-1014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 251] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 46. | Stoltz K, Sinyuk M, Hale JS, Wu Q, Otvos B, Walker K, Vasanji A, Rich JN, Hjelmeland AB, Lathia JD. Development of a Sox2 reporter system modeling cellular heterogeneity in glioma. Neuro Oncol. 2015;17:361-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 47. | Wu G, Wilson G, Zhou G, Hebbard L, George J, Qiao L. Oct4 is a reliable marker of liver tumor propagating cells in hepatocellular carcinoma. Discov Med. 2015;20:219-229. [PubMed] |
| 48. | Brugnoli F, Grassilli S, Al-Qassab Y, Capitani S, Bertagnolo V. CD133 in Breast Cancer Cells: More than a Stem Cell Marker. J Oncol. 2019;2019:7512632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 49. | Kim WT, Ryu CJ. Cancer stem cell surface markers on normal stem cells. BMB Rep. 2017;50:285-298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 219] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 50. | Meisel CT, Porcheri C, Mitsiadis TA. Cancer Stem Cells, Quo Vadis? Cells. 2020;9:1879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 51. | D'Angelo RC, Ouzounova M, Davis A, Choi D, Tchuenkam SM, Kim G, Luther T, Quraishi AA, Senbabaoglu Y, Conley SJ, Clouthier SG, Hassan KA, Wicha MS, Korkaya H. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol Cancer Ther. 2015;14:779-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 98] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 52. | Hassan KA, Wang L, Korkaya H, Chen G, Maillard I, Beer DG, Kalemkerian GP, Wicha MS. Notch pathway activity identifies cells with cancer stem cell-like properties and correlates with worse survival in lung adenocarcinoma. Clin Cancer Res. 2013;19:1972-1980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 53. | Yu L, Liu S, Zhang C, Zhang B, Simões BM, Eyre R, Liang Y, Yan H, Wu Z, Guo W, Clarke RB. Enrichment of human osteosarcoma stem cells based on hTERT transcriptional activity. Oncotarget. 2013;4:2326-2338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 54. | De Kumar B, Parker HJ, Parrish ME, Lange JJ, Slaughter BD, Unruh JR, Paulson A, Krumlauf R. Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc Natl Acad Sci USA. 2017;114:5838-5845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 55. | Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 849] [Cited by in RCA: 855] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 56. | Chen Y, Xu L, Lin RY, Müschen M, Koeffler HP. Core transcriptional regulatory circuitries in cancer. Oncogene. 2020;39:6633-6646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 57. | Lu CS, Shieh GS, Wang CT, Su BH, Su YC, Chen YC, Su WC, Wu P, Yang WH, Shiau AL, Wu CL. Chemotherapeutics-induced Oct4 expression contributes to drug resistance and tumor recurrence in bladder cancer. Oncotarget. 2017;8:30844-30858. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 58. | Gwak JM, Kim M, Kim HJ, Jang MH, Park SY. Expression of embryonal stem cell transcription factors in breast cancer: Oct4 as an indicator for poor clinical outcome and tamoxifen resistance. Oncotarget. 2017;8:36305-36318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 59. | Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, Cao X, Ling EA, Hao A. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 127] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 60. | Zhao X, Lu H, Sun Y, Liu L, Wang H. Prognostic value of octamer binding transcription factor 4 for patients with solid tumors: A meta-analysis. Medicine (Baltimore). 2020;99:e22804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 61. | Li C, Yan Y, Ji W, Bao L, Qian H, Chen L, Wu M, Chen H, Li Z, Su C. OCT4 positively regulates Survivin expression to promote cancer cell proliferation and leads to poor prognosis in esophageal squamous cell carcinoma. PLoS One. 2012;7:e49693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 62. | Santini R, Pietrobono S, Pandolfi S, Montagnani V, D'Amico M, Penachioni JY, Vinci MC, Borgognoni L, Stecca B. SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene. 2014;33:4697-4708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 63. | Bora-Singhal N, Mohankumar D, Saha B, Colin CM, Lee JY, Martin MW, Zheng X, Coppola D, Chellappan S. Novel HDAC11 inhibitors suppress lung adenocarcinoma stem cell self-renewal and overcome drug resistance by suppressing Sox2. Sci Rep. 2020;10:4722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 72] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 64. | Chen J, Ge X, Zhang W, Ding P, Du Y, Wang Q, Li L, Fang L, Sun Y, Zhang P, Zhou Y, Zhang L, Lv X, Zhang X, Zhang Q, Xue K, Gu H, Lei Q, Wong J, Hu W. PI3K/AKT inhibition reverses R-CHOP resistance by destabilizing SOX2 in diffuse large B cell lymphoma. Theranostics. 