Published online Apr 26, 2021. doi: 10.4252/wjsc.v13.i4.281
Peer-review started: February 1, 2021
First decision: February 28, 2021
Revised: March 11, 2021
Accepted: March 29, 2021
Article in press: March 29, 2021
Published online: April 26, 2021
Processing time: 79 Days and 17.5 Hours
Among inherited cardiac conditions, a special place is kept by cardiomyopathies (CMPs) and channelopathies (CNPs), which pose a substantial healthcare burden due to the complexity of the therapeutic management and cause early mortality. Like other inherited cardiac conditions, genetic CMPs and CNPs exhibit incomplete penetrance and variable expressivity even within carriers of the same pathogenic deoxyribonucleic acid variant, challenging our understanding of the underlying pathogenic mechanisms. Until recently, the lack of accurate physiological preclinical models hindered the investigation of fundamental cellular and molecular mechanisms. The advent of induced pluripotent stem cell (iPSC) technology, along with advances in gene editing, offered unprecedented opportunities to explore hereditary CMPs and CNPs. Hallmark features of iPSCs include the ability to differentiate into unlimited numbers of cells from any of the three germ layers, genetic identity with the subject from whom they were derived, and ease of gene editing, all of which were used to generate “disease-in-a-dish” models of monogenic cardiac conditions. Functionally, iPSC-derived cardiomyocytes that faithfully recapitulate the patient-specific phenotype, allowed the study of disease mechanisms in an individual-/allele-specific manner, as well as the customization of therapeutic regimen. This review provides a synopsis of the most important iPSC-based models of CMPs and CNPs and the potential use for modeling disease mechanisms, personalized therapy and deoxyribonucleic acid variant functional annotation.
Core Tip: Induced pluripotent stem cell (iPSC) technology holds a great potential for medical research. Patient-specific iPSC-derived cardiomyocytes offer a unique framework for various applications, such as cardiotoxicity screening, drug discovery, disease modeling, and cell therapy. In the particular case of inherited cardiomyopathies and channelopathies, iPSC-based models have prompted study of disease mechanisms in an individual-/allele-specific manner, as well as the customization of therapeutic regimens. Herein, we present and critically discuss the current knowledge and key experimental approaches that support patient-specific iPSCs as robust “disease-in-a-dish” models for genetic cardiomyopathies and channelopathies after 15 years of research.
- Citation: Micheu MM, Rosca AM. Patient-specific induced pluripotent stem cells as “disease-in-a-dish” models for inherited cardiomyopathies and channelopathies – 15 years of research. World J Stem Cells 2021; 13(4): 281-303
- URL: https://www.wjgnet.com/1948-0210/full/v13/i4/281.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i4.281
Stem cell technology is one of the most dynamic areas in modern research, holding a great potential to alleviate or even cure various diseases. Particularly, induced pluripotent stem cells (iPSCs) are of a great interest given that they share the benefits of embryonic stem cells but lack their downsides. First, similar to embryonic stem cells, iPSCs are able to differentiate into tissues derived from all three germ layers, both in vitro and in vivo. Secondly, iPSCs-derived cells will be immunologically identical to the host, making the use of immunosuppression unnecessary. Thirdly, there are no bioethical issues with the use of iPSCs. These unique features endorse them an excellent candidate for a wide array of applications such as cardiotoxicity screening, drug discovery, disease modeling, and cell therapy.
Ever since their first mention in 2006[1], we have witnessed a mounting body of data related to this rapidly growing field. Progress has been made in reprogramming and differentiation methods. Strategies for improving the maturity of iPSC-derived cardiomyocytes (iPSC-CMs) have been tested, and new applications to manage cardiac diseases have been tested. A recent Scientific Statement from the American Heart Association acknowledges disease modeling as possibly the most productive use of iPSCs[2]. Several key characteristics endorse iPSCs as an ideal candidate for generating “disease-in-a-dish” models, particularly with regard to monogenic conditions. First of all, each iPSC line has a donor-specific genetic profile. Secondly, when collected, iPSCs are devoid of many of the epigenetic modifications caused by environmental and lifestyle factors, thus enabling the study of the genetic contribution to the disease. This aspect is of a particular importance in the case of Mendelian cardiac maladies, which are characterized by variable clinical expression and incomplete penetrance as a consequence of complex interactions between genetic backgrounds and environmental disease modifiers[3]. Thirdly, iPSCs are quite malleable to genetic modification; accordingly, by using appropriate genome editing tools such as TALENs and CRISPR-Cas9, the deoxyribonucleic acid (DNA) sequence can be altered either by introducing causal DNA mutations into wild-type iPSC lines, or by repairing the causative factor to achieve phenotypic rescue in differentiated cells[2,4].
Inherited cardiac conditions (ICCs) include a variety of genetic disorders that primarily affect the heart. Among ICCs, a special place is kept by cardiomyopathies (CMPs) and arrhythmic diseases (i.e. channelopathies), which pose a substantial healthcare burden due to the complexity of therapeutic management and occurrence early mortality. Importantly, sudden cardiac death is frequently the first expression of the disease. Understanding the underlying genetic cause is the centerpiece of a timely diagnosis and targeted treatment[5].
CMPs are characterized by both structural and functional abnormalities of the ventricular myocardium that are not explained by flow-limiting coronary artery disease or abnormal loading conditions, each entity having particular characteristics at macroscopic and molecular level[6]. Based on morphology, hereditary CMPs comprise the following types: hypertrophic cardiomyopathy (HCM), dilated cardiomyopathy (DCM), restrictive cardiomyopathy (RCM), arrhythmogenic cardiomyopathy (ACM), and left ventricular noncompaction (LVNC).
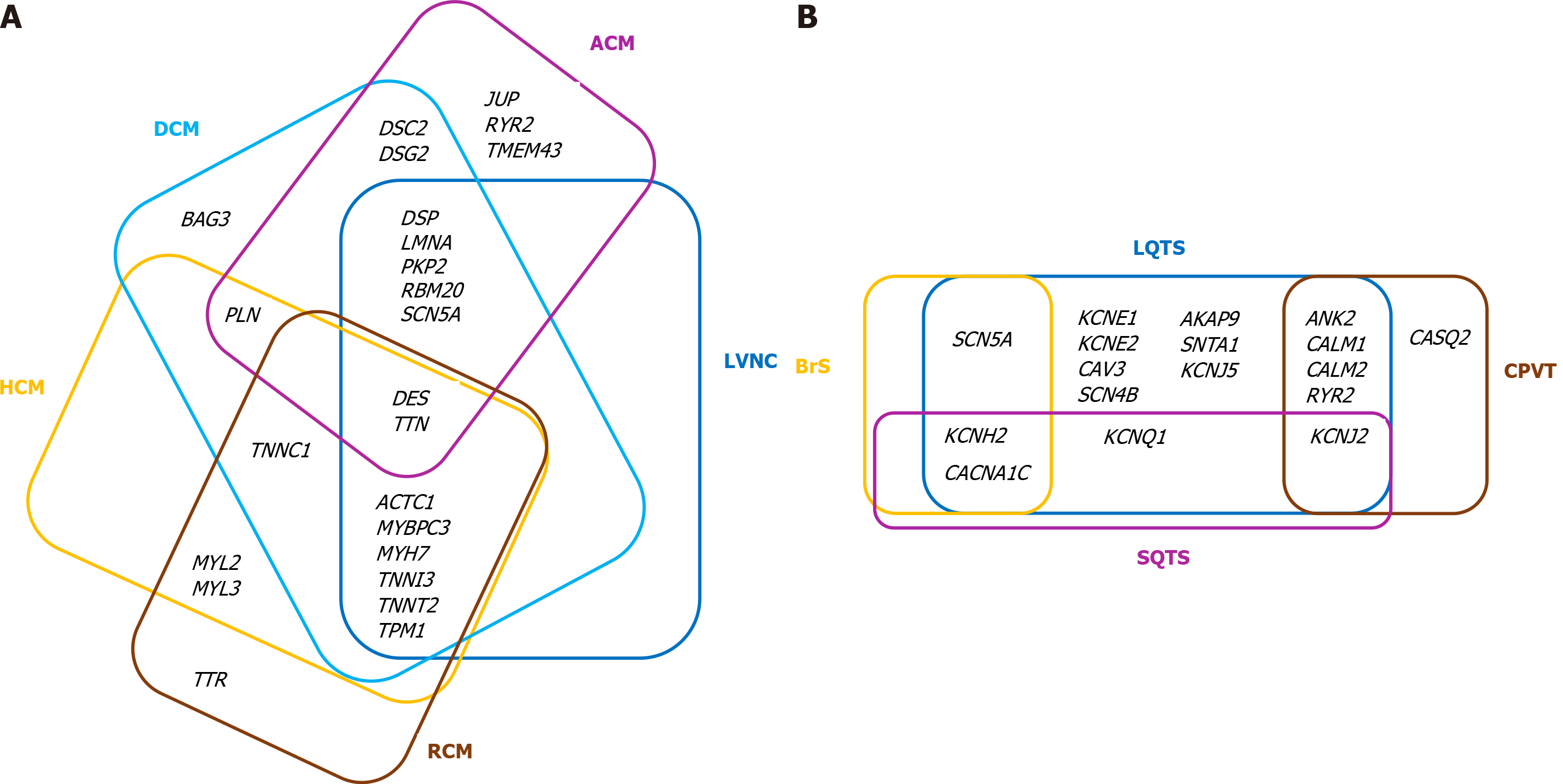
Inherited channelopathies (CNPs) are primary electrical disorders caused by mutations in genes encoding cardiac ion channels or associated proteins. As a result, malfunction of specific ion channels or of intracellular calcium handling occur, leading to electrical instability and predisposition to malignant arrhythmias in the absence of structural heart disease[7,8]. The main cardiac channelopathies associated with increased risk of sudden cardiac death are long QT syndrome (LQTS), short QT syndrome (SQTS), Brugada syndrome (BrS), and catecholaminergic polymorphic ventricular tachycardia (CPVT). As comprehensive reviews of the genetics and clinical presentation of various ICCs have been written by our group[3,9] and other groups[10-12], we briefly point out the core genes associated with the CMPs and CNPs discussed in the present paper (see Tables 1 and 2)[12-19]. It is to be noted that there is considerable genetic overlap among different CMPs and CNPs (Figure 1A and B, respectively).
Due to the potential to differentiate into functional cardiomyocytes (CMs) that recapitulate patient-specific phenotypes, human iPSCs provide an excellent in vitro platform to decipher the underlying disease-specific mechanisms and efficiently study inherited CMPs and CNPs in an individualized manner.
HCM and DCM are the most frequently encountered genetic CMPs in daily clinical practice, therefore, unsurprisingly, they have been the most studied iPSC-CM-based models. In a recent report, Eschenhagen et al[19] comparatively analyzed 38 original papers that reported the characteristics of iPSC-CMs obtained from patients with HCM/DCM or generated from iPSC lines in which a HCM or DCM mutation had been genetically introduced[19]. In summary, compared with their respective controls, the main features exhibited by HCM iPSC-CMs were the following: larger cell size, increased nuclear localization of nuclear factor of activated T cells (NFAT, a transcription factor) and increased MYH7 (or MYH7/MYH6 ratio) expression[20-24]. The most constant aberration identified in DCM lines was reduced peak force development[25], the molecular mechanisms of which are discussed, and explain the main clinical presentation of the disease. As for similarities between the two considered diseases, three anomalies were the most documented: sarcomere disarray, increased NPPA/NPPB gene expression, and arrhythmic behavior[20,24,26,27].
Although not constantly associated with alterations in contractile force or kinetics, abnormal calcium handling appears to be a key pathological mechanism observed in iPSC-CM models of HCM[20,21,24,28]. Valuable insights related to the molecular mechanisms of HCM pathogenesis have been provided by Seeger et al[29] in a model of patient-derived iPSC-CMs harboring a premature stop codon in the MYBPC3 gene. When compared with the isogenic mutation-corrected iPSC lines, in addition to aberrant calcium signaling, patient-derived iPSC-CMs displayed molecular dysregulations without haploinsufficiency of the MYBPC3 protein. This observation could challenge the existing dogma of haploinsufficiency as the underlying mechanisms for HCM caused by MYBPC3 premature termination codon mutations. The specific molecular signature included dysregulation of genes involved in calcium handling (ATP2A2, ATP2B2, and CASQ1), cardiac hypertrophy (GP130, JAK2, RRAS, MEK1, TWEAKR, and NPPB), stress response (HSPB1, HSPB6, HSPB7, IGF1, and IGF2), and structural organization of sarcomeres and mechanosensors (CSRP3 and TCAP).
