Published online Aug 26, 2019. doi: 10.4252/wjsc.v11.i8.535
Peer-review started: February 18, 2019
First decision: May 9, 2019
Revised: June 15, 2019
Accepted: July 29, 2019
Article in press: July 29, 2019
Published online: August 26, 2019
Processing time: 192 Days and 5.6 Hours
Human hepatocyte-like cells (HLCs) derived from human pluripotent stem cells (hPSCs) promise a valuable source of cells with human genetic background, physiologically relevant liver functions, and unlimited supply. With over 10 years’ efforts in this field, great achievements have been made. HLCs have been successfully derived and applied in disease modeling, toxicity testing and drug discovery. Large cohorts of induced pluripotent stem cells-derived HLCs have been recently applied in studying population genetics and functional outputs of common genetic variants in vitro. This has offered a new paradigm for genome-wide association studies and possibly in vitro pharmacogenomics in the nearly future. However, HLCs have not yet been successfully applied in bioartificial liver devices and have only displayed limited success in cell transplantation. HLCs still have an immature hepatocyte phenotype and exist as a population with great heterogeneity, and HLCs derived from different hPSC lines display variable differentiation efficiency. Therefore, continuous improvement to the quality of HLCs, deeper investigation of relevant biological processes, and proper adaptation of recent advances in cell culture platforms, genome editing technology, and bioengineering systems are required before HLCs can fulfill the needs in basic and translational research. In this review, we summarize the discoveries, achievements, and challenges in the derivation and applications of HLCs.
Core tip: Hepatocyte-like cells (HLCs) derived from human pluripotent stem cells (hPSCs) have a great application prospect as an unlimited supply of human hepatocytes in disease modeling, toxicity testing and drug discovery. In this review, we summarize the derivation of HLCs from hPSCs, and the limitations and optimization of current differentiation protocols. We also discuss progress in the application of HLCs, and reveal the exciting future of HLCs for use in the study of rare diseases, population genetics, and in vitro pharmacogenomics.
- Citation: Li S, Huang SQ, Zhao YX, Ding YJ, Ma DJ, Ding QR. Derivation and applications of human hepatocyte-like cells. World J Stem Cells 2019; 11(8): 535-547
- URL: https://www.wjgnet.com/1948-0210/full/v11/i8/535.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v11.i8.535
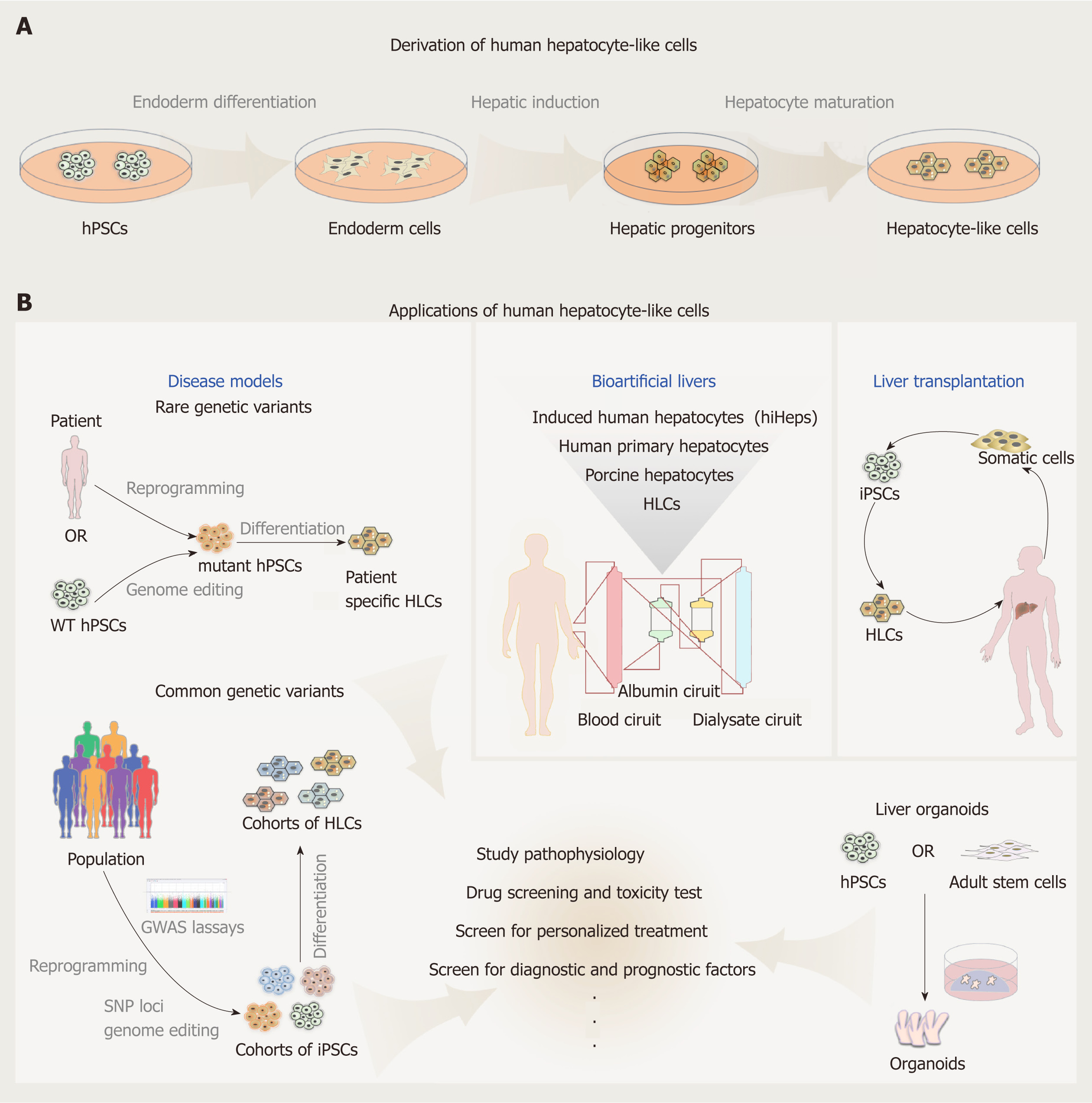
The liver represents one of the most pivotal organs of the human body in regulating glucose homeostasis, lipid metabolism, detoxification and many other physiological processes. As liver diseases, including fatty liver diseases, hepatic carcinoma, and viral hepatitis, continue to increase in prevalence, there is an urgent need for development of effective treatments, and sufficiently cell or tissue sources for transplantation. Primary human hepatocytes and liver donors offer immediate resources for studying liver diseases and transplantation. However, both primary cells and available donor transplants are in persistent shortage. Although different culture systems have been identified recently that enable long-term culture and expansion of both rodent and human primary hepatocytes[1-4], the capacity of expansion is still limited and has donor-dependent variability. As stem cells are known to have potent self-renewal ability as well as the capacity to differentiate into different somatic cell types, they have been proposed as an ideal alternative cell source for large or even unlimited supplies of hepatocytes and even liver tissues. Human hepatocytes can be derived from embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), mesenchymal stem cells and hepatic progenitor cells[5]. As the cells derived from stem cells often have incomplete function and exhibit characteristics of fetal liver cells, they are generally defined as hepatocyte-like cells (HLCs). The discovery made by Gurdon and Yamanaka that mature cells from individual patients can be reprogrammed to iPSCs, opened up the possibility that these cells can be applied to disease modeling and organ transplantation. Furthermore, intense efforts have been made in recent years in generating better HLCs and liver organoids from PSCs, and in applications of these cells in various fields. Therefore, in this review, we focus on HLCs derived from human pluripotent stem cells (hPSCs) and discuss recent progress in the derivation and applications of HLCs in biomedical research.
hPSCs include human ESCs, mostly derived from the inner cell mass of the fertilized eggs, and iPSCs reprogramed from terminally differentiated somatic cells. hPSCs promise an unlimited supply of human somatic cells, due to their theoretical capacity for self-renewal and differentiation into any kind of somatic cell types in human body. To date, many protocols have been established to generate human hepatocytes derived from hPSCs. Most induction methods are based on the understanding of the embryonic development processes of the liver, and aimed to imitate in Petri dishes the endoderm development, endoderm hepatic specification and hepatic maturation stages. The directed differentiation protocols either rely on the use of embryoid body (EB) formation[6,7] or start with monolayer culture, with the latter more frequently adapted currently in laboratories. EB formation means to mimic the blastocyst and epiblast architecture; however, it can be easily disturbed by suboptimal culture conditions and sources of reagents, for example, different batches of fetal bovine serum can affect to a large degree the quality of generated EBs. Most protocols currently in use apply similar strategies with contributions from individual laboratories by improving inducers of differentiation and optimizing their combinations (Table 1). These protocols can be largely specified to three consecutive steps: endoderm differentiation, hepatic induction, and liver maturation.
| Ref. | EB / monolayer | Protocol features | In vitro functional assays | In vivo assay | |
| Endoderm induction | Hepatic specification and maturation | ||||
| Cai et al[18], 2007 | Monolayer | Activin A, ITS | FGF4, BMP2, HGF, OSM, Dex | ALB, Glycogen, ICG, LDL, CYP450 | Yes |
| Hay et al[19], 2008 | Monolayer | Activin A, Wnt3a | Serum, DMSO, Insulin, HGF, OSM | Urea, Gluconeogenesis, AFP | Yes |
| Agarwal et al[24], 2008 | Monolayer | Activin A, low serum | FGF4, HGF, OSM, Dex | ALB, Glycogen, ICG | Yes |
| Basma et al[7], 2009 | EB/monolayer | Activin A, bFGF | FGF, DMSO, Dex | ALB, Urea, AAT, CYP450 | Yes |
| Song et al[20], 2009 | Monolayer | Activin A | FGF4, BMP2, HGF, KGF, OSM, Dex | ALB, Urea, Glycogen, CYP450 | No |
| Si-Tayeb et al[21], 2010 | Monolayer | Activin A | BMP4 FGF2, HGF, OSM | Glycogen, LDL, oil red O storage, ICG, Urea | Yes |
| Sullivan et al[22], 2010 | Monolayer | Activin A, Wnt3a | β-ME, DMSO, Insulin, HGF, OSM | CYP450, Fibrinogen, Fibronectin, Transthyretin, AFP | No |
| Touboul et al[23], 2010 | Monolayer | Activin A, FGF2, BMP4, LY294002 | FGF10, RA, SB431542, FGF4, HGF, EGF | Glycogen, CYP450, ICG, LDL | Yes |
| Borowiak et al[35], 2009 | Monolayer | Activin A, Wnt3a, HGF | OSM, Dex, ITS | CYP450, Urea, LDL, Glycogen | Yes |
| Ogawa et al[34], 2013 | EB/monolayer | BMP4, Activin A, Wnt3a | FGF10, bFGF, BMP4, HGF, OSM, Dex, cAMP | ICG, Glycogen, ALB, CYP450 | No |
| Siller et al[37], 2015 | Monolayer | CHIR99021 | DMSO, dihexa, Dex | ALB, Glycogen, ICG, CYP450 | No |
Transforming growth factor (TGF) β family member Nodal is vital in endoderm formation, based on studies in developmental biology in models including frogs, zebrafish, and mice[8-10]. Although Nodal is an attractive candidate for inducing hPSCs to differentiate into definitive endoderm (DE), it is difficult to get highly active protein. Activin is another TGFβ family member, which mimics Nodal activity in triggering similar intracellular signaling events[11], thus is often used as a substitution of Nodal in vitro[12]. In 2005, D’Amour et al[12] demonstrated efficient endoderm induction from monolayers of hPSCs by applying activin A, which was subsequently reproduced by many other groups. The monolayer culture here seems important to the endoderm differentiation in that cells can be exposed evenly to the endodermal inducer, activin A, and can better synchronize development of the endodermal cell fate[13]. Levels of Nodal signaling comprise key elements in cell fate determination, with high level promotes endoderm differentiation, whereas low level initiates mesoderm specification[14-17]. Therefore, high concentrations of activin A are now widely utilized for endoderm induction in hPSC culture[18-22]. Besides, activation of fibroblast growth factor (FGF), bone morphogenetic protein (BMP) and Wnt signaling pathways also promote endoderm development[7,19,23]. Phosphatidylinositol 3-kinase (PI3K) inhibitors, such as LY 294002 and AKT1-II, also promote activin-A-induced endoderm development[17]. Several studies have shown that low doses of serum are necessary for activin A to induce an efficient endoderm program[12,17,24].
In early embryo development, FGF signals and BMP signals initiate the liver gene program and simultaneously block that for pancreas development[25]. Consistent with the in vivo discoveries, the signaling molecules FGF and BMP have also been demonstrated to be important in generating hepatic cells from DE cells in vitro. The combination of FGFs and BMPs are thus widely used to induce hepatic endoderm programs[18,21,23]. Dimethylsulfoxide (DMSO) can assist in promoting hPSC differentiation and specific generation of hepatic progenitors, and is usually used in hepatic differentiation[19,22,26,27].
As for further liver maturation, hepatic progenitors are mostly treated by hepatocyte growth factor (HGF), oncostatin M (OSM), and glucocorticoid dexamethasone (Dex). HGF binds to its tyrosine kinase receptor c-Met, promoting hepatoblast proliferation, increasing cell migration and improving cell survival[28,29]. OSM produced by hematopoietic cells is an interleukin(IL)-6 family cytokine, which induces hepatic maturation by the phosphorylation of signal transducer and activator of transcription[28,30]. The glucocorticoid dexamethasone has also been implicated in the maturation of the hepatocytes[31,32]. After the maturation stage, obtained HLCs display many hepatocyte features, such as albumin expression and secretion, urea secretion, low-density lipoprotein (LDL) uptake, indocyanine green (ICG) uptake, and glycogen storage (Table 1). However, those cells express fetal liver markers, such as α-fetoprotein (AFP), and have lower activities of CYP450 enzymes when compared to primary liver tissue. With comparison of a set of human adult and fetal liver markers, it is roughly estimated that the HLCs have the characteristics of fetal hepatocytes at < 20 wk gestation[33].
Different strategies have been adopted with the aim to promote maturation and to reduce the large heterogeneity of HLCs. One strategy is to use 3D culture, mimicking liver development in the body, thus promoting further maturation. Indeed, it has been shown that cells demonstrate more matured phenotypes in 3D than other culture systems. For example, it has been demonstrated that cAMP signaling within the 3D hepatoblast aggregates can promote further maturation of HLCs that display comparable metabolic enzyme levels to those of primary human hepatocytes[34] . The other main strategy is to optimize the current protocols through screening for molecules that can improve differentiation, and to understand better the molecular mechanisms underlying liver development. Towards this aim, by screening 4000 compounds, the Melton group identified IDE1 and IDE2, which can efficiently promote differentiation of mouse and human ESCs into DE cells[35]. Other groups have also identified other small molecules, and demonstrated their effects in improving hPSC differentiation toward endoderm[36]. In 2015, the Siller et al[37] group developed a new method for HLC differentiation with a combination of small molecules without the inclusion of growth factors in a defined minimum medium. Shan et al[38] developed a high-throughput chemical screening platform and identified two different classes of small molecules, which are able to induce functional proliferation of human primary hepatocytes in vitro and improve HLC maturation. By utilizing an established hepatic lineage hPSC reporter line, our laboratory performed genetic and chemical screenings, and identified several modulators involved in hepatic differentiation, and CI-994 compound (histone deacetylase 3 inhibitor) that can promote HLC differentiation at a late stage[39].
Human PSCs offer a unique in vitro cellular model system for disease modeling. Induced PSCs derived from patients or hPSCs engineered with specific disease-causing mutations using genome editing technologies allow researchers to study the consequences of genetic mutations with a human- and patient-specific genetic background; whereas the differentiation processes in vitro often recapitulate aspects of normal development, thus providing the opportunity to investigate the developmental and degenerative processes of certain human diseases. Furthermore, as hPSCs possess great capacity in self-renewal, they can offer large-scale cellular materials with identical genetic background for disease modeling and for possible compound screenings to develop potential treatments.
For modeling liver diseases with rare mutations in Mendelian diseases, patient-specific iPSCs carrying certain genetic mutations are often derived and differentiated to HLCs. Many disease models of inborn liver metabolic disorders, such as α1-antitrypsin deficiency, familial hypercholesterolemia, glycogen storage disease type 1a, and Wilson’s disease, have been generated[40-42]. Upon differentiation to HLCs, these cells with genetic mutations displayed certain disease phenotypes that are reflected in patients, highlighting potential utility of these models for studying diseases or screening for therapeutic interventions. In situations in which patients are not available, disease mutations of interest can be engineered using genome editing technologies into wild-type hPSCs to create mutant hPSCs for disease study[43,44]. Drug screening with these disease models can highlight novel discoveries for disease treatment. In a study by the Duncan and Rader groups[45], HLCs derived from familial hypercholesterolemia iPSCs were applied to drug screening to identify potential LDL–cholesterol (LDL-C)-lowering drugs, which has successfully revealed cardiac glycosides as a candidate treatment for hypercholesterolemia. Other than studying diseases harboring genetic mutations, hPSC-derived HLCs are also powerful in providing cellular models for studying the lifecycle of hepatitis viruses. hPSC-derived HLCs have be used in hepatitis C virus (HCV) infection and screening for anti-HCV drugs[46], as well as modeling hepatitis B virus infection[47].
As a remarkable improvement in the recent iPSC disease modeling fields, large, diverse population cohorts of iPSCs have been generated and differentiated in parallel to HLCs as well as other cell types, offering valuable tissue substitutes for studies to reveal the relationship between genotype and phenotype; for example, expression quantitative trait locus (eQTL) analysis[48,49]. Two independent cohorts of iPSCs have been generated from healthy donors (68 iPSC lines from 34 donors in one study and 91 iPSC lines from 91 donors in the other study) and used for subsequent hepatic differentiation and genetic analysis. Studies either successfully confirmed eQTLs previously characterized in vivo[49], or identified a number of loci controlling hepatic gene expression with these in vitro HLCs[48]. In one study, the cohort of iPSC-derived HLCs were also subjected to metabolite abundance quantitative trait locus (mQTL) analysis, leading to the discovery of a strong association between a lipid-dysregulating phenotype and the minor allele at the 1p13 locus[49]. For the first time, these two studies demonstrated the capacity for iPSCs-derived cells to reproduce in vivo phenotypes driven by common genetic variants, and uncovered a potentially unlimited supply of human cells that allow to discover cell-type-specific QTL phenotypes (eQTL, mQTL and potentially others) that would be inaccessible using in vivo tissues. Together with several other studies that have performed genome-wide QTL analyses and identified a number of loci that contribute to interline heterogeneity using hundreds of undifferentiated iPSC lines[50-52], these studies have offered a new paradigm for human research, with iPSC-driven disease modeling being applied to study population genetics in vitro.
Aside from drug discovery with iPSC-derived disease models with small cohorts, large cohorts of iPSCs and iPSC-derived cells have been proposed to perform trials-in-dish, to assist in translating the discoveries of genome-wide association studies (GWASs) into improved treatment regimens and drug discovery; that is, to apply genotype analysis to patient stratification and design of individual treatment plans[53]. In possible scenarios, iPSC-derived cells may provide an important link between drug development and Phase I trials, where iPSC-derived hepatocytes, cardiomyocytes or neurons can be used for preliminary safety screens with candidate drugs that might induce hepatotoxicity, cardiotoxicity, neurotoxicity or other off-target effects. Furthermore, between Phase I and Phase II trials, drug target cells derived from large cohorts of iPSCs can serve as the surrogate human population and be used in testing for drug efficacy; results from which can be applied to classify patients into responder and non-responder groups, thus increasing the relevance and successfully rate of further Phase 2 and 3 trials. Altogether, small or large cohorts of iPSCs and iPSC-derived function cell types are revolutionizing the field of drug discovery.
The liver is a highly specialized organ consisting of mostly hepatocytes, but also several other cell types, such as Kupffer cells, endothelia cells, bile duct cells, and hepatic stellate cells. These cells all contribute to the highly organized architecture and functions of liver tissue. Compared to HLCs in 2D culture, liver tissue organoids constitute more than one cell type, can resemble part of the architecture of liver tissue, and possess some functions that may not exist in HLCs. Liver organoids can either be derived from adult stem cells[54,55] or hPSCs[56-59]. Other than HLCs, development of protocols to obtain other cell types derived from hPSCs that constitute the liver tissue are important. To date, protocols of directed differentiation to obtain cholangiocytes[56,57], endothelia cells[60] and hepatic stellate cells[61] have been established, which may further aid the generation of functional liver tissue organoids. Other reviews discuss the generation and application of tissue organoids, which can assist in better understanding the opportunities as well as challenges in this field[62,63].
Artificial liver support systems have been developed to provide an alternative to orthotopic liver transplantation (OLT). Artificial livers use nonbiological components to perform hepatic detoxification, removing toxins and drugs that accumulate in the blood during liver failure[64]. However, artificial livers do not have the capacity to adequately replicate the physiological liver function. The incorporation of live cells harboring liver functions into these artificial liver systems, which establishes the bioartificial livers (BALs) systems, offers a solution to overcome these limitations[64]. BAL support systems are extracorporeal bioreactors in which whole livers or liver cells are cultured in a 3D manner within a network of hollow fibers for blood plasma perfusion. BAL systems provide both biotransformation and hepatic synthetic functions[65]. To date, different sources of liver cells have been tested in BAL devices, for example, human primary hepatocytes, immortalized human hepatoma cell lines, porcine hepatocytes[66], as well as induced human hepatocytes transdifferentiated from human fibroblasts (hiHeps)[67]. While human hepatocytes are the preferred cells, obtaining sufficient human hepatocytes faces the same difficulty of organ shortage. Porcine hepatocytes are close to human hepatocytes, but have potential risk of xenozoonosis and immunological response. Hepatoma cells can provide large amounts of materials, but suffer from incompetent metabolism and ammonia clearance[68]. HiHeps representing a new invaluable cell source for BAL devices, and have been successful in pigs[67] as well as in primary tests in patients. While we have not seen reports of HLCs being applied in BAL devices, we envisage that HLCs will be a potential cell source for the treatment of liver failure in BAL support systems in the future. The advantages of HLCs are obvious: human or patient-specific genetic background, normal karyotype, potentially unlimited supply, and better liver functions. However, to obtain a large amount of functional and homogeneous hepatocytes from hPSCs still depends on continuous improvement to the differentiation protocols and development of optimal large-scale culture systems.
OLT remains the most effective treatment for end-stage liver diseases. However, liver donor shortage and life-long need for immunosuppression are the main limitations to liver transplantation. A potential alternative to liver transplantation is hepatocyte transplantation[69-71]. However, cell transplantation is also limited by the availability of effective cell sources, generation of alternative hepatocytes is thus an urgent problem. The ideal cell source should at least meet the following requirements: (1) Available in large quantity. Similar to hepatocytes needed in BAL devices, a large number of cells (> 109) may be needed for transplantation to every adult patient; (2) High efficiency of in vivo homing and repopulation. Transplanted cells can home and adapt to the microenvironment in recipient and successfully repopulate the liver; (3) Low immunogenicity. Cells have no or low immunogenic responses, which can be su-ppressed by low doses of immunosuppressant; (4) No tumorigenic risk. Transplanted cells should have normal karyotype and be free of potential tumorigenic modulations, such as modifications in oncogenic or tumor suppressor genes. To date, several mouse models have been adopted in testing the transplantation efficiency of human hepatocytes, which in general can be divided into two categories[72]. One is a mouse model with a genetic disorder that causes depletion of the host hepatocytes, such as mice expressing urinary plasminogen activator (uPA) driven by the albumin or Mup promoter[73,74], and immunodeficient FRG [Fah(-/-) Rag2(-/-) Il2rg (-/-)] mice[75]; another is a mouse model with drug- or surgery-induced liver damage, including mice receiving treatment with retrorsine[7], CCl4[24,76], diethylnitrosamine[77] or partial hepatectomy[7,78] (Table 2). Transplantation using primary human hepatocytes has been successful in mouse models, for example, with the FRG mouse model, the ratio of human hepatocytes in a mouse liver can be up to 90%[75]. However, there are no definitive conclusions so far regarding whether the maturity of transplanted liver cells affects the efficiency of transplantation when HLCs are used. Cells in endoderm, hepatoblasts, and mature hepatocyte stages along the HLC differentiation process all have possibilities as donor cells in cell transplantation[7,24,73,76,77] (Table 2). The microenvironment in recipient liver is thought to supply necessary signals to promote further maturation of transplanted cells, although direct evidence and the underlying mechanism are lacking. However, the overall HLC transplantation efficiency is lower compared to that of human primary hepatocytes[75] (Table 2). Furthermore, transplantation with HLCs may suffer tumorigenic risks due to remnant undifferentiated hPSCs, and the immunogenicity has not been addressed so far, as most studies were performed with immunocompromised animals.
| Ref. | Animal model | Route | Proliferative stimulus | Type and number of cells | Donor / recipient (% engrafted) | Donor / recipient (% repopulated) | Time post-transplantation |
| Agarwal et al[24], 2008 | NOD-SCID mice | Portal vein | CCl4-injured | 106 hES-DEs | < 1% | NA | 28 d |
| Basma et al[7], 2009 | NOD-SCID mice | Spleen | Retrorsine and partial hepatectomy | 1 × 106 hES-HLCs | NA | NA | 21 d |
| Liu et al[77], 2011 | NSG mice | Tail vein | dimethylnitrosamine -injured | 0.1 - 2 × 106 hiPSC- multistage hepatic cells | 2%–17% | 8%–15% | 56 d |
| Asgari et al[76], 2013 | Normal mouse | Tail vein | CCl4-injured | 1 × 106 hiPSC- HLCs | 2 ± 0.7% | NA | 35 d |
| Carpentier et al[73], 2014 | MUP-uPA/ SCID/Bg mice | Spleen | NA | 4 × 106 hiPSC-HLCs | 1%-7% | < 1 to up to 20% | 100 d |
| Song et al[20], 2015 | Immunocompetent mice | Intraperitoneal cavity | NA | 4.4×105 hiPSC-HLCs in capsules | NA | NA | 24 d |
| Nagamoto et al[74], 2015 | uPA/SCID mice | Spleen | NA | 1 × 106 Ad-FNK-transduced hiPSC-HLCs | NA | NA | 28 d |
| Nagamoto et al[78], 2016 | Mice | hiPS-HLC sheet transplantation | 2/3 partial hepatectomy and CCl4-injured | 8 × 105 hiPSC-HLCs | NA | NA | 14 d |
To improve the transplantation efficiency, several ectopic sites have been investigated, including spleen, peritoneal cavity, kidney, lung, pancreas and fat pads. Bioengineering approaches have also been applied in cell transplantation. For example, Song et al[20] transplanted hPSC-derived HLCs in immunocompetent mice via 3D cell coaggregates with stromal cells and encapsulation. This study demonstrated an improved approach for the engraftment of hPSC-derived HLCs[79]. In a different study, Nagamoto et al[78]. used a cell sheet engineering technology by attaching HLC sheets onto the surface of mouse liver with acute liver failure, which showed improved hepatocyte engraftment and animal survival in contrast, genetic modification to HLCs represents another approach to improve transplantation efficiency. For example, Nagamoto et al[74] demonstrated higher transplantation efficiency using HLCs transduced with an adenovirus vector expressing FNK (Ad-FNK), by inhibiting apoptosis in the process of integration into liver. However, there is still a long way to go before HLCs can be used in clinical liver transplantation. Strenuous efforts are needed to understand the complex processes of cell transplantation, for example, the donor–host interactions, to improve the quality of HLCs and optimize the transplantation strategy. Plus, the potential tumorigenic risk of transplanted HLCs had to be carefully considered. Specifically, tumor cells can arise from cells with residual expression of factors in iPSC reprogramming process (e.g., the myc expression), undifferentiated iPSCs remaining in the culture, and cells with mutations or karyotype abnormalities caught in the rather long in vitro culture and differentiation processes. Several approaches can be adopted to reduce the tumorigenic risk: (1) Use integrating-free viruses or small molecules for iPSC reprogramming[80,81]; (2) Improve the in vitro culture conditions and enhance the differentiation efficiency of hPSC-derived HLCs[82]; (3) Remove undifferentiated iPSCs, e.g. through treatments with small molecules or antibodies that can specifically target iPSCs[83,84]; or enrich HLCs using HLC specific surface markers before transplantation[85]; (4) Monitor the genome integrity of cells at the iPSC stage and the HLC stage, through karyotype analysis and whole-genome sequencing; (5) Engineer a self-killing circuit in cells that would allow the trigger of cell death in vivo to remove tumorigenic cells, if necessary, to further assure safety[86]. Nonetheless, hPSC-derived HLCs provide a potential valuable cell source to OLT for liver diseases that is worth pursuing.
The generation of iPSCs has revolutionized the whole field of cell biology. It is truly inspiring to imagine that we can grow any person’s pluripotent cells indefinitely in a dish and turn them into any cell type. With this capability of iPSCs, the approach to the study of human biology has been profoundly changed. HLCs were among the first batch of adult cell types that have been derived from iPSCs, and have been tested ever since for disease modeling, toxicity screening, and drug discovery, and as donor cells for transplantation (Figure 1). Complexities and difficulties in the derivation and applications of these HLCs seem beyond our initial expectations. More than 10 years have passed, but HLCs derived from hPSCs remain a largely heterogeneous population with incompetent liver cell function and low transplantation efficiency. Protocols to grow HLCs from hPSCs need to be substantially and continuously improved and standardized on the basis of deeper understanding of liver development. Despite the gap between the reality and ideal conditions, efforts have paid off well and the field has made tremendous achievements in recent years, such as generation of functional liver organoids, successful modeling of certain liver diseases, identification of candidate treatments, and application of large cohorts of HLCs for human genetic studies, to name a few (Figure 1). With advances in cell culture systems including 3D culture platforms[87], coculturing conditions[88], tissue-on-a-chip approaches[89], and invention of new technologies including genome editing tools and bioengineering systems, HLCs obtained from hPSCs will eventually be able to fulfill the needs in biomedical research and clinical translation.
Manuscript source: Invited Manuscript
Specialty type: Cell and tissue engineering
Country of origin: China
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Grawish ME, Li SC, Politi LE S-Editor: Cui LJ L-Editor: A E-Editor: Xing YX
| 1. | Zhang K, Zhang L, Liu W, Ma X, Cen J, Sun Z, Wang C, Feng S, Zhang Z, Yue L, Sun L, Zhu Z, Chen X, Feng A, Wu J, Jiang Z, Li P, Cheng X, Gao D, Peng L, Hui L. In Vitro Expansion of Primary Human Hepatocytes with Efficient Liver Repopulation Capacity. Cell Stem Cell. 2018;23:806-819.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 2. | Hu H, Gehart H, Artegiani B, LÖpez-Iglesias C, Dekkers F, Basak O, van Es J, Chuva de Sousa Lopes SM, Begthel H, Korving J, van den Born M, Zou C, Quirk C, Chiriboga L, Rice CM, Ma S, Rios A, Peters PJ, de Jong YP, Clevers H. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell. 2018;175:1591-1606.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 553] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 3. | Fu GB, Huang WJ, Zeng M, Zhou X, Wu HP, Liu CC, Wu H, Weng J, Zhang HD, Cai YC, Ashton C, Ding M, Tang D, Zhang BH, Gao Y, Yu WF, Zhai B, He ZY, Wang HY, Yan HX. Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell Res. 2019;29:8-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 115] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 4. | Peng WC, Logan CY, Fish M, Anbarchian T, Aguisanda F, Álvarez-Varela A, Wu P, Jin Y, Zhu J, Li B, Grompe M, Wang B, Nusse R. Inflammatory Cytokine TNFα Promotes the Long-Term Expansion of Primary Hepatocytes in 3D Culture. Cell. 2018;175:1607-1619.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 226] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 5. | Hu C, Li L. In vitro culture of isolated primary hepatocytes and stem cell-derived hepatocyte-like cells for liver regeneration. Protein Cell. 2015;6:562-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 6. | Baharvand H, Hashemi SM, Kazemi Ashtiani S, Farrokhi A. Differentiation of human embryonic stem cells into hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol. 2006;50:645-652. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 286] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 7. | Basma H, Soto-Gutiérrez A, Yannam GR, Liu L, Ito R, Yamamoto T, Ellis E, Carson SD, Sato S, Chen Y, Muirhead D, Navarro-Alvarez N, Wong RJ, Roy-Chowdhury J, Platt JL, Mercer DF, Miller JD, Strom SC, Kobayashi N, Fox IJ. Differentiation and transplantation of human embryonic stem cell-derived hepatocytes. Gastroenterology. 2009;136:990-999. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 416] [Cited by in RCA: 364] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 8. | Tam PP, Kanai-Azuma M, Kanai Y. Early endoderm development in vertebrates: Lineage differentiation and morphogenetic function. Curr Opin Genet Dev. 2003;13:393-400. [PubMed] |
| 9. | Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell. 2001;1:605-617. [PubMed] |
| 10. | Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 481] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 11. | de Caestecker M. The transforming growth factor-β superfamily of receptors. Cytokine Growth Factor Rev. 2004;15:1-11. [PubMed] |
| 12. | D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534-1541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1288] [Cited by in RCA: 1285] [Article Influence: 64.3] [Reference Citation Analysis (0)] |
| 13. | Han S, Bourdon A, Hamou W, Dziedzic N, Goldman O, Gouon-Evans V. Generation of functional hepatic cells from pluripotent stem cells. J Stem Cell Res Ther. 2012;Suppl 10:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327-342. [PubMed] |
| 15. | Stainier DY. A glimpse into the molecular entrails of endoderm formation. Genes Dev. 2002;16:893-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 137] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 16. | Thisse B, Wright CV, Thisse C. Activin- and Nodal-related factors control antero-posterior patterning of the zebrafish embryo. Nature. 2000;403:425-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 166] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 17. | McLean AB, D'Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 337] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 18. | Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, Guo Y, Ding M, Deng H. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229-1239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 471] [Cited by in RCA: 456] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 19. | Hay DC, Fletcher J, Payne C, Terrace JD, Gallagher RC, Snoeys J, Black JR, Wojtacha D, Samuel K, Hannoun Z, Pryde A, Filippi C, Currie IS, Forbes SJ, Ross JA, Newsome PN, Iredale JP. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci USA. 2008;105:12301-12306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 345] [Cited by in RCA: 320] [Article Influence: 18.8] [Reference Citation Analysis (1)] |
| 20. | Song Z, Cai J, Liu Y, Zhao D, Yong J, Duo S, Song X, Guo Y, Zhao Y, Qin H, Yin X, Wu C, Che J, Lu S, Ding M, Deng H. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009;19:1233-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 365] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 21. | Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 944] [Cited by in RCA: 949] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 22. | Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, Dalgetty D, Black JR, Ross JA, Samuel K, Wang G, Daley GQ, Lee JH, Church GM, Forbes SJ, Iredale JP, Wilmut I. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 312] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 23. | Touboul T, Hannan NR, Corbineau S, Martinez A, Martinet C, Branchereau S, Mainot S, Strick-Marchand H, Pedersen R, Di Santo J, Weber A, Vallier L. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 369] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 24. | Agarwal S, Holton KL, Lanza R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells. 2008;26:1117-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 300] [Cited by in RCA: 271] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 25. | Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 356] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 26. | Rambhatla L, Chiu CP, Kundu P, Peng Y, Carpenter MK. Generation of hepatocyte-like cells from human embryonic stem cells. Cell Transplant. 2003;12:1-11. [PubMed] |
| 27. | Soto-Gutierrez A, Navarro-Alvarez N, Rivas-Carrillo JD, Chen Y, Yamatsuji T, Tanaka N, Kobayashi N. Differentiation of human embryonic stem cells to hepatocytes using deleted variant of HGF and poly-amino-urethane-coated nonwoven polytetrafluoroethylene fabric. Cell Transplant. 2006;15:335-341. [PubMed] |
| 28. | Kamiya A, Kinoshita T, Miyajima A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett. 2001;492:90-94. [PubMed] |
| 29. | Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1076] [Cited by in RCA: 1032] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 30. | Kinoshita T, Sekiguchi T, Xu MJ, Ito Y, Kamiya A, Tsuji K, Nakahata T, Miyajima A. Hepatic differentiation induced by oncostatin M attenuates fetal liver hematopoiesis. Proc Natl Acad Sci USA. 1999;96:7265-7270. [PubMed] |
| 31. | Kinoshita T, Miyajima A. Cytokine regulation of liver development. Biochim Biophys Acta. 2002;1592:303-312. [PubMed] |
| 32. | Kamiya A, Kinoshita T, Ito Y, Matsui T, Morikawa Y, Senba E, Nakashima K, Taga T, Yoshida K, Kishimoto T, Miyajima A. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J. 1999;18:2127-2136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 328] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 33. | Funakoshi N, Duret C, Pascussi JM, Blanc P, Maurel P, Daujat-Chavanieu M, Gerbal-Chaloin S. Comparison of hepatic-like cell production from human embryonic stem cells and adult liver progenitor cells: CAR transduction activates a battery of detoxification genes. Stem Cell Rev. 2011;7:518-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 34. | Ogawa S, Surapisitchat J, Virtanen C, Ogawa M, Niapour M, Sugamori KS, Wang S, Tamblyn L, Guillemette C, Hoffmann E, Zhao B, Strom S, Laposa RR, Tyndale RF, Grant DM, Keller G. Three-dimensional culture and camp signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development. 2013;140:3285-3296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 35. | Borowiak M, Maehr R, Chen S, Chen AE, Tang W, Fox JL, Schreiber SL, Melton DA. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell. 2009;4:348-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 355] [Cited by in RCA: 327] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 36. | Tahamtani Y, Azarnia M, Farrokhi A, Sharifi-Zarchi A, Aghdami N, Baharvand H. Treatment of human embryonic stem cells with different combinations of priming and inducing factors toward definitive endoderm. Stem Cells Dev. 2013;22:1419-1432. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Siller R, Greenhough S, Naumovska E, Sullivan Gareth J. Small-molecule-driven hepatocyte differentiation of human pluripotent stem cells. Stem Cell Reports. 2015;4:939-952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 38. | Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA, North TE, Goessling W, Carpenter AE, Bhatia SN. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514-520. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 197] [Cited by in RCA: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 39. | Li S, Li M, Liu X, Yang Y, Wei Y, Chen Y, Qiu Y, Zhou T, Feng Z, Ma D, Fang J, Ying H, Wang H, Musunuru K, Shao Z, Zhao Y, Ding Q. Genetic and Chemical Screenings Identify HDAC3 as a Key Regulator in Hepatic Differentiation of Human Pluripotent Stem Cells. Stem Cell Reports. 2018;11:22-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 40. | Rashid ST, Corbineau S, Hannan N, Marciniak SJ, Miranda E, Alexander G, Huang-Doran I, Griffin J, Ahrlund-Richter L, Skepper J, Semple R, Weber A, Lomas DA, Vallier L. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127-3136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 498] [Cited by in RCA: 440] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 41. | Zhang S, Chen S, Li W, Guo X, Zhao P, Xu J, Chen Y, Pan Q, Liu X, Zychlinski D, Lu H, Tortorella MD, Schambach A, Wang Y, Pei D, Esteban MA. Rescue of ATP7B function in hepatocyte-like cells from Wilson's disease induced pluripotent stem cells using gene therapy or the chaperone drug curcumin. Hum Mol Genet. 2011;20:3176-3187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 114] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 42. | Cayo MA, Cai J, DeLaForest A, Noto FK, Nagaoka M, Clark BS, Collery RF, Si-Tayeb K, Duncan SA. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012;56:2163-2171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 109] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 43. | Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, Trevisan M, Gupta RM, Moisan A, Banks E, Friesen M, Schinzel RT, Xia F, Tang A, Xia Y, Figueroa E, Wann A, Ahfeldt T, Daheron L, Zhang F, Rubin LL, Peng LF, Chung RT, Musunuru K, Cowan CA. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12:238-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 401] [Cited by in RCA: 387] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 44. | Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 401] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 45. | Cayo MA, Mallanna SK, Di Furio F, Jing R, Tolliver LB, Bures M, Urick A, Noto FK, Pashos EE, Greseth MD, Czarnecki M, Traktman P, Yang W, Morrisey EE, Grompe M, Rader DJ, Duncan SA. A Drug Screen using Human iPSC-Derived Hepatocyte-like Cells Reveals Cardiac Glycosides as a Potential Treatment for Hypercholesterolemia. Cell Stem Cell. 2017;20:478-489.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 84] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 46. | Yoshida T, Takayama K, Kondoh M, Sakurai F, Tani H, Sakamoto N, Matsuura Y, Mizuguchi H, Yagi K. Use of human hepatocyte-like cells derived from induced pluripotent stem cells as a model for hepatocytes in hepatitis C virus infection. Biochem Biophys Res Commun. 2011;416:119-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 47. | Zhou XL, Sullivan GJ, Sun P, Park IH. Humanized murine model for HBV and HCV using human induced pluripotent stem cells. Arch Pharm Res. 2012;35:261-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 48. | Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, Li W, Lian Z, Liu Y, Lv W, Lytle-Gabbin SL, Marchadier DH, Rogov P, Shi J, Slovik KJ, Stylianou IM, Wang L, Yan R, Zhang X, Kathiresan S, Duncan SA, Mikkelsen TS, Morrisey EE, Rader DJ, Brown CD, Musunuru K. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell. 2017;20:558-570.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 121] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 49. | Warren CR, O'Sullivan JF, Friesen M, Becker CE, Zhang X, Liu P, Wakabayashi Y, Morningstar JE, Shi X, Choi J, Xia F, Peters DT, Florido MHC, Tsankov AM, Duberow E, Comisar L, Shay J, Jiang X, Meissner A, Musunuru K, Kathiresan S, Daheron L, Zhu J, Gerszten RE, Deo RC, Vasan RS, O'Donnell CJ, Cowan CA. Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Metabolic Disease. Cell Stem Cell. 2017;20:547-557.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 50. | Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D'Souza SL, Knowles JW, Patel A, Papatsenko D, Abbasi F, Reaven GM, Whalen S, Lee P, Shahbazi M, Henrion MYR, Zhu K, Wang S, Roussos P, Schadt EE, Pandey G, Chang R, Quertermous T, Lemischka I. Analysis of Transcriptional Variability in a Large Human iPSC Library Reveals Genetic and Non-genetic Determinants of Heterogeneity. Cell Stem Cell. 2017;20:518-532.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 199] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 51. | DeBoever C, Li H, Jakubosky D, Benaglio P, Reyna J, Olson KM, Huang H, Biggs W, Sandoval E, D'Antonio M, Jepsen K, Matsui H, Arias A, Ren B, Nariai N, Smith EN, D'Antonio-Chronowska A, Farley EK, Frazer KA. Large-Scale Profiling Reveals the Influence of Genetic Variation on Gene Expression in Human Induced Pluripotent Stem Cells. Cell Stem Cell. 2017;20:533-546.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 52. | Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Danecek P, Faulconbridge A, Harrison PW, Kathuria A, McCarthy D, McCarthy SA, Meleckyte R, Memari Y, Moens N, Soares F, Mann A, Streeter I, Agu CA, Alderton A, Nelson R, Harper S, Patel M, White A, Patel SR, Clarke L, Halai R, Kirton CM, Kolb-Kokocinski A, Beales P, Birney E, Danovi D, Lamond AI, Ouwehand WH, Vallier L, Watt FM, Durbin R, Stegle O, Gaffney DJ. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370-375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 473] [Cited by in RCA: 401] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 53. | Warren CR, Cowan CA. Humanity in a Dish: Population Genetics with iPSCs. Trends Cell Biol. 2018;28:46-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 54. | Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A, Ringnalda F, Begthel H, Hamer K, Mulder J, van Es JH, de Koning E, Vries RG, Heimberg H, Clevers H. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708-2721. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 460] [Cited by in RCA: 535] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 55. | Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299-312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1153] [Cited by in RCA: 1130] [Article Influence: 113.0] [Reference Citation Analysis (0)] |
| 56. | Ogawa M, Ogawa S, Bear CE, Ahmadi S, Chin S, Li B, Grompe M, Keller G, Kamath BM, Ghanekar A. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat Biotechnol. 2015;33:853-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 224] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 57. | Sampaziotis F, de Brito MC, Madrigal P, Bertero A, Saeb-Parsy K, Soares FAC, Schrumpf E, Melum E, Karlsen TH, Bradley JA, Gelson WT, Davies S, Baker A, Kaser A, Alexander GJ, Hannan NRF, Vallier L. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015;33:845-852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 292] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 58. | Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1763] [Cited by in RCA: 1511] [Article Influence: 125.9] [Reference Citation Analysis (0)] |
| 59. | Takebe T, Zhang RR, Koike H, Kimura M, Yoshizawa E, Enomura M, Koike N, Sekine K, Taniguchi H. Generation of a vascularized and functional human liver from an iPSC-derived organ bud transplant. Nat Protoc. 2014;9:396-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 273] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 60. | Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O'Sullivan JF, Grainger SJ, Kapp FG, Sun L, Christensen K, Xia Y, Florido MH, He W, Pan W, Prummer M, Warren CR, Jakob-Roetne R, Certa U, Jagasia R, Freskgård PO, Adatto I, Kling D, Huang P, Zon LI, Chaikof EL, Gerszten RE, Graf M, Iacone R, Cowan CA. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol. 2015;17:994-1003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 455] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 61. | Coll M, Perea L, Boon R, Leite SB, Vallverdú J, Mannaerts I, Smout A, El Taghdouini A, Blaya D, Rodrigo-Torres D, Graupera I, Aguilar-Bravo B, Chesne C, Najimi M, Sokal E, Lozano JJ, van Grunsven LA, Verfaillie CM, Sancho-Bru P. Generation of Hepatic Stellate Cells from Human Pluripotent Stem Cells Enables In Vitro Modeling of Liver Fibrosis. Cell Stem Cell. 2018;23:101-113.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 163] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 62. | Sun Y, Ding Q. Genome engineering of stem cell organoids for disease modeling. Protein Cell. 2017;8:315-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 63. | Solanas E, Pla-Palacín I, Sainz-Arnal P, Almeida M, Lue A, Serrano T, Baptista PM. Tissue organoids: Liver. Soker S and Skardal A Tumor organoids. Cham: Springer International Publishing 2018; 17-33. |
| 64. | Mito M. Hepatic assist: present and future. Artif Organs. 1986;10:214-218. [PubMed] |
| 65. | Sakiyama R, Blau BJ, Miki T. Clinical translation of bioartificial liver support systems with human pluripotent stem cell-derived hepatic cells. World J Gastroenterol. 2017;23:1974-1979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 34] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (3)] |
| 66. | Struecker B, Raschzok N, Sauer IM. Liver support strategies: cutting-edge technologies. Nat Rev Gastroenterol Hepatol. 2014;11:166-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 67. | Shi XL, Gao Y, Yan Y, Ma H, Sun L, Huang P, Ni X, Zhang L, Zhao X, Ren H, Hu D, Zhou Y, Tian F, Ji Y, Cheng X, Pan G, Ding YT, Hui L. Improved survival of porcine acute liver failure by a bioartificial liver device implanted with induced human functional hepatocytes. Cell Res. 2016;26:206-216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 68. | Mavri-Damelin D, Damelin LH, Eaton S, Rees M, Selden C, Hodgson HJ. Cells for bioartificial liver devices: the human hepatoma-derived cell line C3A produces urea but does not detoxify ammonia. Biotechnol Bioeng. 2008;99:644-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 67] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 69. | Allen KJ, Soriano HE. Liver cell transplantation: the road to clinical application. J Lab Clin Med. 2001;138:298-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 70. | Dhawan A, Mitry RR, Hughes RD. Hepatocyte transplantation for liver-based metabolic disorders. J Inherit Metab Dis. 2006;29:431-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 139] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 71. | Fisher RA, Strom SC. Human hepatocyte transplantation: worldwide results. Transplantation. 2006;82:441-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 324] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 72. | Roy-Chowdhury N, Wang X, Guha C, Roy-Chowdhury J. Hepatocyte-like cells derived from induced pluripotent stem cells. Hepatol Int. 2017;11:54-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 73. | Carpentier A, Tesfaye A, Chu V, Nimgaonkar I, Zhang F, Lee SB, Thorgeirsson SS, Feinstone SM, Liang TJ. Engrafted human stem cell-derived hepatocytes establish an infectious HCV murine model. J Clin Invest. 2014;124:4953-4964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 74. | Nagamoto Y, Takayama K, Tashiro K, Tateno C, Sakurai F, Tachibana M, Kawabata K, Ikeda K, Tanaka Y, Mizuguchi H. Efficient Engraftment of Human Induced Pluripotent Stem Cell-Derived Hepatocyte-Like Cells in uPA/SCID Mice by Overexpression of FNK, a Bcl-xL Mutant Gene. Cell Transplant. 2015;24:1127-1138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 75. | Azuma H, Paulk N, Ranade A, Dorrell C, Al-Dhalimy M, Ellis E, Strom S, Kay MA, Finegold M, Grompe M. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903-910. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 712] [Cited by in RCA: 647] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 76. | Asgari S, Moslem M, Bagheri-Lankarani K, Pournasr B, Miryounesi M, Baharvand H. Differentiation and transplantation of human induced pluripotent stem cell-derived hepatocyte-like cells. Stem Cell Rev. 2013;9:493-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 77. | Liu H, Kim Y, Sharkis S, Marchionni L, Jang YY. In vivo liver regeneration potential of human induced pluripotent stem cells from diverse origins. Sci Transl Med. 2011;3:82ra39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 181] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 78. | Nagamoto Y, Takayama K, Ohashi K, Okamoto R, Sakurai F, Tachibana M, Kawabata K, Mizuguchi H. Transplantation of a human iPSC-derived hepatocyte sheet increases survival in mice with acute liver failure. J Hepatol. 2016;64:1068-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 110] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 79. | Song W, Lu YC, Frankel AS, An D, Schwartz RE, Ma M. Engraftment of human induced pluripotent stem cell-derived hepatocytes in immunocompetent mice via 3D co-aggregation and encapsulation. Sci Rep. 2015;5:16884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 80. | Lemaigre FP. Mechanisms of liver development: concepts for understanding liver disorders and design of novel therapies. Gastroenterology. 2009;137:62-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 175] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 81. | Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1459] [Cited by in RCA: 1328] [Article Influence: 78.1] [Reference Citation Analysis (0)] |
| 82. | Takayama K, Akita N, Mimura N, Akahira R, Taniguchi Y, Ikeda M, Sakurai F, Ohara O, Morio T, Sekiguchi K, Mizuguchi H. Generation of safe and therapeutically effective human induced pluripotent stem cell-derived hepatocyte-like cells for regenerative medicine. Hepatol Commun. 2017;1:1058-1069. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 83. | Ben-David U, Gan QF, Golan-Lev T, Arora P, Yanuka O, Oren YS, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, Gromo G, Benvenisty N. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 255] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 84. | Tang C, Lee AS, Volkmer JP, Sahoo D, Nag D, Mosley AR, Inlay MA, Ardehali R, Chavez SL, Pera RR, Behr B, Wu JC, Weissman IL, Drukker M. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29:829-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 291] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 85. | Peters DT, Henderson CA, Warren CR, Friesen M, Xia F, Becker CE, Musunuru K, Cowan CA. Asialoglycoprotein receptor 1 is a specific cell-surface marker for isolating hepatocytes derived from human pluripotent stem cells. Development. 2016;143:1475-1481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 86. | Kiuru M, Boyer JL, O'Connor TP, Crystal RG. Genetic control of wayward pluripotent stem cells and their progeny after transplantation. Cell Stem Cell. 2009;4:289-300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 87. | Choi SH, Kim YH, Quinti L, Tanzi RE, Kim DY. 3D culture models of Alzheimer's disease: a road map to a "cure-in-a-dish". Mol Neurodegener. 2016;11:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 88. | Giacomelli E, Bellin M, Sala L, van Meer BJ, Tertoolen LG, Orlova VV, Mummery CL. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development. 2017;144:1008-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 198] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 89. | Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang DZ, Li K, Wang J, Wanders RJ, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616-623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 612] [Cited by in RCA: 655] [Article Influence: 59.5] [Reference Citation Analysis (0)] |