2020;10:3151-3163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 65. | Yu X, Cates JM, Morrissey C, You C, Grabowska MM, Zhang J, DeGraff DJ, Strand DW, Franco OE, Lin-Tsai O, Hayward SW, Matusik RJ. SOX2 expression in the developing, adult, as well as, diseased prostate. Prostate Cancer Prostatic Dis. 2014;17:301-309. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 66. | Gao J, Zhang JY, Li YH, Ren F. Decreased expression of SOX9 indicates a better prognosis and inhibits the growth of glioma cells by inducing cell cycle arrest. Int J Clin Exp Pathol. 2015;8:10130-10138. [PubMed] |
| 67. | Lei B, Zhang Y, Liu T, Li Y, Pang D. Sox9 upregulation in breast cancer is correlated with poor prognosis and the CD44 + /CD24-/Low phenotype. Int J Clin Exp Pathol. 2016;9:7345-7351. |
| 68. | Grimm D, Bauer J, Wise P, Krüger M, Simonsen U, Wehland M, Infanger M, Corydon TJ. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2020;67:122-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 268] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 69. | Zhang S, Xiong X, Sun Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct Target Ther. 2020;5:135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 70. | Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, Repass J, Zaehres H, Shen JJ, Tang DG. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833-3845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 295] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 71. | Mahalaxmi I, Devi SM, Kaavya J, Arul N, Balachandar V, Santhy KS. New insight into NANOG: A novel therapeutic target for ovarian cancer (OC). Eur J Pharmacol. 2019;852:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 72. | Ling K, Jiang L, Liang S, Kwong J, Yang L, Li Y, PingYin, Deng Q, Liang Z. Nanog interaction with the androgen receptor signaling axis induce ovarian cancer stem cell regulation: studies based on the CRISPR/Cas9 system. J Ovarian Res. 2018;11:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 73. | Rasti A, Mehrazma M, Madjd Z, Abolhasani M, Saeednejad Zanjani L, Asgari M. Co-expression of Cancer Stem Cell Markers OCT4 and NANOG Predicts Poor Prognosis in Renal Cell Carcinomas. Sci Rep. 2018;8:11739. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 74. | You L, Guo X, Huang Y. Correlation of Cancer Stem-Cell Markers OCT4, SOX2, and NANOG with Clinicopathological Features and Prognosis in Operative Patients with Rectal Cancer. Yonsei Med J. 2018;59:35-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 75. | Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ, Zhou J, Ding YQ, Li J. Over-expression of Nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;9:295-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 176] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 76. | Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 448] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 77. | Tang H, Zhu H, Wang X, Hua L, Li J, Xie Q, Chen X, Zhang T, Gong Y. KLF4 is a tumor suppressor in anaplastic meningioma stem-like cells and human meningiomas. J Mol Cell Biol. 2017;9:315-324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 78. | Chang YL, Zhou PJ, Wei L, Li W, Ji Z, Fang YX, Gao WQ. MicroRNA-7 inhibits the stemness of prostate cancer stem-like cells and tumorigenesis by repressing KLF4/PI3K/Akt/p21 pathway. Oncotarget. 2015;6:24017-24031. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 79. | Qi XT, Li YL, Zhang YQ, Xu T, Lu B, Fang L, Gao JQ, Yu LS, Zhu DF, Yang B, He QJ, Ying MD. KLF4 functions as an oncogene in promoting cancer stem cell-like characteristics in osteosarcoma cells. Acta Pharmacol Sin. 2019;40:546-555. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 80. | Yu F, Li J, Chen H, Fu J, Ray S, Huang S, Zheng H, Ai W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161-2172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 369] [Cited by in RCA: 364] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 81. | Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 176] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 82. | Liu Z, He Q, Ding X, Zhao T, Zhao L, Wang A. SOD2 is a C-myc target gene that promotes the migration and invasion of tongue squamous cell carcinoma involving cancer stem-like cells. Int J Biochem Cell Biol. 2015;60:139-146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 83. | Cheng CC, Shi LH, Wang XJ, Wang SX, Wan XQ, Liu SR, Wang YF, Lu Z, Wang LH, Ding Y. Stat3/Oct-4/c-Myc signal circuit for regulating stemness-mediated doxorubicin resistance of triple-negative breast cancer cells and inhibitory effects of WP1066. Int J Oncol. 2018;53:339-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 84. | Munro MJ, Wickremesekera SK, Peng L, Marsh RW, Itinteang T, Tan ST. Cancer stem cell subpopulations in primary colon adenocarcinoma. PLoS One. 2019;14:e0221963. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 85. | Upadhyay VA, Shah KA, Makwana DP, Raval AP, Shah FD, Rawal RM. Putative stemness markers octamer-binding transcription factor 4, sex-determining region Y-box 2, and NANOG in non-small cell lung carcinoma: A clinicopathological association. J Cancer Res Ther. 2020;16:804-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 86. | Sodja E, Rijavec M, Koren A, Sadikov A, Korošec P, Cufer T. The prognostic value of whole blood SOX2, NANOG and OCT4 mRNA expression in advanced small-cell lung cancer. Radiol Oncol. 2016;50:188-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 87. | van Schaijik B, Davis PF, Wickremesekera AC, Tan ST, Itinteang T. Subcellular localisation of the stem cell markers OCT4, SOX2, NANOG, KLF4 and c-MYC in cancer: a review. J Clin Pathol. 2018;71:88-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 137] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 88. | Kim JW, Chung JY, Ylaya K, Park Y, Jun SY, Hong SM, Hewitt SM. Prognostic implication of SOX2 expression in small intestinal adenocarcinoma. Virchows Arch. 2020;478:1049-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 89. | Wiechert A, Saygin C, Thiagarajan PS, Rao VS, Hale JS, Gupta N, Hitomi M, Nagaraj AB, DiFeo A, Lathia JD, Reizes O. Cisplatin induces stemness in ovarian cancer. Oncotarget. 2016;7:30511-30522. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 90. | Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3173] [Cited by in RCA: 3049] [Article Influence: 169.4] [Reference Citation Analysis (0)] |
| 91. | Koual M, Tomkiewicz C, Cano-Sancho G, Antignac JP, Bats AS, Coumoul X. Environmental chemicals, breast cancer progression and drug resistance. Environ Health. 2020;19:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 106] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 92. | Gammaitoni L, Giraudo L, Leuci V, Todorovic M, Mesiano G, Picciotto F, Pisacane A, Zaccagna A, Volpe MG, Gallo S, Caravelli D, Giacone E, Venesio T, Balsamo A, Pignochino Y, Grignani G, Carnevale-Schianca F, Aglietta M, Sangiolo D. Effective activity of cytokine-induced killer cells against autologous metastatic melanoma including cells with stemness features. Clin Cancer Res. 2013;19:4347-4358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 93. | Adorno-Cruz V, Kibria G, Liu X, Doherty M, Junk DJ, Guan D, Hubert C, Venere M, Mulkearns-Hubert E, Sinyuk M, Alvarado A, Caplan AI, Rich J, Gerson SL, Lathia J, Liu H. Cancer stem cells: targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015;75:924-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 192] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 94. | Ishii T, Yasuchika K, Suemori H, Nakatsuji N, Ikai I, Uemoto S. Alpha-fetoprotein producing cells act as cancer progenitor cells in human cholangiocarcinoma. Cancer Lett. 2010;294:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 95. | Sasaki N, Ishii T, Kamimura R, Kajiwara M, Machimoto T, Nakatsuji N, Suemori H, Ikai I, Yasuchika K, Uemoto S. Alpha-fetoprotein-producing pancreatic cancer cells possess cancer stem cell