Disturbed calcium signaling has been shown to be a central pathological mechanism of diastolic dysfunction in familial HCM lines with variations in the MYH7, MYBPC3, and TNNT2 genes[30]. Using comprehensive functional imaging analysis (i.e. calcium imaging and traction force microscopy), Wu et al[30] revealed that diastolic Ca2+ overload and increased myofilament Ca2+ sensitivity contribute to diastolic dysfunction and demonstrated for the first time that patient-specific iPSC-CMs can recapitulate diastolic dysfunction characteristics at the single-cell level. Furthermore, calcium homeostasis was restored by Ca2+ and late Na+ blockers (verapamil, dilitiazem, ranolazine, and electlazine), which was reflected in diastolic function improvement in HCM iPSC-CMs.
More recently, it has been shown that molecular signaling differs within HCM iPSC-CMs with diverse gene mutations. Isogenic models of HCM revealed differential phenotypes and mechanism-driven possible therapeutic targets in MYH7 and ACTC1 cell lines, respectively[31]. In spite of sharing key disease hallmarks, such as intracellular calcium overload and calcium transient arrhythmias, which were common in both models, modifications in contractility were entirely divergent, namely decreased contractility of MYH7 cells and gain of hypercontractility of ACTC1 cells. Notably, the expression of Ca2+-binding proteins and hypertrophy-associated transcription factor activation also showed opposing behavior. Accordingly, compared with their respective controls, MYH7-R453C mutants were characterized by upregulation of CASQ2, CALM1 and CAMK2D along with MEF and NFAT nuclear translocation prompted by IRF8 downregulation. Reversed expression patterns of those genes were found in ACTC1-E99K iPSC-CMs. The study offered clinically-relevant data, given that the arrhythmogenic phenotype was rescued in both models following treatment with a mixture of dantrolene and ranolazine (a ryanodine receptor antagonist that inhibits sarcoplasmic Ca2+ release into the cytosol and a late sodium current blocker promoting intracellular Ca2+ efflux). In addition, the enhanced contractility displayed in ACTC1 iPSC-CMs was rescued by mavacamten (a selective allosteric inhibitor of cardiac myosin ATPase). Of note, an earlier study reported differences in the phenotypic features of iPSC-CMs obtained from a family with ACTC1-E99K mutation. These differences varied depending on the source subject, highlighting the value of isogenic iPSC-CMs in genotype-phenotype correlations. Thus, while arrhythmogenesis was manifested in all ACTC1-E99K iPSC-CM lines, it was more common in cells obtained from the father, and less apparent in those derived from the two sons. This suggests that, in addition to the causative mutation, other factors (either genetic or epigenetic) might contribute to the disease phenotype[32].
The contractility consequences seem to be dependent on the specific DNA change rather than the affected gene. In contrast to the hypo-contractile phenotype described in MYH7-R453C mutants[31], MYH7-R403Q, MYH7-V606, and MYH7-R719 iPSC-CMs exhibited hyperdynamic contraction resulting from an increased proportion of myosin molecules in a disordered relaxed state conformation[33]. Imbalance of myosin configurations (i.e. super and disordered relaxed states) led to the destabilization of interacting-heads motif interactions that were followed, in addition to increased contractility, by impaired relaxation, hypertrophic remodeling, excessive energy consumption, and metabolic stress. Mavacamten treatment restored the physiological super relaxed state/disordered relaxed state ratio and relieved downstream functional abnormalities, suggesting that chronic dysregulation of myosin conformations is a central mechanism of HCM.
Other reports have used iPSCs to decode genetic DCM physiopathology. The most frequently mutated gene in DCM is TTN, with truncating variants explaining up to 30% of familial cases[34,35]. Other genes involved in DCM etiology are shown in Table 1. The most commonly encountered genes responsible for DCM are LMNA, DES, MYH7, MYH6, SCN5A, MYBPC3, and TNNT2, although at a lower prevalence than TTN[36]. Patient-derived iPSC-CMs have characteristics consistent with DCM, including sarcomere disorganization, reduced contractile function, and altered calcium handling[25,37-39]. Hinson et al[38] investigated the disease phenotypes of iPSCs engineered from DCM subjects with either truncating or missense mutations in TTN, and showing sarcomere insufficiency, impaired responses to mechanical and βadrenergic stress, and attenuated growth factor and cell signaling activation.
| Condition | Genotype | Ref. |
| HCM1 | MYBPC3, MYH7, TNNT2, TNNI3, TPM1, ACTC1, MYL2, MYL3 | [13] |
| DCM | TTN, LMNA, MYH7, TNNT2, BAG3, RBM20, TNNC1, TNNI3, TPM1, SCN5A, PLN | [14,15] |
| LVNC | Overlap with HCM and DCM | [14,15] |
| ACM | DES, DSC2, DSG2, DSP, JUP, LMNA, PKP2, PLN, RYR2, SCN5A, TMEM43, TTN | [14,15] |
| RCM | TTR, TNNI3, DES. Overlap with HCM and DCM | [14,15] |
In a series of studies that focused on TNNT2-R173W missense mutation, iPSCCMs lines generated from affected family members exhibited a compromised ability to regulate calcium flux, reduced contraction force, and heterogeneous myofilament organization that were exacerbated by βadrenergic stimulation[25]. In-depth analysis uncovered the epigenetic activation of phosphodiesterase genes PDE2A and PDE3A as the underlying mechanism responsible for the defective βadrenergic signaling and contractile dysfunction[40]. The latest data indicates that TNNT2-R173W hampers molecular interactions between troponin and tropomyosin and restricts the binding of PKA to local sarcomere microdomains, resulting in diminished troponin phospho-rylation and misalignment of sarcomeric proteins[41]. An R173W variant also altered the interaction between sarcomere microdomain and cytoskeleton filaments via MYH7 and AMPK, with consequent disturbance of sarcomere protein alignment and impaired contractility. In terms of phenotype rescue, AMPK activation by small molecules, such as A-769662, improved sarcomere-cytoskeleton attachment and partially recovered sarcomere protein misalignment and subsequent impaired contractility in mutated iPSC-CMs[41].
The phenotypes and the causal disease mechanisms linked to LMNA variants have also been intensively studied. In iPSC-CMs with either a nonsense or missense mutation in LMNA, Siu et al[39] noticed enhanced nuclear senescence and augmented electrical stress-induced apoptosis that was significantly attenuated by pharmacological inhibition of the ERK1/2 pathway with the MEK1/2 inhibitors U0126 and selumetinib. Other investigators analyzed iPSC-CMs from three patients with distinctive LMNA mutations (R225X, Q354, and T518fs)[42]. Although all three types of diseased cells recapitulated the pathophysiological hallmarks of LMNA-related DCM, the positive effect of ataluren treatment on the expression of full-length Lamin A/C, nuclear blebbing, apoptosis, and contractility was detected only in the LMNA-R225X mutant, suggesting that the effect might be codon selective[42]. Other traits expressed by LMNA-mutated iPSC-CMs were bradyarrhythmia, beat rate variability, abnormal calcium handling, stress hypersensitivity, disorganized sarcomeres, and increased cell size[43,44]. The incriminating mechanisms were aberrant activation of PDGF-signaling, which was rescued by pharmacological and molecular inhibition of PDGF receptor B[43], or epigenetic inhibition of SCN5A that was rescued by SCN5A overexpression[45]. Mutations in other DCM-related genes such as DES[37], MYH7[46], DMD[47,48], PLN[26], or even compound mutations have been modeled[49].
Characterization of iPSC-CMs from family members with LVNC who carried a nonsense variant in cardiac transcription factor TBX20, compared with iPSCCMs from unaffected relatives, revealed abnormal activation of transforming growth factor-β signaling leading to decreased cell proliferation capability. Moreover, inhibition of the transforming growth factor-β signaling pathway and correction of TBX20 alteration by CRISPR-Cas9 technology, successfully rectified the pathological phenotype[50].
Impairments in contractility, calcium handling, and metabolic activity have been nominated as key features in another model of iPSC-CMs generated from patients with delayed-onset LVNC caused by a missense mutation in the GATA4 gene[51].
In an iPSC model with a TPM1-R178H mutation, it was shown that mislocalization of tropomyosin 1 was a central pathological change, triggering disturbance of the sarcomere structure and impaired calcium handling. Comprehensive analysis found involvement of complex molecular pathways centered on downregulation of the expression of numerous genes controlling heart development and positive regulation of cellular processes, including transcription factors (GATA4, GATA6) and sarcomeric proteins (MYBPC3, MYH6, TTN, TNNI3, TNNT2), thus linking sarcomeric dysfunction to LVNC[52].
ACM is another inherited CMP studied in iPSC models. In a series of reports published in 2013, CMs engineered from subjects having mutations in the PKP2 gene efficiently recapitulated key disease features, including reduced cell surface localization of desmosomal proteins with altered desmosomal structure and a more adipogenic phenotype[53]. These phenotypical changes were accompanied by upregulation of the pro-adipogenic transcription factor peroxisome proliferator-activated receptor (PPAR)-γ and enhanced activation of respective signaling pathways[54,55]. Furthermore, lipid droplets accumulation was prevented by admini-stration of a specific inhibitor of glycogen synthase kinase 3β (6-bromoindirubin-3'-oxime)[55]. Subsequent work revealed novel mechanistic insights in ACM pathogenesis or confirmed those already cited. Wen et al[56] reported that coactivation of normal PPAR-α and abnormal PPAR-γ pathways in ACM iPSC-CMs triggered markedly increased lipogenesis, apoptosis, Na+ channel downregulation and defective intracellular calcium handling[56]. In another iPSC-based model it was found that RhoA/ROCK signaling at the intercalated disc was essential for cardiomyocyte homeostasis[57]. Using patient-derived iPSC-CMs with impaired cell-cell adhesion due to a PKP2 frameshift mutation, or disturbed RhoA signaling caused by a nonsense MYH10 mutation, Dorn et al[57] elegantly demonstrated that cardiomyocyte identity was lost following disruption of the RhoA/MRTF/SRF-signaling circuit. RhoA recruitment to cell-cell junctions was abridged in diseased cells, prompting increased levels of cytosolic G-actin and successive cytoplasmic sequestration of transcription factors such as MRTF that are involved in myocyte identity, preventing their entry into the nucleus. Finally, when exposed to an adipogenic environment, the mutated cells were poised to switch to a brown/beige adipocyte lineage, providing a possible molecular explanation of the clinical phenotype observed in ACM. Interestingly, a recent study reported lipid accumulation, increased pleomorphism, irregular Z-bands, and increased L-type calcium currents in iPSC-CMs carrying a novel frameshift mutation (L5218fs) in the OBSCN gene[58]. The phenotypic alterations were accompanied by activation of adipocytokines and PPAR signaling pathways, diminished expression of the mutant protein and its anchor protein Ank1.5, in addition to downregulation of other desmosomal coding genes (PKP2, JUP, DSP)[58].
IPSC-CMs generated from an ACM patient with a DSG2 mutation exhibited complex ion channel dysfunctions and abnormal cellular electrophysiology as well as increased sensitivity to adrenergic stimulation, indicating involvement of ion channel dysfunctions in arrhythmogenesis, independent of structural abnormalities[59]. Subsequent work conducted by the same group established for the first time that enhanced NDPK-B expression, via activating SK4 channels, contributed to arrhythmogenesis in DSG2-related ACM, suggesting that NDPK-B could be a specific therapeutic target in selected patients[60].
Ng et al[61] reported for the first time that some desmoplakin missense variants, such as DSP-R451G, are functionally equivalent to truncating alleles by promoting pathological vulnerability to calpain proteolysis and subsequent desmoplakin insufficiency.
There are few data related to restrictive cardiomyopathy modeling by iPSC. CMs harboring homozygous DES-Y122H mutation were reported to display abnormal desmin cytoplasmic aggregates responsible for the pathological phenotype[62].
A second category of genetic cardiac conditions extensively modeled using iPSC technology is represented by CNPs, for which electrophysiology studies exposed alterations of action potential, field potential, or Ca2+ transients in engineered CMs. LQTS, in particular the first three types (LQTS1, LQTS2 and LQTS3), benefit from the most well-characterized iPSC-CMs models. Types LQTS that differ according to the underlying channel or gene mutation are shown in Table 1.
The first model of an arrhythmic syndrome using patient-specific iPSC-CMs was reported in 2010 by Moretti et al[63], who generated iPSCs from two affected members of a family with LQTS1 caused by a missense mutation (R190Q) in the KCNQ1 gene. Mutant iPSC-CMs effectively reproduced the relevant features of the disease, namely prolongation of the action potential duration into atrial-like and ventricular-like cells, and increased occurrence of arrhythmic events when exposed to β-adrenergic agonists. Voltage clamp analysis revealed a substantial 70% to 80% reduction in the slowly activating delayed rectifier potassium currents (IKs) of LQTS1-iPSC-derived ventricular CMs due to a decreased number of functional KCNQ1 channels in the sarcolemma compared with the healthy counterpart. Beta-blocker treatment of LQTS1 CMs had a protective effect against catecholamine-induced arrhythmia. Similar findings have been reported in subsequent models of iPSC-CMs from LQTS1 with KCNQ1 missense or frameshift mutations[64,65]. Additionally, Wang et al[66] identified abnormalities in Ca2+ handling linked to three distinct KCNQ1 variants (R190Q, G269S, and G345E). CMs derived from all three edited iPSC lines displayed a characteristic LQTS phenotype and significant prolongation of the action potential duration compared with isogenic controls, which were corrected by treatment with L-type calcium channel antagonists. Similar results have been reported recently following an increase in the number of iPSC-CMs models of autosomal dominant, recessive, and compound heterozygous LQTS1[67-73], including analysis of models derived from specific populations[67,72]. A plethora of LQTS2 iPSC-CMs models developed from patients harboring missense mutations in KCNH2 have reproducibly shown prolonged action potential, increased arrhythmogenicity, and reduced rapidly activating delayed rectifier potassium current (IKr), compared with healthy control lines. The first analyzes of iPSC-based LQTS2 models were published in 2011 by two independent groups[74,75]. Itzhaki et al[74] generated iPSC lines from a patient with a KCNH2-A614V variant. As expected, the derived-CMs exhibited substantial prolongation of the action potential, diminished IKr, early after depolarizations (EADs), and triggered arrhythmias. Specific pharmacological inhibition of IKr worsened the cellular phenotype, while administration of other pharmacological agents such as Ca2+ channel blockers, or KATP-channel openers alleviated the pathological features[74]. The in vitro model developed by the second group effectively replicated the variation in clinical phenotypes of two family members carrying the same KCNH2-A561V mutation. Although the action potential duration was increased in the iPSC-CMs derived from both the clinically symptomatic patient and the clinically asymptomatic mother, an increased sensitivity (appearance of EADs) to stress (i.e. β-adrenoreceptor stimulation) was detected only in the symptomatic patient-derived cells[75]. By comparing the electrophysiological properties of spontaneously beating CMs produced from LQTS2 cases and controls, it was suggested that cell-to-cell contacts in the syncytium result in compensatory mechanisms with a tendency to protect the repolarization system from major aberrations of physiological parameters. Although a considerable signal difference was detected between LQTS2 and control iPSC-CMs on single-cell patch-clamp recordings (a 66% increase in action potential in LQTS2 cells), the differences were more modest (10%-20%) when using a microelectrode array technique on cell aggregates, similar to the surface electrocardiogram in respective patients[76]. In later studies, the diseased phenotype was rescued either by genetic correction of the KCNH2 mutation[77], or by allele-specific ribonucleic acid (RNA) interference, which selectively destroyed the mutant mRNA while leaving the wild-type mRNA undamaged[78]. Various pharmacological agents were also shown to correct the electrophysiological anomalies[79-81], although for some molecules the effect was mutation-specific[82]. An interesting observation was noted by Spencer et al[83], who established that in iPSC-CMs with a KCNH2-A422T mutation, the action potentials and intracellular calcium transients were prolonged in parallel. Furthermore, exposure to a Ca2+ antagonist such as nifedipine, abbreviated the action potentials despite the IKr deficit.
Although abnormal calcium handling is common in both LQTS1 and LQTS2, there are major differences in this regard. Joutsijoki et al[84] used an innovative approach to differentiate the Ca2+ transient statistics between these two LQTS-mutated iPSC-CMs. By combining machine learning and iPSC technology, the authors analyzed 90 LQTS1 transient signals from two cell lines and 138 LQTS2 signals from four cell lines, resulting in classification accuracies of up to 100%. The findings support the hypothesis that Ca2+ transients are disease-specific or even mutation-specific.
Patient-specific iPSC-CMs models harboring gain-of-function mutations in the SCN5A gene efficaciously summarized LQTS3 pathognomonic electrophysiological traits, such as abnormal sodium currents and prolonged APD[85-88]. A study by Malan at al[89] complemented prior findings by also showing a high incidence of EADs, a recognized trigger mechanism for arrhythmia, in disease cells. Treatment with mexiletine, specific sodium channel inhibitors, reduced action and field potential durations in LQT3 iPSC-CMs and alleviated EADs in a dose-dependent manner. Other types of LQTS, including LQTS7[90], LQTS8[91], LQTS14, and LQTS15[92-94] have been successfully investigated with iPSC technology.
CPVT comprises two main subtypes, CPTV1 (caused by mutations in the RYR2 gene) and CPTV2 (determined by mutations in the CASQ2 gene). Both genes are key regulators of CM calcium homeostasis, and dysfunction of either gene prompts abnormal intracellular Ca2+ handling and signaling, and increased arrhythmogenicity under adrenergic stimulation. To date, numerous CPVT models have been developed using the iPSC platform, successfully recapitulating the arrhythmogenic phenotype seen in patients[95-104]. In a study published in 2011, Fatima et al[95] analyzed the functional properties of iPSC-CMs from healthy donors and a patient with CPVT1 carrying a novel heterozygous autosomal dominant mutation (RYR2-F2483I). Compared with healthy CMs, the mutated cells displayed arrhythmias and delayed afterdepolarizations (DADs), higher amplitudes and longer duration of spontaneous Ca2+ release events in the basal state, as revealed by patch-clamp recordings and calcium imaging studies. Additionally, in CPVT-iPSC-CMs the Ca2+-induced Ca2+-release events continued after repolarization and were eliminated by increasing cytosolic cAMP levels with forskolin. In another CPVT1 model of iPSC-CMs harboring the RYR2-M4109R mutation, intracellular electrophysiological recordings evidenced increased development of DADs in CPVT-iPSCs-CMs compared with healthy CMs, which were further exacerbated by β-adrenergic stimulation; and, as opposed to previous findings, by forskolin. In contrast, thapsigargin (a specific inhibitor of the sarcoplasmic reticulum calcium ATPase pump) eradicated all afterdepolarizations in those cells, indicating that internal Ca2+ stores play a central role in the pathogenesis of DADs. Indeed, laser-confocal Ca2+ imaging revealed extensive whole-cell Ca2+ anomalies (such as frequent local and large-storage Ca2+-release events, broad and double-humped transients, and triggered activity) that were aggravated under catecholaminergic stress and alleviated by beta-blockers. Also, RYR2-M4109R mutations significantly reduced the threshold for store-overload-induced Ca2+ release[96]. Dantrolene[97], flecainide[105], and S107 (an RYR stabilizer[104]) were other pharmacological agents shown to ameliorate the disease phenotypes.
More recently, the combined iPSC and CRISPR/Cas9 gene editing technics were used to validate preliminary data and, more importantly, to gain further insight into dysfunction produced by variations in RYR2 gene. Wei et al[106], introduced a point mutation into wild-type RYR2 iPSCs by CRISPR/Cas9 gene editing. Similar to CMs generated from CPVT1 patient harboring F2483I-RyR2 mutation[101], edited iPSC-CMs carrying the same CPVT1-associated variant showed abnormal intracellular Ca2+ handling, including longer and wandering Ca2+ sparks, elevated diastolic Ca2+ leaks, reduced sarcoplasmic reticulum (SR) Ca2+ content, and increased susceptibility to arrhythmias caused by isoproterenol[106], suggesting that F2483I-RyR2 mutation produces leaky RyR2 channels. The same approach was used by Zhang et al[107] to assess aberrant Ca2+ signaling and pharmacological sensitivity to three distinct CPVT1-associated mutations. While all three diseased iPSC-CM lines exhibited some abnormalities in calcium handling (i.e. irregular, long-lasting, spatially wandering Ca2+ sparks and aberrant Ca2+ releases), enhanced SR Ca2+ leaks and diminished SR Ca2+ contents were seen only in cells with Q4201R and F2483I, but not R420Q. Moreover, fractional Ca2+ release and calcium-induced calcium release gain were higher in Q4201R than in R420Q and F2483I iPSC-CMs, emphasizing that Ca2+ signaling abnormalities may vary depending on the mutation site. Several potential therapeutic interventions, including flecainide, dantrolene, and JTV519 (a Ca2+-dependent blocker of SERCA) were tested, indicating that drug sensitivities may also be mutation dependent. Using a wide-ranging methodology integrating optogenetics, tissue engineering, lab-on-a-chip technology, gene editing, and iPSC technology, Park et al[108] identified calmodulin-dependent protein kinase II activation as a key molecular pathway underlying exercise-triggered arrhythmia in CPVT patients, suggesting that its inhibition might be an effective therapeutic strategy in selected cases.
Recent data indicate that RYR2 screening should not be indicated only in subjects with stress- or exercise-induced symptoms. Using patient-specific iPSC-CMs, it has been shown that RYR2-H29D variants elicit alteration of calcium homeostasis and molecular modifications such as aberrant SR Ca2+ leak under physiological pacing, pro-arrhythmic electrical phenotypes, impaired and asynchronous contractile properties, and aberrant RyR2 post-translational modifications that occur only at rest[109]. Furthermore, the authors hypothesized that the uncommon location of RYR2-H29D mutations outside the four hot-spot regions linked to CPVT1, might be responsible for the distinct phenotypic expression. iPSC-based platforms have also been used to explore functional abnormalities in CMs generated from CPVT2 patients[99,102,110,111]. Under beta-adrenergic stimulation, patient-derived iPSC-CMs carrying the CASQ2-D307H variant demonstrated disease-specific arrhythmogenic characteristics due to Ca2+-transient anomalies, afterdepolarization, reduced threshold for store overload-induced Ca2+-release, and upsurge of diastolic intracellular calcium concentration[99,102,110].
SQTS is a rare inheritable, autosomal dominant cardiac condition characterized by abnormally short QT intervals and an increased risk of atrial and ventricular tachyarrhythmias. The causal ion channel genes are shown in Table 2, variation in KCNH2 being the most frequently observed in genotyped cases[7,112]. The first SQTS model utilizing the iPSC platform was reported in 2018 by El-Battrawy et al[113]. The authors generated iPSC-CMs from a patient with a KCNH2-N588K mutation and two healthy control subjects. Mutated cardiac myocytes exhibited enhanced IKr density and shortened APD compared with control cells, along with abnormal calcium transients and arhythmic propensity. Carbachol, an acetylcholine receptor agonist, increased the occurrence of arrhythmic events in diseased iPSC-CMs, while quinidine, and not sotalol or metoprolol, prolonged the APD and alleviated carbachol-prompted arrhythmias. In subsequent studies, patient-specific and gene-corrected iPSC-CMs were used to elucidate the SQTS phenotype either at single-cell[114] or tissue level[115]. When compared with healthy control and gene-corrected CMs, KCNH2-T618I iPSC-CMs were shown to display shortened APDs and increased beat-beat interval variability. Although no significant differences in total KCNH2 expression in control, gene-corrected, and SQTS iPSC-CMs were seen, membrane expression of KCNH2 was approximately 8-fold higher in mutated iPSC-CMs than in isogenic cells, suggesting that the aforesaid variant results in enhanced membrane expression of KCNH2, which may contribute to the increased IKr density. Moreover, the phenotype was successfully rescued by BmKKx2, a short-peptide scorpion toxin acting as a selective IKr blocker[114]. Shinnawi et al[115] examined conduction and arrhythmogenesis in confluent 2-dimensional iPSC-derived cardiac cell monolayers generated from a symptomatic SQTS patient also with KCNH2-N588K mutation. SQTS-iPSC-CM monolayers were characterized by abnormal repolarization properties and induced sustained re-entrant arrhythmias, while retaining a normal conduction appearance.
| Condition | Genotype | Ref. |
| LQTS1 | KCNQ1 | [16] |
| LQTS2 | KCNH2 | [16] |
| LQTS3 | SCN5A | [16] |
| LQTS4 | ANK2 | [17] |
| LQTS5 | KCNE1 | [17] |
| LQTS6 | KCNE2 | [17] |
| LQTS7 | KCNJ2 | [17] |
| LQTS8 | CACNA1C | [17] |
| LQTS9 | CAV3 | [17] |
| LQTS10 | SCN4B | [17] |
| LQTS11 | AKAP9 | [17] |
| LQTS12 | SNTA1 | [17] |
| LQTS13 | KCNJ5 | [17] |
| LQTS14 | CALM1 | [17] |
| LQTS15 | CALM2 | [17] |
| JLN1 | KCNQ1 | [17] |
| JLN2 | KCNE1 | [17] |
| CPVT1 | RYR2 | [16] |
| CPVT2 | CASQ2 | [16] |
| SQTS | KCNH2, KCNQ1, KCNJ2 | [16] |
| BrS | SCN5A | [18] |
BrS is another cardiac channelopathy that has been modeled using iPSC technology. Various genes encoding either sodium, potassium, or calcium channels have been linked to BrS[116]. Among them, the SCN5A gene was found to be most commonly mutated (Table 2). That gene encodes the alpha subunit of the main cardiac sodium channel (Nav1.5); loss of function variants result in reduced availability of functional Nav1.5 channels either through decreased plasma membrane channel expression or through altered channel gating properties[117]. iPSC-CMs generated from BrS patients were shown to reflect the pro-arrhythmic phenotype associated with the disease and caused by blunted inward sodium currents, increased triggered activity, and calcium transient abnormalities. Davis et al[118] were the first to describe the molecular mechanisms that underlie BrS by using patient-specific iPSC-CMs harboring SCN5A_1795insD mutation, which effectively recapped the INa peak reduction and persistent INa associated with overlapped LQTS3/BrS[118]. Another group investigated sodium currents, action potentials and calcium dynamics in iPSC-CMs derived from patients with type 1 BrS carrying two different SCN5A variants and in healthy control subjects[119]. Mutated cardiac cells showed reductions in inward sodium current density, reduced maximal upstroke velocity of the action potential (AP), increased burden of triggered activity, abnormal calcium transients, and beating interval variation compared with control iPSC-CMs from healthy controls. Further analysis revealed markedly reduced expression of KCNJ2, which encodes Kir2.1 inwardly rectifying potassium channel, an observation not previously described in BrS. Correction of the causative variant by CRISPR/Cas9-mediated genome editing prompted efficient resolution of triggered activity and abnormal Ca2+ transients.
Additional data were provided by the work of Ma et al[120], who reported that a repolarization deficit was involved in BrS. By comparing electrophysiological properties of iPSC-CMs generated from a patient carrying a compound SCN5A mutation (A226V and R1629X) and a healthy sibling control, they observed an over 75% loss of sodium current in diseased cells. The decline in INa was reflected by altered action potential morphology with reduced maximum upstroke velocity and action potential amplitude at normal 1.0 Hz pacing frequency. At slow a slow pacing 0.1 Hz pacing frequency, an increased phase-1 repolarization action potential pattern characterized by a marked reduction of action potential duration and increased resting membrane potential occurred in a fraction of BrS CMs. Furthermore, disparities in the levels of transient outward K+ currents (Ito) among the iPSC-CMs from either compound carriers or healthy controls were noticed, with 19% to 23% of the studied cells displaying high Ito densities. Importantly, in patient-derived iPSC-CMs, treatment with 4-Aminopyridine, an Ito blocker, completely reversed the increased phase-1 repolarization and restored the APD, indicating a coordinated role of INa and Ito in the arrhythmogenic mechanism of BrS. In-depth analysis of iPSC-CMs derived from two BrS subjects with an SCN5A-S1812X variant revealed reduced INa, amplified Ito, and increased ICaL window current probability along with conduction slowing, demonstrating coexistence of repolarization and depolarization impairment in diseased cells[121].
At present, it is widely acknowledged that patient-specific genetic background is a key determinant of the phenotypical manifestation of BrS, as was reported by a team of researchers from Spain and United Kingdom[122]. As expected, iPSC-CMs from a patient with a SCN5A variation recapitulated the loss of function of Nav1.5 associated with the syndrome. Also, a shift in both activation and inactivation voltage-dependence curves and faster recovery from inactivation were reported. Remarkably, conventional heterologous expression systems (i.e. immortalized HEK293 cells co-expressing wild-type and mutant channels) failed to exhibit pro-arrhythmic changes in channel function, showing only a reduction in sodium current density, highlighting once again the need to assess the pathophysiological mechanisms of sodium channel mutations in a cardiac- and patient-specific model.
IPSC technology has also been used to model BrS caused by mutations in genes other than SCN5A. Cerrone et al[123] were the first to describe the association of BrS and genetic variation in PKP2. They analyzed iPSC-CMs derived from five index cases carrying missense mutations in PKP2 and perceived reduced sodium channel expression and current. The phenotype was rescued by transfection of wild-type PKP2, demonstrating that not only loss of PKP2, but also single amino acid mutations, can interfere with INa.
In the vast majority of clinically-diagnosed BrS cases (85%), the genetic cause is not known despite extensive use of NGS[124]. Few research groups have used iPSC technology to uncover disease mechanisms at the cellular level in phenotype-positive genotype-negative patients[125,126]. Notably, no clear cellular electrophysiological differences between the iPSC-CMs obtained from BrS patients without identified pathogenic mutations and control-derived cells were seen. That finding indicated that alternative pathophysiological mechanisms may be involved in those specific cases, such as right ventricular fibrosis or diminished cardiomyocyte coupling through gap junctions. Last but not least, BrS may be a multifactorial disorder, caused by an interaction of common genetic variations and environmental factors[125].
The right dose of the right drug for the right patient at the right time is not only the mantra of personalized or precision medicine, but a common challenge faced daily by clinicians all over the world[2,127]. With the advent of iPSC technology to guide therapeutic decisions in a patient-specific manner, tailoring treatment to a patient’s genetic background is yet to become a reality.
Prondzynski et al[128] employed patient-specific iPSC-CMs to define disease-related mechanisms and also to guide treatment in an HCM-affected family carrying a novel ACTN2 missense mutation[128]. Apart from previously described hallmarks of HCM, such as myofibrillar disarray, cell hypertrophy, increased myofilament Ca2+ sensitivity, hypercontractility, and prolonged relaxation, iPSC-CMs demonstrated enhanced L-type calcium channel current and prolonged action potential duration compared with isogenic controls. Following the beneficial results of improved contractile and electrophysiological in vitro phenotype with diltiazem, an L-type Ca2+ channel blocker, the findings were translated into clinical settings where standard-dose diltiazem reverted the LQT phenotype in the son and sister of the index patient.
Although still in early stages, patient-derived iPSCs have been shown to facilitate optimal treatment in arrhythmic disorders. In a stepwise study, Terrenoire et al[88] established a patient-specific therapeutic regimen in a LQTS child with complex genetics and only partially-controlled arrhythmia with high-dose mexiletine[88]. The index patient had a de novo mutation in the sodium channel SCN5A and a common polymorphism in the potassium channel KCNH2. First, electrophysiological analysis of the iPSC-CMs revealed that the SCN5A mutation was responsible for the patient’s symptoms. Furthermore, the authors found that mexiletine inhibited the IKr potassium channels in iPSC-CMs from both the father and the proband, irrespective of KCNH2 polymorphism, which explained the limited ability of mexiletine to completely correct the repolarization defect. Hence, alternative strategies to control INaL have been tested on patient-derived iPSC-CMs, such as changes in pacing rate or the addition of a second Na+ channel blocker. The experimental data recommended mexiletine alone and an increased pacemaker rate as the best therapeutic option, which was further confirmed by the patient’s clinical evolution. In another LQTS3 model, mexiletine rescued the abnormal electrophysiology in iPSC-CMs from a patient harboring a SCN5A mutation (p.V1763M)[87].
Specific drug screening using patient-derived iPSC models has also been performed in CPVT, where β-blockers are the drugs of choice, but often fail to avoid malignant arrhythmias. In symptomatic CPVT patients under standard β-blocker treatment, it was shown that individual-specific iPSC-CMs had a subadequate antiarrhythmic response to β-blockers, while both patient and iPSC-CMs responded more effectively to flecainide[105,110]. Clearly, the antiarrhythmic efficacy of different drugs is dependent on the underlying genetic variation. By patch-clamp analysis alone or by simultaneous patch-clamp and video imaging, Pölönen et al[129,130] assessed the antiarrhythmic effects of carvedilol and flecainide in CPVT patient-specific iPSC-CMs carrying diverse RYR2 variants. They found mutation-specific differences in arrhythmias and drug responses, suggesting that proper treatment may vary even among subjects with mutations in the same genes[129,130]. Evidence from earlier studies indicated that dantrolene was able to restore normal Ca2+ spark properties and rescue the arrhythmogenic phenotype in a patient-specific iPSC model[97]. Subsequent study revealed that not only that the location of the RYR2 mutation was critical for a favorable effect of dantrolene, but also suggested that the drug effect was dependent on the specific DNA alteration. Specifically, the antiarrhythmic effect was detected only in cases carrying mutations in the NH2-terminal or central regions of RYR2 protein. No effect was seen in subjects carrying mutations in the transmembrane region. Moreover, the effect of dantrolene was only minimal in iPSC-CMs with a Q4201R variant despite being located in the central region of RYR2 protein, even if at its end[131]. Indeed, the dantrolene binding site is located in the NH2-terminal region of RYR2 between amino acid 601 and 620. After specific binding, the drug restores normal channel gating and prevents uncontrolled Ca2+ release by stabilizing interdomain interactions between the NH2-terminal and central regions of RYR2, as previously reported[132,133].
Sequencing of wide-ranging gene panels by high-throughput techniques on a daily basis has increased the rate of positive genetic testing, and it has also increased the detection of variants of uncertain significance (VUS). Recently, our group reported the yield of DNA testing in a cohort of HCM probands[134,135]. Nearly half (45%) of the rare variants identified in our study were novel, and thus classified as of VUS. All but two were found only once in our cohort. Similar results were obtained in other studies, which reported a prevalence of 35%-40% of new mutations, half of which were unique for a family[136]. The conclusive classification of VUS is encumbered by challenges, particularly in cases of “private” mutations, as it involves computational and population-based studies, not rarely misleading[137,138]. Combined use of recent technologies such as iPSC and gene editing have enabled functional annotation in specific cases.
Lv et al[139] used a dual-integrase cassette exchange platform to rapidly and efficiently generate iPSCs with the TNNT2-E251D variant harbored by a woman with severe HCM and otherwise negative genetic testing. Although the mutation was generally predicted to be pathogenic by in silico analysis, the allele frequency of 0.03% in the Exome Aggregation Consortium database was inconsistent with the disease incidence (i.e. too high), and the ClinVar archive included conflicting interpretations of clinical significance, but mostly VUS. TNNT2-E251D iPSC-CMs had normal responses to isoproterenol, suggesting that the variant might not be pathogenic. To exclude the possibility that the failure in attaining a pathological phenotype was due to lack of a permissive genetic background in the studied cells, the authors introduced an E251D point mutation into an edited iPSC line known to be vulnerable to cardiomyopathy with CRISPR-Cas9. Comprehensive investigation of the E251D iPSC-CMs showed normal responses to isoproterenol and no significant increase in cell size or expression of genes previously reported to be upregulated in HCM iPSC-CMs (e.g., TNNT2, MYL2, MYL4, and MYH7). This approach allowed specific recommendations to be made to relatives, namely not to undergo cascade genetic screening for the E251D variant.
In another study, iPSC-CMs were produced from an asymptomatic subject with a HCM associated mutation in MYL3, and reported by the ClinVar database to be likely pathogenic[23]. Extensive assays, including measurement of gene expression, sarcomere structure, cell size, contractility, action potentials, and calcium handling, were performed on isogenic iPSC-CMs that were either corrected or carrying homozygous alleles found that the VUS was benign.
With the goal of functional prediction of pathogenicity, Pettinato et al[140] developed a scalable human cardiomyocyte platform to interrogate TNNT2 variants previously identified in the human population. Using iPSC-CMs in cardiac microtissue and single-cell assays, they examined 51 TNNT2 variants, including 30 pathogenic/likely pathogenic variants associated with HCM/DCM, and 21 VUS. Experimental evidence including transcriptomic changes and cardiac microtissue contraction, supported the reclassification of two pathogenic/likely pathogenic variants and two VUSs. These findings are of a great interest given that most TNNT2 variants identified in the human population are classified as of VUS. therefore definite reclassification would enable specific clinical decision making for individuals harboring these variants.
In a similar manner, iPSC models were used to decipher the pathogenicity of variants detected in patients with inherited CNPs. By combining patient-specific iPSCs and genome editing, Garg et al[141] demonstrated the pathogenicity of a novel VUS in the KCNH2 gene. Compared with healthy control cells, VUS iPSC-CMs displayed electrophysiological abnormalities consistent with LQTS2 phenotype (prolongation of action potential duration and reduced IKr density), which were rescued by VUS correction by CRISPR/Cas9. Furthermore, the introduction of the homozygous KCNH2-T983I variant in a healthy control line recapitulated the hallmark LQTS phenotype, confirming that the mutation was sufficient to prompt the disease.
Generation of iPSC lines from every single individual with a VUS in a CMP-/CNP-related gene, followed by allele correction, and functional assessment is laborious and virtually impossible. Hence alternative approaches exploiting already existing and functionally characterized human iPSC lines has been considered. For example, commercially available human iPSC-CMs were used to screen a KCNJ2 VUS detected in a LQTS7 proband by whole exome sequencing[142]. VUS overexpression was associated with a substantial prolongation of APD with evidence of arrhythmic activity, emulating the clinical phenotype, and thus supporting causality of the variant.
Chavali et al[143] established a patient-independent human iPSC model as a new tool for rapid determination of genetic variant pathogenicity in LQTS. The authors used CRISPR/Cas9 to introduce a CACNA1C VUS from an unrelated healthy volunteer into a previously established iPSC line. Functional changes detected in gene-edited iPSC-CMs allowed reclassification of CACNA1C-N639T variant to ‘’likely pathogenic”.
Considering all the available evidence, it can be easily seen that a screening platform based on edited human iPSC lines might be more informative than currently used procedures for variant classification, such as computational and population-based methods.
Due to various genetic and environmental modulatory factors, Mendelian CMPs and CNPs are characterized by variable expressivity and incomplete penetrance, which often delays the clinical management of such patients. One issue to be addressed by upcoming studies is whether iPSCs can be used to identify genetic modifiers and to unveil the protective or aggravating underlying regulatory mechanisms. As a proof of concept, Chai et al[144] used complementary physiological and genomic analyses to identify genetic modifiers explaining the variable expressivity observed in a large LQTS2 family.
Although the feasibility of this new technology for disease modeling and drug testing has been demonstrated, there are currently some limitations that should be addressed in order to further recommend the use of iPSC-CMs in clinical practice. Thus, the main setbacks in using this approach on a large scale are the reproducibility of results among multiple laboratories and the immature phenotype displayed by these cells.
The first is due to the use of various methods for inducing pluripotency, chromosomal instability throughout the reprogramming process and in vitro manipulation, the purity in myocyte composition, and batch disparities in differentiated CMs[145]. Therefore, implementation of standardized protocols for patient-specific lines is important. Second, most iPSC-CMs have an immature structural and functional phenotype, with fetal gene expression, disorganized sarcomeres, primarily relying on glycolysis, and having contractile features different from those of adult CMs, such as spontaneous beating[146]. Those properties could negatively impact the interpretation of the cellular responses to various drugs and the prediction of the clinical value of the respective compounds. Consequently, it is imperative to develop methods to generate CMs with a more mature phenotype in order to improve the predictive value of in vitro studies. Recently, important progress in the maturation of iPSC-CMs has been made by using small molecules[147], environmental manipulation[148] and three-dimensional culture[149]. IPSC-based research is still at an early stage. Nevertheless, one can undoubtedly see its boundless potential for advancing personalized clinical management of individuals with inherited CMPs and CNPs.
Manuscript source: Invited manuscript
Specialty type: Cardiac and cardiovascular systems
Country/Territory of origin: Romania
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Thummer RP, Wu HY S-Editor: Zhang L L-Editor: Filipodia P-Editor: Wang LL
| 1. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18157] [Article Influence: 955.6] [Reference Citation Analysis (0)] |
| 2. | Musunuru K, Sheikh F, Gupta RM, Houser SR, Maher KO, Milan DJ, Terzic A, Wu JC; American Heart Association Council on Functional Genomics and Translational Biology; Council on Cardiovascular Disease in the Young; and Council on Cardiovascular and Stroke Nursing. Induced Pluripotent Stem Cells for Cardiovascular Disease Modeling and Precision Medicine: A Scientific Statement From the American Heart Association. Circ Genom Precis Med. 2018;11:e000043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 145] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 3. | Popa-Fotea NM, Micheu MM, Bataila V, Scafa-Udriste A, Dorobantu L, Scarlatescu AI, Zamfir D, Stoian M, Onciul S, Dorobantu M. Exploring the Continuum of Hypertrophic Cardiomyopathy-From DNA to Clinical Expression. Medicina (Kaunas). 2019;55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 4. | Musunuru K, Arora P, Cooke JP, Ferguson JF, Hershberger RE, Hickey KT, Lee JM, Lima JAC, Loscalzo J, Pereira NL, Russell MW, Shah SH, Sheikh F, Wang TJ, MacRae CA; American Heart Association Council on Genomic and Precision Medicine; Council on Cardiovascular Disease in the Young; Council on Cardiovascular and Stroke Nursing; Council on Cardiovascular Radiology and Intervention; Council on Peripheral Vascular Disease; Council on Quality of Care and Outcomes Research; and Stroke Council. Interdisciplinary Models for Research and Clinical Endeavors in Genomic Medicine: A Scientific Statement From the American Heart Association. Circ Genom Precis Med. 2018;11:e000046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Micheu MM, Popescu I, Capatana CO, Barbarii LE, Dorobantu M. Molecular autopsy in sudden cardiac death – Ethical issues and clinical implication for relatives. Rom J Leg Med. 2016;24:157-163. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, Dubourg O, Kühl U, Maisch B, McKenna WJ, Monserrat L, Pankuweit S, Rapezzi C, Seferovic P, Tavazzi L, Keren A. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29:270-276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1766] [Cited by in RCA: 1889] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 7. | Fernández-Falgueras A, Sarquella-Brugada G, Brugada J, Brugada R, Campuzano O. Cardiac Channelopathies and Sudden Death: Recent Clinical and Genetic Advances. Biology (Basel). 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 8. | Garcia-Elias A, Benito B. Ion Channel Disorders and Sudden Cardiac Death. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Popa-Fotea NM, Cojocaru C, Scafa-Udriste A, Micheu MM, Dorobantu M. The Multifaced Perspectives of Genetic Testing in Pediatric Cardiomyopathies and Channelopathies. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 10. | Kline J, Costantini O. Inherited Cardiac Arrhythmias and Channelopathies. Med Clin North Am. 2019;103:809-820. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Wilde AAM, Nannenberg E, van der Werf C. Cardiogenetics, 25 years a growing subspecialism. Neth Heart J. 2020;28:39-43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 12. | Yogasundaram H, Alhumaid W, Dzwiniel T, Christian S, Oudit GY. Cardiomyopathies and Genetic Testing in Heart Failure: Role in Defining Phenotype-Targeted Approaches and Management. Can J Cardiol. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 13. | Ingles J, Goldstein J, Thaxton C, Caleshu C, Corty EW, Crowley SB, Dougherty K, Harrison SM, McGlaughon J, Milko LV, Morales A, Seifert BA, Strande N, Thomson K, Peter van Tintelen J, Wallace K, Walsh R, Wells Q, Whiffin N, Witkowski L, Semsarian C, Ware JS, Hershberger RE, Funke B. Evaluating the Clinical Validity of Hypertrophic Cardiomyopathy Genes. Circ Genom Precis Med. 2019;12:e002460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 304] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 14. | Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, Morales A, Taylor MRG, Vatta M, Ware SM. Genetic Evaluation of Cardiomyopathy-A Heart Failure Society of America Practice Guideline. J Card Fail. 2018;24:281-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 301] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 15. | Hershberger RE, Givertz MM, Ho CY, Judge DP, Kantor PF, McBride KL, Morales A, Taylor MRG, Vatta M, Ware SM; ACMG Professional Practice and Guidelines Committee. Genetic evaluation of cardiomyopathy: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2018;20:899-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 185] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 16. | Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le Marec H, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DP. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm. 2011;8:1308-1339. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 761] [Cited by in RCA: 763] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 17. | Schwartz PJ, Ackerman MJ, George AL Jr, Wilde AAM. Impact of genetics on the clinical management of channelopathies. J Am Coll Cardiol. 2013;62:169-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 236] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 18. | Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, Jamal SM, Szybowska M, Morel CF, Bowdin S, Garcia J, Care M, Sturm AC, Novelli V, Ackerman MJ, Ware JS, Hershberger RE, Wilde AAM, Gollob MH; National Institutes of Health Clinical Genome Resource Consortium. Reappraisal of Reported Genes for Sudden Arrhythmic Death: Evidence-Based Evaluation of Gene Validity for Brugada Syndrome. Circulation. 2018;138:1195-1205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 262] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 19. | Eschenhagen T, Carrier L. Cardiomyopathy phenotypes in human-induced pluripotent stem cell-derived cardiomyocytes-a systematic review. Pflugers Arch. 2019;471:755-768. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 20. | Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 472] [Cited by in RCA: 520] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 21. | Han L, Li Y, Tchao J, Kaplan AD, Lin B, Mich-Basso J, Lis A, Hassan N, London B, Bett GC, Tobita K, Rasmusson RL, Yang L. Study familial hypertrophic cardiomyopathy using patient-specific induced pluripotent stem cells. Cardiovasc Res. 2014;104:258-269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 158] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 22. | Prondzynski M, Krämer E, Laufer SD, Shibamiya A, Pless O, Flenner F, Müller OJ, Münch J, Redwood C, Hansen A, Patten M, Eschenhagen T, Mearini G, Carrier L. Evaluation of MYBPC3 trans-Splicing and Gene Replacement as Therapeutic Options in Human iPSC-Derived Cardiomyocytes. Mol Ther Nucleic Acids. 2017;7:475-486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 23. | Ma N, Zhang JZ, Itzhaki I, Zhang SL, Chen H, Haddad F, Kitani T, Wilson KD, Tian L, Shrestha R, Wu H, Lam CK, Sayed N, Wu JC. Determining the Pathogenicity of a Genomic Variant of Uncertain Significance Using CRISPR/Cas9 and Human-Induced Pluripotent Stem Cells. Circulation. 2018;138:2666-2681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 24. | Mosqueira D, Mannhardt I, Bhagwan JR, Lis-Slimak K, Katili P, Scott E, Hassan M, Prondzynski M, Harmer SC, Tinker A, Smith JGW, Carrier L, Williams PM, Gaffney D, Eschenhagen T, Hansen A, Denning C. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. Eur Heart J. 2018;39:3879-3892. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 177] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 25. | Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Pavlovic A, Lin S, Chen R, Hajjar RJ, Snyder MP, Dolmetsch RE, Butte MJ, Ashley EA, Longaker MT, Robbins RC, Wu JC. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 535] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 26. | Karakikes I, Stillitano F, Nonnenmacher M, Tzimas C, Sanoudou D, Termglinchan V, Kong CW, Rushing S, Hansen J, Ceholski D, Kolokathis F, Kremastinos D, Katoulis A, Ren L, Cohen N, Gho JMIH, Tsiapras D, Vink A, Wu JC, Asselbergs FW, Li RA, Hulot JS, Kranias EG, Hajjar RJ. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 128] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 27. | Judge LM, Perez-Bermejo JA, Truong A, Ribeiro AJ, Yoo JC, Jensen CL, Mandegar MA, Huebsch N, Kaake RM, So PL, Srivastava D, Pruitt BL, Krogan NJ, Conklin BR. A BAG3 chaperone complex maintains cardiomyocyte function during proteotoxic stress. JCI Insight. 2017;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 28. | Wang L, Kim K, Parikh S, Cadar AG, Bersell KR, He H, Pinto JR, Kryshtal DO, Knollmann BC. Hypertrophic cardiomyopathy-linked mutation in troponin T causes myofibrillar disarray and pro-arrhythmic action potential changes in human iPSC cardiomyocytes. J Mol Cell Cardiol. 2018;114:320-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 29. | Seeger T, Shrestha R, Lam CK, Chen C, McKeithan WL, Lau E, Wnorowski A, McMullen G, Greenhaw M, Lee J, Oikonomopoulos A, Lee S, Yang H, Mercola M, Wheeler M, Ashley EA, Yang F, Karakikes I, Wu JC. A Premature Termination Codon Mutation in MYBPC3 Causes Hypertrophic Cardiomyopathy via Chronic Activation of Nonsense-Mediated Decay. Circulation. 2019;139:799-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 30. | Wu H, Yang H, Rhee JW, Zhang JZ, Lam CK, Sallam K, Chang ACY, Ma N, Lee J, Zhang H, Blau HM, Bers DM, Wu JC. Modelling diastolic dysfunction in induced pluripotent stem cell-derived cardiomyocytes from hypertrophic cardiomyopathy patients. Eur Heart J. 2019;40:3685-3695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 31. | Bhagwan JR, Mosqueira D, Chairez-Cantu K, Mannhardt I, Bodbin SE, Bakar M, Smith JGW, Denning C. Isogenic models of hypertrophic cardiomyopathy unveil differential phenotypes and mechanism-driven therapeutics. J Mol Cell Cardiol. 2020;145:43-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 32. | Smith JGW, Owen T, Bhagwan JR, Mosqueira D, Scott E, Mannhardt I, Patel A, Barriales-Villa R, Monserrat L, Hansen A, Eschenhagen T, Harding SE, Marston S, Denning C. Isogenic Pairs of hiPSC-CMs with Hypertrophic Cardiomyopathy/LVNC-Associated ACTC1 E99K Mutation Unveil Differential Functional Deficits. Stem Cell Reports. 2018;11:1226-1243. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 33. | Toepfer CN, Garfinkel AC, Venturini G, Wakimoto H, Repetti G, Alamo L, Sharma A, Agarwal R, Ewoldt JF, Cloonan P, Letendre J, Lun M, Olivotto I, Colan S, Ashley E, Jacoby D, Michels M, Redwood CS, Watkins HC, Day SM, Staples JF, Padrón R, Chopra A, Ho CY, Chen CS, Pereira AC, Seidman JG, Seidman CE. Myosin Sequestration Regulates Sarcomere Function, Cardiomyocyte Energetics, and Metabolism, Informing the Pathogenesis of Hypertrophic Cardiomyopathy. Circulation. 2020;141:828-842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 195] [Cited by in RCA: 208] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 34. | Gerull B, Gramlich M, Atherton J, McNabb M, Trombitás K, Sasse-Klaassen S, Seidman JG, Seidman C, Granzier H, Labeit S, Frenneaux M, Thierfelder L. Mutations of TTN, encoding the giant muscle filament titin, cause familial dilated cardiomyopathy. Nat Genet. 2002;30:201-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 425] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 35. | Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1072] [Cited by in RCA: 1053] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 36. | Hershberger RE, Morales A. Dilated Cardiomyopathy Overview. 2007 Jul 27 [updated 2018 Aug 23]. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2021. |
| 37. | Tse HF, Ho JC, Choi SW, Lee YK, Butler AW, Ng KM, Siu CW, Simpson MA, Lai WH, Chan YC, Au KW, Zhang J, Lay KW, Esteban MA, Nicholls JM, Colman A, Sham PC. Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing. Hum Mol Genet. 2013;22:1395-1403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 38. | Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, Gorham J, Yang L, Schafer S, Sheng CC, Haghighi A, Homsy J, Hubner N, Church G, Cook SA, Linke WA, Chen CS, Seidman JG, Seidman CE. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 456] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 39. | Siu CW, Lee YK, Ho JC, Lai WH, Chan YC, Ng KM, Wong LY, Au KW, Lau YM, Zhang J, Lay KW, Colman A, Tse HF. Modeling of lamin A/C mutation premature cardiac aging using patient‐specific induced pluripotent stem cells. Aging (Albany NY). 2012;4:803-822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 40. | Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, Churko JM, Sallam KI, Matsa E, Sharma A, Gold JD, Engler AJ, Xiang YK, Bers DM, Wu JC. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised β-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell Stem Cell. 2015;17:89-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 41. | Dai Y, Amenov A, Ignatyeva N, Koschinski A, Xu H, Soong PL, Tiburcy M, Linke WA, Zaccolo M, Hasenfuss G, Zimmermann WH, Ebert A. Troponin destabilization impairs sarcomere-cytoskeleton interactions in iPSC-derived cardiomyocytes from dilated cardiomyopathy patients. Sci Rep. 2020;10:209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 42. | Lee YK, Lau YM, Cai ZJ, Lai WH, Wong LY, Tse HF, Ng KM, Siu CW. Modeling Treatment Response for Lamin A/C Related Dilated Cardiomyopathy in Human Induced Pluripotent Stem Cells. J Am Heart Assoc. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 43. | Lee J, Termglinchan V, Diecke S, Itzhaki I, Lam CK, Garg P, Lau E, Greenhaw M, Seeger T, Wu H, Zhang JZ, Chen X, Gil IP, Ameen M, Sallam K, Rhee JW, Churko JM, Chaudhary R, Chour T, Wang PJ, Snyder MP, Chang HY, Karakikes I, Wu JC. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 2019;572:335-340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 44. | Shah D, Virtanen L, Prajapati C, Kiamehr M, Gullmets J, West G, Kreutzer J, Pekkanen-Mattila M, Heliö T, Kallio P, Taimen P, Aalto-Setälä K. Modeling of LMNA-Related Dilated Cardiomyopathy Using Human Induced Pluripotent Stem Cells. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 45. | Salvarani N, Crasto S, Miragoli M, Bertero A, Paulis M, Kunderfranco P, Serio S, Forni A, Lucarelli C, Dal Ferro M, Larcher V, Sinagra G, Vezzoni P, Murry CE, Faggian G, Condorelli G, Di Pasquale E. The K219T-Lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat Commun. 2019;10:2267. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 46. | Yang KC, Breitbart A, De Lange WJ, Hofsteen P, Futakuchi-Tsuchida A, Xu J, Schopf C, Razumova MV, Jiao A, Boucek R, Pabon L, Reinecke H, Kim DH, Ralphe JC, Regnier M, Murry CE. Novel Adult-Onset Systolic Cardiomyopathy Due to MYH7 E848G Mutation in Patient-Derived Induced Pluripotent Stem Cells. JACC Basic Transl Sci. 2018;3:728-740. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 47. | Lin B, Li Y, Han L, Kaplan AD, Ao Y, Kalra S, Bett GC, Rasmusson RL, Denning C, Yang L. Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy. Dis Model Mech. 2015;8:457-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 97] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 48. | Dick E, Kalra S, Anderson D, George V, Ritso M, Laval SH, Barresi R, Aartsma-Rus A, Lochmüller H, Denning C. Exon skipping and gene transfer restore dystrophin expression in human induced pluripotent stem cells-cardiomyocytes harboring DMD mutations. Stem Cells Dev. 2013;22:2714-2724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 49. | Deacon DC, Happe CL, Chen C, Tedeschi N, Manso AM, Li T, Dalton ND, Peng Q, Farah EN, Gu Y, Tenerelli KP, Tran VD, Chen J, Peterson KL, Schork NJ, Adler ED, Engler AJ, Ross RS, Chi NC. Combinatorial interactions of genetic variants in human cardiomyopathy. Nat Biomed Eng. 2019;3:147-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 50. | Kodo K, Ong SG, Jahanbani F, Termglinchan V, Hirono K, InanlooRahatloo K, Ebert AD, Shukla P, Abilez OJ, Churko JM, Karakikes I, Jung G, Ichida F, Wu SM, Snyder MP, Bernstein D, Wu JC. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol. 2016;18:1031-1042. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 145] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 51. | Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, Tippens ND, Li M, Narasimha A, Radzinsky E, Moon-Grady AJ, Yu H, Pruitt BL, Snyder MP, Srivastava D. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 2016; 167: 1734-1749. e22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 191] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 52. | Takasaki A, Hirono K, Hata Y, Wang C, Takeda M, Yamashita JK, Chang B, Nakaoka H, Okabe M, Miyao N, Saito K, Ibuki K, Ozawa S, Sekine M, Yoshimura N, Nishida N, Bowles NE, Ichida F. Sarcomere gene variants act as a genetic trigger underlying the development of left ventricular noncompaction. Pediatr Res. 2018;84:733-742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 53. | Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, Oh Y, Tan SH, Ng ML, Shim W, Wong P, Liew R. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2013;34:1122-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 156] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 54. | Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, Kan NG, Forcales S, Puri PL, Leone TC, Marine JE, Calkins H, Kelly DP, Judge DP, Chen HS. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105-110. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 388] [Cited by in RCA: 412] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 55. | Caspi O, Huber I, Gepstein A, Arbel G, Maizels L, Boulos M, Gepstein L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet. 2013;6:557-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 56. | Wen JY, Wei CY, Shah K, Wong J, Wang C, Chen HS. Maturation-Based Model of Arrhythmogenic Right Ventricular Dysplasia Using Patient-Specific Induced Pluripotent Stem Cells. Circ J. 2015;79:1402-1408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 57. | Dorn T, Kornherr J, Parrotta EI, Zawada D, Ayetey H, Santamaria G, Iop L, Mastantuono E, Sinnecker D, Goedel A, Dirschinger RJ, My I, Laue S, Bozoglu T, Baarlink C, Ziegler T, Graf E, Hinkel R, Cuda G, Kääb S, Grace AA, Grosse R, Kupatt C, Meitinger T, Smith AG, Laugwitz KL, Moretti A. Interplay of cell-cell contacts and RhoA/MRTF-A signaling regulates cardiomyocyte identity. EMBO J. 2018;37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 58. | Chen P, Xiao Y, Wang Y, Zheng Z, Chen L, Yang X, Li J, Wu W, Zhang S. Intracellular calcium current disorder and disease phenotype in OBSCN mutant iPSC-based cardiomyocytes in arrhythmogenic right ventricular cardiomyopathy. Theranostics. 2020;10:11215-11229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | El-Battrawy I, Zhao Z, Lan H, Cyganek L, Tombers C, Li X, Buljubasic F, Lang S, Tiburcy M, Zimmermann WH, Utikal J, Wieland T, Borggrefe M, Zhou XB, Akin I. Electrical dysfunctions in human-induced pluripotent stem cell-derived cardiomyocytes from a patient with an arrhythmogenic right ventricular cardiomyopathy. Europace. 2018;20:f46-f56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 60. | Buljubasic F, El-Battrawy I, Lan H, Lomada SK, Chatterjee A, Zhao Z, Li X, Zhong R, Xu Q, Huang M, Liao Z, Lang S, Cyganek L, Zhou X, Wieland T, Borggrefe M, Akin I. Nucleoside Diphosphate Kinase B Contributes to Arrhythmogenesis in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes from a Patient with Arrhythmogenic Right Ventricular Cardiomyopathy. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 61. | Ng R, Manring H, Papoutsidakis N, Albertelli T, Tsai N, See CJ, Li X, Park J, Stevens TL, Bobbili PJ, Riaz M, Ren Y, Stoddard CE, Janssen PM, Bunch TJ, Hall SP, Lo YC, Jacoby DL, Qyang Y, Wright N, Ackermann MA, Campbell SG. Patient mutations linked to arrhythmogenic cardiomyopathy enhance calpain-mediated desmoplakin degradation. JCI Insight. 2019;5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 62. | Brodehl A, Pour Hakimi SA, Stanasiuk C, Ratnavadivel S, Hendig D, Gaertner A, Gerull B, Gummert J, Paluszkiewicz L, Milting H. Restrictive Cardiomyopathy is Caused by a Novel Homozygous Desmin (DES) Mutation p.Y122H Leading to a Severe Filament Assembly Defect. Genes (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 63. | Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Seyfarth M, Sinnecker D, Schömig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 927] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 64. | Egashira T, Yuasa S, Suzuki T, Aizawa Y, Yamakawa H, Matsuhashi T, Ohno Y, Tohyama S, Okata S, Seki T, Kuroda Y, Yae K, Hashimoto H, Tanaka T, Hattori F, Sato T, Miyoshi S, Takatsuki S, Murata M, Kurokawa J, Furukawa T, Makita N, Aiba T, Shimizu W, Horie M, Kamiya K, Kodama I, Ogawa S, Fukuda K. Disease characterization using LQTS-specific induced pluripotent stem cells. Cardiovasc Res. 2012;95:419-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 65. | Ma D, Wei H, Lu J, Huang D, Liu Z, Loh LJ, Islam O, Liew R, Shim W, Cook SA. Characterization of a novel KCNQ1 mutation for type 1 Long QT syndrome and assessment of the therapeutic potential of a novel IKs activator using patient-specific induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Res Ther. 2015;6:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 66. | Wang Y, Liang P, Lan F, Wu H, Lisowski L, Gu M, Hu S, Kay MA, Urnov FD, Shinnawi R, Gold JD, Gepstein L, Wu JC. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014;64:451-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 67. | Wang Z, Wang L, Liu W, Hu D, Gao Y, Ge Q, Liu X, Li L, Wang Y, Wang S, Li C. Pathogenic mechanism and gene correction for LQTS-causing double mutations in KCNQ1 using a pluripotent stem cell model. Stem Cell Res. 2019;38:101483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 68. | Ge N, Liu M, Krawczyk J, McInerney V, Galvin J, Shen S, O'Brien T, Prendiville T. Generation of eight human induced pluripotent stem cell (iPSC) lines from familial Long QT Syndrome type 1 (LQT1) patients carrying KCNQ1 c.1697C>A mutation (NUIGi005-A, NUIGi005-B, NUIGi005-C, NUIGi006-A, NUIGi006-B, NUIGi006-C, NUIGi007-A, and NUIGi007-B). Stem Cell Res. 2019;39:101502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 69. | Ge N, Liu M, Ding Y, Krawczyk J, McInerney V, Galvin J, Shen S, Prendiville T, O'Brien T. Generation and characterization of twelve human induced pluripotent stem cell (iPSC) lines from four familial long QT syndrome type 1 (LQT1) patients carrying KCNQ1 c.1201dupC mutation. Stem Cell Res. 2019;41:101650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 70. | Mura M, Lee YK, Pisano F, Ginevrino M, Boni M, Calabrò F, Crotti L, Valente EM, Schwartz PJ, Tse HF, Gnecchi M. Generation of the human induced pluripotent stem cell (hiPSC) line PSMi004-A from a carrier of the KCNQ1-R594Q mutation. Stem Cell Res. 2019;37:101431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 71. | Mura M, Lee YK, Pisano F, Ginevrino M, Boni M, Calabrò F, Crotti L, Valente EM, Schwartz PJ, Tse HF, Gnecchi M. Generation of the human induced pluripotent stem cell (hiPSC) line PSMi005-A from a patient carrying the KCNQ1-R190W mutation. Stem Cell Res. 2019;37:101437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 72. | Mura M, Pisano F, Stefanello M, Ginevrino M, Boni M, Calabrò F, Crotti L, Valente EM, Schwartz PJ, Brink PA, Gnecchi M. Generation of two human induced pluripotent stem cell (hiPSC) lines from a long QT syndrome South African founder population. Stem Cell Res. 2019;39:101510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 73. | Mura M, Bastaroli F, Corli M, Ginevrino M, Calabrò F, Boni M, Crotti L, Valente EM, Schwartz PJ, Gnecchi M. Generation of the human induced pluripotent stem cell (hiPSC) line PSMi006-A from a patient affected by an autosomal recessive form of long QT syndrome type 1. Stem Cell Res. 2020;42:101658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 74. | Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 794] [Cited by in RCA: 777] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 75. | Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, Denning C. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32:952-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 328] [Cited by in RCA: 285] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 76. | Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkelä E, Hyttinen J, Kontula K, Swan H, Conklin BR, Yamanaka S, Silvennoinen O, Aalto-Setälä K. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 216] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 77. | Bellin M, Casini S, Davis RP, D'Aniello C, Haas J, Ward-van Oostwaard D, Tertoolen LG, Jung CB, Elliott DA, Welling A, Laugwitz KL, Moretti A, Mummery CL. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32:3161-3175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 153] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 78. | Matsa E, Dixon JE, Medway C, Georgiou O, Patel MJ, Morgan K, Kemp PJ, Staniforth A, Mellor I, Denning C. Allele-specific RNA interference rescues the long-QT syndrome phenotype in human-induced pluripotency stem cell cardiomyocytes. Eur Heart J. 2014;35:1078-1087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 79. | Smith JL, Anderson CL, Burgess DE, Elayi CS, January CT, Delisle BP. Molecular pathogenesis of long QT syndrome type 2. J Arrhythm. 2016;32:373-380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 80. | Schwartz PJ, Gnecchi M, Dagradi F, Castelletti S, Parati G, Spazzolini C, Sala L, Crotti L. From patient-specific induced pluripotent stem cells to clinical translation in long QT syndrome Type 2. Eur Heart J. 2019;40:1832-1836. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 81. | Perry MD, Ng CA, Mangala MM, Ng TYM, Hines AD, Liang W, Xu MJO, Hill AP, Vandenberg JI. Pharmacological activation of IKr in models of long QT Type 2 risks overcorrection of repolarization. Cardiovasc Res. 2020;116:1434-1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 82. | Mehta A, Ramachandra CJA, Singh P, Chitre A, Lua CH, Mura M, Crotti L, Wong P, Schwartz PJ, Gnecchi M, Shim W. Identification of a targeted and testable antiarrhythmic therapy for long-QT syndrome type 2 using a patient-specific cellular model. Eur Heart J. 2018;39:1446-1455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 83. | Spencer CI, Baba S, Nakamura K, Hua EA, Sears MA, Fu CC, Zhang J, Balijepalli S, Tomoda K, Hayashi Y, Lizarraga P, Wojciak J, Scheinman MM, Aalto-Setälä K, Makielski JC, January CT, Healy KE, Kamp TJ, Yamanaka S, Conklin BR. Calcium transients closely reflect prolonged action potentials in iPSC models of inherited cardiac arrhythmia. Stem Cell Reports. 2014;3:269-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 84. | Joutsijoki H, Penttinen K, Juhola M, Aalto-Setälä K. Separation of HCM and LQT Cardiac Diseases with Machine Learning of Ca2+ Transient Profiles. Methods Inf Med. 2019;58:167-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 85. | Fatima A, Kaifeng S, Dittmann S, Xu G, Gupta MK, Linke M, Zechner U, Nguemo F, Milting H, Farr M, Hescheler J, Sarić T. The disease-specific phenotype in cardiomyocytes derived from induced pluripotent stem cells of two long QT syndrome type 3 patients. PLoS One. 2013;8:e83005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 86. | Navarrete EG, Liang P, Lan F, Sanchez-Freire V, Simmons C, Gong T, Sharma A, Burridge PW, Patlolla B, Lee AS, Wu H, Beygui RE, Wu SM, Robbins RC, Bers DM, Wu JC. Screening drug-induced arrhythmia [corrected] using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation. 2013;128:S3-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 233] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 87. | Ma D, Wei H, Zhao Y, Lu J, Li G, Sahib NB, Tan TH, Wong KY, Shim W, Wong P, Cook SA, Liew R. Modeling type 3 Long QT syndrome with cardiomyocytes derived from patient-specific induced pluripotent stem cells. Int J Cardiol. 2013;168:5277-5286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 123] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 88. | Terrenoire C, Wang K, Tung KW, Chung WK, Pass RH, Lu JT, Jean JC, Omari A, Sampson KJ, Kotton DN, Keller G, Kass RS. Induced pluripotent stem cells used to reveal drug actions in a long QT syndrome family with complex genetics. J Gen Physiol. 2013;141:61-72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 89. | Malan D, Zhang M, Stallmeyer B, Müller J, Fleischmann BK, Schulze-Bahr E, Sasse P, Greber B. Human iPS cell model of type 3 Long QT syndrome recapitulates drug-based phenotype correction. Basic Res Cardiol. 2016;111:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 90. | Kuroda Y, Yuasa S, Watanabe Y, Ito S, Egashira T, Seki T, Hattori T, Ohno S, Kodaira M, Suzuki T, Hashimoto H, Okata S, Tanaka A, Aizawa Y, Murata M, Aiba T, Makita N, Furukawa T, Shimizu W, Kodama I, Ogawa S, Kokubun N, Horigome H, Horie M, Kamiya K, Fukuda K. Flecainide ameliorates arrhythmogenicity through NCX flux in Andersen-Tawil syndrome-iPS cell-derived cardiomyocytes. Biochem Biophys Rep. 2017;9:245-256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 91. | Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230-234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 567] [Cited by in RCA: 499] [Article Influence: 35.6] [Reference Citation Analysis (0)] |
| 92. | Limpitikul WB, Dick IE, Tester DJ, Boczek NJ, Limphong P, Yang W, Choi MH, Babich J, DiSilvestre D, Kanter RJ, Tomaselli GF, Ackerman MJ, Yue DT. A Precision Medicine Approach to the Rescue of Function on Malignant Calmodulinopathic Long-QT Syndrome. Circ Res. 2017;120:39-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 127] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 93. | Rocchetti M, Sala L, Dreizehnter L, Crotti L, Sinnecker D, Mura M, Pane LS, Altomare C, Torre E, Mostacciuolo G, Severi S, Porta A, De Ferrari GM, George AL Jr, Schwartz PJ, Gnecchi M, Moretti A, Zaza A. Elucidating arrhythmogenic mechanisms of long-QT syndrome CALM1-F142L mutation in patient-specific induced pluripotent stem cell-derived cardiomyocytes. Cardiovasc Res. 2017;113:531-541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 94. | Yamamoto Y, Makiyama T, Harita T, Sasaki K, Wuriyanghai Y, Hayano M, Nishiuchi S, Kohjitani H, Hirose S, Chen J, Yokoi F, Ishikawa T, Ohno S, Chonabayashi K, Motomura H, Yoshida Y, Horie M, Makita N, Kimura T. Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum Mol Genet. 2017;26:1670-1677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 95. | Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnáiz-Cot JJ, Rosa AO, Nguemo F, Matzkies M, Dittmann S, Stone SL, Linke M, Zechner U, Beyer V, Hennies HC, Rosenkranz S, Klauke B, Parwani AS, Haverkamp W, Pfitzer G, Farr M, Cleemann L, Morad M, Milting H, Hescheler J, Saric T. In vitro modeling of ryanodine receptor 2 dysfunction using human induced pluripotent stem cells. Cell Physiol Biochem. 2011;28:579-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 96. | Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, Miller L, Belhassen B, Nof E, Glikson M, Gepstein L. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012;60:990-1000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 97. | Jung CB, Moretti A, Mederos y Schnitzler M, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, Dirschinger RJ, Seyfarth M, Lam JT, Sinnecker D, Gudermann T, Lipp P, Laugwitz KL. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Mol Med. 2012;4:180-191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 249] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 98. | Kujala K, Paavola J, Lahti A, Larsson K, Pekkanen-Mattila M, Viitasalo M, Lahtinen AM, Toivonen L, Kontula K, Swan H, Laine M, Silvennoinen O, Aalto-Setälä K. Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed afterdepolarizations. PLoS One. 2012;7:e44660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 99. | Novak A, Barad L, Zeevi-Levin N, Shick R, Shtrichman R, Lorber A, Itskovitz-Eldor J, Binah O. Cardiomyocytes generated from CPVTD307H patients are arrhythmogenic in response to β-adrenergic stimulation. J Cell Mol Med. 2012;16:468-482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 133] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 100. | Di Pasquale E, Lodola F, Miragoli M, Denegri M, Avelino-Cruz JE, Buonocore M, Nakahama H, Portararo P, Bloise R, Napolitano C, Condorelli G, Priori SG. CaMKII inhibition rectifies arrhythmic phenotype in a patient-specific model of catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis. 2013;4:e843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 101. | Zhang XH, Haviland S, Wei H, Sarić T, Fatima A, Hescheler J, Cleemann L, Morad M. Ca2+ signaling in human induced pluripotent stem cell-derived cardiomyocytes (iPS-CM) from normal and catecholaminergic polymorphic ventricular tachycardia (CPVT)-afflicted subjects. Cell Calcium. 2013;54:57-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 102. | Novak A, Barad L, Lorber A, Gherghiceanu M, Reiter I, Eisen B, Eldor L, Itskovitz-Eldor J, Eldar M, Arad M, Binah O. Functional abnormalities in iPSC-derived cardiomyocytes generated from CPVT1 and CPVT2 patients carrying ryanodine or calsequestrin mutations. J Cell Mol Med. 2015;19:2006-2018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 103. | Lodola F, Morone D, Denegri M, Bongianino R, Nakahama H, Rutigliano L, Gosetti R, Rizzo G, Vollero A, Buonocore M, Napolitano C, Condorelli G, Priori SG, Di Pasquale E. Adeno-associated virus-mediated CASQ2 delivery rescues phenotypic alterations in a patient-specific model of recessive catecholaminergic polymorphic ventricular tachycardia. Cell Death Dis. 2016;7:e2393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 104. | Sasaki K, Makiyama T, Yoshida Y, Wuriyanghai Y, Kamakura T, Nishiuchi S, Hayano M, Harita T, Yamamoto Y, Kohjitani H, Hirose S, Chen J, Kawamura M, Ohno S, Itoh H, Takeuchi A, Matsuoka S, Miura M, Sumitomo N, Horie M, Yamanaka S, Kimura T. Patient-Specific Human Induced Pluripotent Stem Cell Model Assessed with Electrical Pacing Validates S107 as a Potential Therapeutic Agent for Catecholaminergic Polymorphic Ventricular Tachycardia. PLoS One. 2016;11:e0164795. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 105. | Preininger MK, Jha R, Maxwell JT, Wu Q, Singh M, Wang B, Dalal A, Mceachin ZT, Rossoll W, Hales CM, Fischbach PS, Wagner MB, Xu C. A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses. Dis Model Mech. 2016;9:927-939. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 106. | Wei H, Zhang XH, Clift C, Yamaguchi N, Morad M. CRISPR/Cas9 Gene editing of RyR2 in human stem cell-derived cardiomyocytes provides a novel approach in investigating dysfunctional Ca2+ signaling. Cell Calcium. 2018;73:104-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 107. | Zhang XH, Wei H, Xia Y, Morad M. Calcium signaling consequences of RyR2 mutations associated with CPVT1 introduced via CRISPR/Cas9 gene editing in human-induced pluripotent stem cell-derived cardiomyocytes: Comparison of RyR2-R420Q, F2483I, and Q4201R. Heart Rhythm. 2021;18:250-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 108. | Park SJ, Zhang D, Qi Y, Li Y, Lee KY, Bezzerides VJ, Yang P, Xia S, Kim SL, Liu X, Lu F, Pasqualini FS, Campbell PH, Geva J, Roberts AE, Kleber AG, Abrams DJ, Pu WT, Parker KK. Insights Into the Pathogenesis of Catecholaminergic Polymorphic Ventricular Tachycardia From Engineered Human Heart Tissue. Circulation. 2019;140:390-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 109. | Sleiman Y, Souidi M, Kumar R, Yang E, Jaffré F, Zhou T, Bernardin A, Reiken S, Cazorla O, Kajava AV, Moreau A, Pasquié JL, Marks AR, Lerman BB, Chen S, Cheung JW, Evans T, Lacampagne A, Meli AC. Modeling polymorphic ventricular tachycardia at rest using patient-specific induced pluripotent stem cell-derived cardiomyocytes. EBioMedicine. 2020;60:103024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 110. | Maizels L, Huber I, Arbel G, Tijsen AJ, Gepstein A, Khoury A, Gepstein L. Patient-Specific Drug Screening Using a Human Induced Pluripotent Stem Cell Model of Catecholaminergic Polymorphic Ventricular Tachycardia Type 2. Circ Arrhythm Electrophysiol. 2017;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 111. | Ross S, Holliday M, Lim S, Semsarian C. Characterization of the first induced pluripotent stem cell line generated from a patient with autosomal dominant catecholaminergic polymorphic ventricular tachycardia due to a heterozygous mutation in cardiac calsequestrin-2. Stem Cell Res. 2019;37:101450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 112. | Mazzanti A, Underwood K, Nevelev D, Kofman S, Priori SG. The new kids on the block of arrhythmogenic disorders: Short QT syndrome and early repolarization. J Cardiovasc Electrophysiol. 2017;28:1226-1236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 113. | El-Battrawy I, Lan H, Cyganek L, Zhao Z, Li X, Buljubasic F, Lang S, Yücel G, Sattler K, Zimmermann WH, Utikal J, Wieland T, Ravens U, Borggrefe M, Zhou XB, Akin I. Modeling Short QT Syndrome Using Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. J Am Heart Assoc. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 114. | Guo F, Sun Y, Wang X, Wang H, Wang J, Gong T, Chen X, Zhang P, Su L, Fu G, Su J, Yang S, Lai R, Jiang C, Liang P. Patient-Specific and Gene-Corrected Induced Pluripotent Stem Cell-Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Short QT Syndrome. Circ Res. 2019;124:66-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 115. | Shinnawi R, Shaheen N, Huber I, Shiti A, Arbel G, Gepstein A, Ballan N, Setter N, Tijsen AJ, Borggrefe M, Gepstein L. Modeling Reentry in the Short QT Syndrome With Human-Induced Pluripotent Stem Cell-Derived Cardiac Cell Sheets. J Am Coll Cardiol. 2019;73:2310-2324. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 116. | Tse G, Liu T, Li KH, Laxton V, Chan YW, Keung W, Li RA, Yan BP. Electrophysiological Mechanisms of Brugada Syndrome: Insights from Pre-clinical and Clinical Studies. Front Physiol. 2016;7:467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 117. | Remme CA. Cardiac sodium channelopathy associated with SCN5A mutations: electrophysiological, molecular and genetic aspects. J Physiol. 2013;591:4099-4116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 131] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 118. | Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, Salvatori D, Oostwaard DW, Wilde AA, Bezzina CR, Verkerk AO, Freund C, Mummery CL. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079-3091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 119. | Liang P, Sallam K, Wu H, Li Y, Itzhaki I, Garg P, Zhang Y, Vermglinchan V, Lan F, Gu M, Gong T, Zhuge Y, He C, Ebert AD, Sanchez-Freire V, Churko J, Hu S, Sharma A, Lam CK, Scheinman MM, Bers DM, Wu JC. Patient-Specific and Genome-Edited Induced Pluripotent Stem Cell-Derived Cardiomyocytes Elucidate Single-Cell Phenotype of Brugada Syndrome. J Am Coll Cardiol. 2016;68:2086-2096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 165] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 120. | Ma D, Liu Z, Loh LJ, Zhao Y, Li G, Liew R, Islam O, Wu J, Chung YY, Teo WS, Ching CK, Tan BY, Chong D, Ho KL, Lim P, Yong RYY, Panama BK, Kaplan AD, Bett GCL, Ware J, Bezzina CR, Verkerk AO, Cook SA, Rasmusson RL, Wei H. Identification of an INa-dependent and Ito-mediated proarrhythmic mechanism in cardiomyocytes derived from pluripotent stem cells of a Brugada syndrome patient. Sci Rep. 2018;8:11246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 121. | Li W, Stauske M, Luo X, Wagner S, Vollrath M, Mehnert CS, Schubert M, Cyganek L, Chen S, Hasheminasab SM, Wulf G, El-Armouche A, Maier LS, Hasenfuss G, Guan K. Disease Phenotypes and Mechanisms of iPSC-Derived Cardiomyocytes From Brugada Syndrome Patients With a Loss-of-Function SCN5A Mutation. Front Cell Dev Biol. 2020;8:592893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 122. | Selga E, Sendfeld F, Martinez-Moreno R, Medine CN, Tura-Ceide O, Wilmut SI, Pérez GJ, Scornik FS, Brugada R, Mills NL. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J Mol Cell Cardiol. 2018;114:10-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 123. | Cerrone M, Lin X, Zhang M, Agullo-Pascual E, Pfenniger A, Chkourko Gusky H, Novelli V, Kim C, Tirasawadichai T, Judge DP, Rothenberg E, Chen HS, Napolitano C, Priori SG, Delmar M. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129:1092-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 262] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 124. | Monasky MM, Micaglio E, Ciconte G, Pappone C. Brugada Syndrome: Oligogenic or Mendelian Disease? Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 125. | Veerman CC, Mengarelli I, Guan K, Stauske M, Barc J, Tan HL, Wilde AA, Verkerk AO, Bezzina CR. hiPSC-derived cardiomyocytes from Brugada Syndrome patients without identified mutations do not exhibit clear cellular electrophysiological abnormalities. Sci Rep. 2016;6:30967. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 126. | Miller DC, Harmer SC, Poliandri A, Nobles M, Edwards EC, Ware JS, Sharp TV, McKay TR, Dunkel L, Lambiase PD, Tinker A. Ajmaline blocks INa and IKr without eliciting differences between Brugada syndrome patient and control human pluripotent stem cell-derived cardiac clusters. Stem Cell Res. 2017;25:233-244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 127. | Abrahams E. Right drug-right patient-right time: personalized medicine coalition. Clin Transl Sci. 2008;1:11-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 128. | Prondzynski M, Lemoine MD, Zech AT, Horváth A, Di Mauro V, Koivumäki JT, Kresin N, Busch J, Krause T, Krämer E, Schlossarek S, Spohn M, Friedrich FW, Münch J, Laufer SD, Redwood C, Volk AE, Hansen A, Mearini G, Catalucci D, Meyer C, Christ T, Patten M, Eschenhagen T, Carrier L. Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol Med. 2019;11:e11115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 129. | Pölönen RP, Penttinen K, Swan H, Aalto-Setälä K. Antiarrhythmic Effects of Carvedilol and Flecainide in Cardiomyocytes Derived from Catecholaminergic Polymorphic Ventricular Tachycardia Patients. Stem Cells Int. 2018;2018:9109503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 130. | Pölönen RP, Swan H, Aalto-Setälä K. Mutation-specific differences in arrhythmias and drug responses in CPVT patients: simultaneous patch clamp and video imaging of iPSC derived cardiomyocytes. Mol Biol Rep. 2020;47:1067-1077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 131. | Penttinen K, Swan H, Vanninen S, Paavola J, Lahtinen AM, Kontula K, Aalto-Setälä K. Antiarrhythmic Effects of Dantrolene in Patients with Catecholaminergic Polymorphic Ventricular Tachycardia and Replication of the Responses Using iPSC Models. PLoS One. 2015;10:e0125366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 132. | Kobayashi S, Bannister ML, Gangopadhyay JP, Hamada T, Parness J, Ikemoto N. Dantrolene stabilizes domain interactions within the ryanodine receptor. J Biol Chem. 2005;280:6580-6587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 133. | Paul-Pletzer K, Yamamoto T, Ikemoto N, Jimenez LS, Morimoto H, Williams PG, Ma J, Parness J. Probing a putative dantrolene-binding site on the cardiac ryanodine receptor. Biochem J. 2005;387:905-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 60] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 134. | Micheu MM, Popa-Fotea NM, Oprescu N, Dorobantu M, Ratiu AC, Ecovoiu AA. NGS data validated by Sanger sequencing reveal a puzzling small deletion of MYBPC3 gene associated with hypertrophic cardiomyopathy. Rom Biotechnol Lett. 2019;24:91-99. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 135. | Micheu MM, Popa-Fotea NM, Oprescu N, Bogdan S, Dan M, Deaconu A, Dorobantu L, Gheorghe-Fronea O, Greavu M, Iorgulescu C, Scafa-Udriste A, Ticulescu R, Vatasescu RG, Dorobanțu M. Yield of Rare Variants Detected by Targeted Next-Generation Sequencing in a Cohort of Romanian Index Patients with Hypertrophic Cardiomyopathy. Diagnostics (Basel). 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 136. | Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, Cox SW, DePalma SR, Ho CY, Seidman JG, Seidman CE, Rehm HL. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880-888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 341] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 137. | Walsh R, Mazzarotto F, Whiffin N, Buchan R, Midwinter W, Wilk A, Li N, Felkin L, Ingold N, Govind R, Ahmad M, Mazaika E, Allouba M, Zhang X, de Marvao A, Day SM, Ashley E, Colan SD, Michels M, Pereira AC, Jacoby D, Ho CY, Thomson KL, Watkins H, Barton PJR, Olivotto I, Cook SA, Ware JS. Quantitative approaches to variant classification increase the yield and precision of genetic testing in Mendelian diseases: the case of hypertrophic cardiomyopathy. Genome Med. 2019;11:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 138. | Campuzano O, Sarquella-Brugada G, Fernandez-Falgueras A, Coll M, Iglesias A, Ferrer-Costa C, Cesar S, Arbelo E, García-Álvarez A, Jordà P, Toro R, Tiron de Llano C, Grassi S, Oliva A, Brugada J, Brugada R. Reanalysis and reclassification of rare genetic variants associated with inherited arrhythmogenic syndromes. EBioMedicine. 2020;54:102732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 139. | Lv W, Qiao L, Petrenko N, Li W, Owens AT, McDermott-Roe C, Musunuru K. Functional Annotation of TNNT2 Variants of Uncertain Significance With Genome-Edited Cardiomyocytes. Circulation. 2018;138:2852-2854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 140. | Pettinato AM, Ladha FA, Mellert DJ, Legere N, Cohn R, Romano R, Thakar K, Chen YS, Hinson JT. Development of a Cardiac Sarcomere Functional Genomics Platform to Enable Scalable Interrogation of Human TNNT2 Variants. Circulation. 2020;142:2262-2275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 141. | Garg P, Oikonomopoulos A, Chen H, Li Y, Lam CK, Sallam K, Perez M, Lux RL, Sanguinetti MC, Wu JC. Genome Editing of Induced Pluripotent Stem Cells to Decipher Cardiac Channelopathy Variant. J Am Coll Cardiol. 2018;72:62-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 142. | Gélinas R, El Khoury N, Chaix MA, Beauchamp C, Alikashani A, Ethier N, Boucher G, Villeneuve L, Robb L, Latour F, Mondesert B, Rivard L, Goyette P, Talajic M, Fiset C, Rioux JD. Characterization of a Human Induced Pluripotent Stem Cell-Derived Cardiomyocyte Model for the Study of Variant Pathogenicity: Validation of a KCNJ2 Mutation. Circ Cardiovasc Genet. 2017;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 143. | Chavali NV, Kryshtal DO, Parikh SS, Wang L, Glazer AM, Blackwell DJ, Kroncke BM, Shoemaker MB, Knollmann BC. Patient-independent human induced pluripotent stem cell model: A new tool for rapid determination of genetic variant pathogenicity in long QT syndrome. Heart Rhythm. 2019;16:1686-1695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 144. | Chai S, Wan X, Ramirez-Navarro A, Tesar PJ, Kaufman ES, Ficker E, George AL Jr, Deschênes I. Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. J Clin Invest. 2018;128:1043-1056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 145. | Pang L. Toxicity testing in the era of induced pluripotent stem cells: A perspective regarding the use of patient-specific induced pluripotent stem cell–derived cardiomyocytes for cardiac safety evaluation. Curr Opin Toxicol. 2020;23:50-55. [RCA] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 146. | Machiraju P, Greenway SC. Current methods for the maturation of induced pluripotent stem cell-derived cardiomyocytes. World J Stem Cells. 2019;11:33-43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 147. | Parikh SS, Blackwell DJ, Gomez-Hurtado N, Frisk M, Wang L, Kim K, Dahl CP, Fiane A, Tønnessen T, Kryshtal DO, Louch WE, Knollmann BC. Thyroid and Glucocorticoid Hormones Promote Functional T-Tubule Development in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ Res. 2017;121:1323-1330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 291] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 148. | Ruan JL, Tulloch NL, Razumova MV, Saiget M, Muskheli V, Pabon L, Reinecke H, Regnier M, Murry CE. Mechanical Stress Conditioning and Electrical Stimulation Promote Contractility and Force Maturation of Induced Pluripotent Stem Cell-Derived Human Cardiac Tissue. Circulation. 2016;134:1557-1567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 330] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 149. | Correia C, Koshkin A, Duarte P, Hu D, Carido M, Sebastião MJ, Gomes-Alves P, Elliott DA, Domian IJ, Teixeira AP, Alves PM, Serra M. 3D aggregate culture improves metabolic maturation of human pluripotent stem cell derived cardiomyocytes. Biotechnol Bioeng. 2018;115:630-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 107] [Article Influence: 13.4] [Reference Citation Analysis (0)] |