characteristics. Cancer Lett. 2011;308:152-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 96. | Bauderlique-Le Roy H, Vennin C, Brocqueville G, Spruyt N, Adriaenssens E, Bourette RP. Enrichment of Human Stem-Like Prostate Cells with s-SHIP Promoter Activity Uncovers a Role in Stemness for the Long Noncoding RNA H19. Stem Cells Dev. 2015;24:1252-1262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 97. | Wu K, Li A, Rao M, Liu M, Dailey V, Yang Y, Di Vizio D, Wang C, Lisanti MP, Sauter G, Russell RG, Cvekl A, Pestell RG. DACH1 is a cell fate determination factor that inhibits cyclin D1 and breast tumor growth. Mol Cell Biol. 2006;26:7116-7129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 105] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 98. | Wu K, Jiao X, Li Z, Katiyar S, Casimiro MC, Yang W, Zhang Q, Willmarth NE, Chepelev I, Crosariol M, Wei Z, Hu J, Zhao K, Pestell RG. Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J Biol Chem. 2011;286:2132-2142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 99. | Hirsch D, Hu Y, Ried T, Moll R, Gaiser T. Transcriptome profiling of LGR5 positive colorectal cancer cells. Genom Data. 2014;2:212-215. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 100. | Thiagarajan PS, Hitomi M, Hale JS, Alvarado AG, Otvos B, Sinyuk M, Stoltz K, Wiechert A, Mulkearns-Hubert E, Jarrar A, Zheng Q, Thomas D, Egelhoff T, Rich JN, Liu H, Lathia JD, Reizes O. Development of a Fluorescent Reporter System to Delineate Cancer Stem Cells in Triple-Negative Breast Cancer. Stem Cells. 2015;33:2114-2125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 101. | Wei F, Rong XX, Xie RY, Jia LT, Wang HY, Qin YJ, Chen L, Shen HF, Lin XL, Yang J, Yang S, Hao WC, Chen Y, Xiao SJ, Zhou HR, Lin TY, Chen YS, Sun Y, Yao KT, Xiao D. Cytokine-induced killer cells efficiently kill stem-like cancer cells of nasopharyngeal carcinoma via the NKG2D-ligands recognition. Oncotarget. 2015;6:35023-35039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 102. | Zhang Y, Eades G, Yao Y, Li Q, Zhou Q. Estrogen receptor α signaling regulates breast tumor-initiating cells by down-regulating miR-140 which targets the transcription factor SOX2. J Biol Chem. 2012;287:41514-41522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 103. | Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E, Brohée S, Salmon I, Dubois C, del Marmol V, Fuks F, Beck B, Blanpain C. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 512] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 104. | Liu XF, Yang WT, Xu R, Liu JT, Zheng PS. Cervical cancer cells with positive Sox2 expression exhibit the properties of cancer stem cells. PLoS One. 2014;9:e87092. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 84] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 105. | Yang WT, Zhao ZX, Li B, Zheng PS. NF-YA transcriptionally activates the expression of SOX2 in cervical cancer stem cells. PLoS One. 2019;14:e0215494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 106. | Lu Y, Zhu Y, Deng S, Chen Y, Li W, Sun J, Xu X. Targeting the Sonic Hedgehog Pathway to Suppress the Expression of the Cancer Stem Cell (CSC)-Related Transcription Factors and CSC-Driven Thyroid Tumor Growth. Cancers (Basel). 2021;13:418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 107. | Vaddi PK, Stamnes MA, Cao H, Chen S. Elimination of SOX2/OCT4-Associated Prostate Cancer Stem Cells Blocks Tumor Development and Enhances Therapeutic Response. Cancers (Basel). 2019;11:1331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 108. | Blum W, Pecze L, Felley-Bosco E, Wu L, de Perrot M, Schwaller B. Stem Cell Factor-Based Identification and Functional Properties of In Vitro-Selected Subpopulations of Malignant Mesothelioma Cells. Stem Cell Reports. 2017;8:1005-1017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 109. | Keysar SB, Le PN, Miller B, Jackson BC, Eagles JR, Nieto C, Kim J, Tang B, Glogowska MJ, Morton JJ, Padilla-Just N, Gomez K, Warnock E, Reisinger J, Arcaroli JJ, Messersmith WA, Wakefield LM, Gao D, Tan AC, Serracino H, Vasiliou V, Roop DR, Wang XJ, Jimeno A. Regulation of Head and Neck Squamous Cancer Stem Cells by PI3K and SOX2. J Natl Cancer Inst. 2017;109:djw189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |