Published online Jul 26, 2019. doi: 10.4252/wjsc.v11.i7.375
Peer-review started: May 10, 2019
First decision: June 5, 2019
Revised: June 12, 2019
Accepted: June 20, 2019
Article in press: June 29, 2019
Published online: July 26, 2019
Processing time: 80 Days and 20.5 Hours
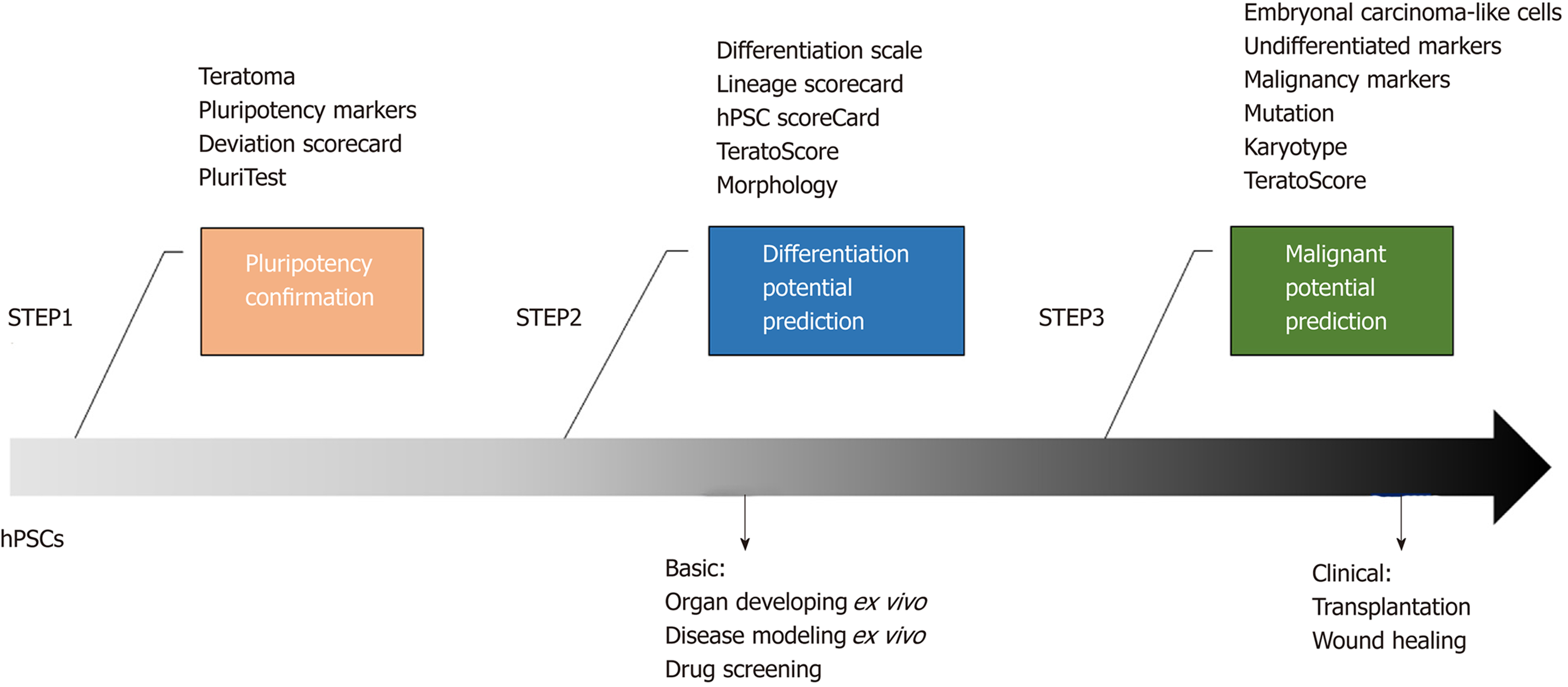
The capability of human pluripotent stem cell (hPSC) lines to propagate indefinitely and differentiate into derivatives of three embryonic germ layers makes these cells be powerful tools for basic scientific research and promising agents for translational medicine. However, variations in differentiation tendency and efficiency as well as pluripotency maintenance necessitate the selection of hPSC lines for the intended applications to save time and cost. To screen the qualified cell lines and exclude problematic cell lines, their pluripotency must be confirmed initially by traditional methods such as teratoma formation or by high-throughput gene expression profiling assay. Additionally, their differentiation potential, particularly the lineage-specific differentiation propensities of hPSC lines, should be predicted in an early stage. As a complement to the teratoma assay, RNA sequencing data provide a quantitative estimate of the differentiation ability of hPSCs in vivo. Moreover, multiple scorecards have been developed based on selected gene sets for predicting the differentiation potential into three germ layers or the desired cell type many days before terminal differentiation. For clinical application of hPSCs, the malignant potential of the cells must also be evaluated. A combination of histologic examination of teratoma with quantitation of gene expression data derived from teratoma tissue provides safety-related predictive information by detecting immature teratomas, malignancy marker expression, and other parameters. Although various prediction methods are available, distinct limitations remain such as the discordance of results between different assays and requirement of a long time and high labor and cost, restricting their wide applications in routine studies. Therefore, simpler and more rapid detection assays with high specificity and sensitivity that can be used to monitor the status of hPSCs at any time and fewer targeted markers that are more specific for a given desired cell type are urgently needed.
Core tip: To save time and costs in basic research and clinical application, it is necessary to predict the differentiation potential of human pluripotent stem cell (hPSC) lines. Multiple methods are available for pluripotency screening, lineage-specific differentiation propensity prediction, and malignancy potential detection, which can be used to select hPSCs. However, simpler and quicker methods using fewer specific targeted markers for the desired cell type are urgently required for routine work.
- Citation: Liu LP, Zheng YW. Predicting differentiation potential of human pluripotent stem cells: Possibilities and challenges. World J Stem Cells 2019; 11(7): 375-382
- URL: https://www.wjgnet.com/1948-0210/full/v11/i7/375.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v11.i7.375
The capability of human pluripotent stem cells (hPSCs) to differentiate into any cell type has revolutionized medical research. Their widely known potential applications include the study of complex diseases, cell-based drug screening, and transplantation therapy[1]. With the development of organoid technology, hPSCs also play a critical role to mimic in vivo tissues and organs at the three-dimensional level and provide a unique opportunity to model human organ development and study various diseases[2]. In the near future, integration of multiple patient-specific hPSC-derived organoids into a dynamic four-dimensional system by organ-on-chip technology will contribute to the study of the systematic interactions among different tissues and organs in the body[3]. All these applications require the selection and characterization of cell lines that reliably, efficiently, and stably differentiate into disease-relevant cell types. However, significant variation has been observed in the differentiation potential and efficiency of various human induced pluripotent stem cell (iPSC) lines[1,4,5] and embryonic stem cells (ESCs)[6-8], and no single cell line can uniformly differentiate into all lineages. Differences among hPSC lines mainly include DNA methylation[9-13] and gene expression[1,8], which have functional implications for both ESC and iPSC lines[1]. Particularly, for iPSCs, the variations may be donor-dependent[14,15] or original cell type-dependent[9], while, in other studies, this relationship was not found[10,16]. Moreover, the characteristics of hPSCs may differ depending on the number of passages[17], culture medium components[18], and feeder conditions[19]. As a result, understanding the variability among different cell lines is necessary for ensuring the efficacy and safety of hPSC applications. Additionally, a long period (up to weeks or months) is typically required to complete hPSC differentiation into a specific cell type, and protocol optimization to improve differentiation efficiency is also time-consuming. Thus, these processes can be considerably accelerated if good-quality hPSC lines are selected, and their differentiation propensities into destination cell types are predicted in an early stage.
Given that large numbers of iPSC lines are currently and will be generated, the pluripotency of these cells must be determined before they are broadly applied. To confirm whether an iPSC line is fully reprogrammed, a teratoma assay is typically performed to reveal the differentiation capacity of hPSCs into three germ layers based on histological analysis. The pluripotency status can also be determined by detecting the expression of a set of marker genes at the molecular level[20]. With the development of high-throughput sequencing techniques, numerous methods have been developed to address this issue. The PluriTest®[21], a rapid test based on microarray and bioinformatics assay, provides quantitative information on hPSC quality. Two summary scores, the pluripotency score and novelty score[22,23], which are generated from global gene expression profiles, can predict whether an hPSC line is pluripotent based on its molecular similarity to other known cell lines and exclude cells that differ substantially from normal hPSC lines. An hPSC line with a high pluripotency score and low novelty score would be regarded as having passed the PluriTest. Another assay, the “deviation scorecard”[1], which combines DNA methylation and gene expression with bioinformatic comparison to an ESC reference, also provides comprehensive information and excludes problematic cell lines that should be avoided for an intended application. However, these methods can only be used to determine whether an hPSC line meets the criteria for pluripotency and do not directly assess the specific differentiation capability of the cells.
As described above, the teratoma assay is the most frequently used method to assess the pluripotency of hPSCs. However, it does not yield quantitative information on lineage differentiation potential[24] nor provide specificity data to support the application-specific selection of the most suitable cell lines[25]. Thus, the “TeratoScore” was developed[26] as a quantitative and unified assessment that analyzes RNA sequencing data within heterogeneous hPSC-derived teratomas. This score weighs differences in tissue-specific expression within a teratoma and provides an estimate of the ability of an hPSC line to differentiate[22,23,26] to overcome some of the limitations of histological analysis.
hPSC differentiation is a complex and multiple-step process, the beginning of which involves a particularly heterogeneous status and diverse developmental mixture. During cell fate commitment to a differentiated lineage, genes are regulated by successive transcriptional programs, and thus, the differentiation status can be determined from transcriptional profiles[27]. A combination of gene expression profiling and bioinformatics assay is invaluable for predicting the trajectory of cells during differentiation. The “differentiation scale”[27] based on mRNA microarray analysis can indicate the staging of differentiation and show how far the hPSCs have departed from the embryonic pluripotent state. However, this method only measures the overall capability of a cell line towards differentiation into any cell type and cannot clearly distinguish between any direction of differentiation into the three germ layers. In contrast, the “lineage scorecard” assay, which combines simple nondirected differentiation with transcript counting of 500 lineage marker genes, can detect the lineage-specific differentiation propensities of an hPSC line[1]. For example, hPSC lines showing high scores for ectoderm and neural differentiation propensities are regarded as well-suited for studying neural function. This prediction has been confirmed in experiments to quantify differentiation efficiencies specific for the ectoderm germ layer. Additionally, other scores calculated based on the expression levels of a set of selected specific gene markers can predict hPSC differentiation potential in similar manners[22,23,28]. These qPCR-based assays are more rapid and accessible than high-throughput methods.
According to the hPSC-derived differentiation protocols for various types of cells, embryoid bodies (EBs) are widely used[29] because they mimic many aspects of cell differentiation during early embryogenesis. Because they consist of tissues containing three germ layers[30], EBs can be utilized as a trigger of not only in vitro differentiation of hPSCs but also in assay predicting differentiation potential. For the latter purpose, EBs are induced to spontaneously differentiate under neutral conditions[1,22] or directed into three germ layer lineages in the presence of specific growth factors[22,28], after which gene expression profiling of EBs is conducted to assess their differen-tiation potential into the ectoderm, mesoderm, or endoderm lineages. In our study to predict the differentiation potential of iPSCs into melanocytes derived from the ectoderm, an EB-based assay showed that this potential could be predicted by the capability of formation and maintenance of optimal EBs under neutral conditions as well as their expression of germ layer-specific markers such as SALL3 (our unpublished data). Thus, EBs are practical tools for use in prediction assays at early stages of differentiation. In addition to EB-based protocols, monolayer differen-tiation[23,31] has been adapted and shown to be applicable for evaluating endoderm and mesoderm direction; however, to predict the ectodermal direction, EB-based protocols may be more effective[23].
Regardless of the detection techniques used, prediction can be achieved many days before the cells exhibit a differentiated phenotype (Table 1). For instance, the efficiency of protocols for directing differentiation to cardiac cells can be predicted at as early as day 2[31], and thus can be utilized to optimize differentiation protocols, particularly for patient-specific iPSC lines by using a high-throughput screening procedure. In contrast to the above methods based on EBs or monolayer diffe-rentiation protocols, which require a couple of days before evaluation, simpler methods with limited specific markers can be used for earlier prediction using undifferentiated hPSCs. The expression level of SALL3 mRNA was used as a diagnostic marker to predict the differentiation tendency of both iPSCs and ESCs into ectodermal cells[32]. hPSCs expressing the highest levels of SALL3 mRNA tend to differentiate into the most ectodermal system, while cells expressing the lowest levels of SALL3 mRNA tend to differentiate into the most mesodermal or endodermal cell types. Specifically, three genes, FGF-1, RHOU, and TYMP, were selected as predictors of hepatic differentiation, with low prediction scores linked to low hepatic diffe-rentiation[33].
| Aim | Techniques | Targets | Cell treatment | Timepoint detection | Ref. |
| Pluripotency | DNA methylation sequencing and microarray | Deviation scorecard: The cell line-specific number of outliers relative to the human ES cell reference | N/A | N/A | [1] |
| Microarray | PluriTest: Pluripotency score (refers to gene expression profiles of a large collection of human PSCs) and novelty score (refers to gene expression patterns not typically associated with human PSCs) | N/A | N/A | [21-23] | |
| qPCR | Level of CHD7 | N/A | N/A | [43] | |
| Differentiation potential | Microarray | Differentiation scale: a subset of the 1000 most informative genes | W/O | At any time of differentiation | [27] |
| Microarray | Lineage scorecard: 500 lineage marker genes to monitor cell state, pluripotency, and differentiation | Nondirected EB differentiation | At 16 d of differentiation | [1] | |
| qPCR | Lineage scorecard: 15 selected marker genes per lineage | Nondirected and directed differentiation into three germ lineages | [22] | ||
| qPCR | hPSC ScoreCard: 9 self-renewal genes and 70 genes representing specific lineages | Monolayer or EB protocol differentiation into three germ lineages | At 5 or 9 d | [23] | |
| Single-cell qPCR | 96 developmental genes; a transcriptional circuit (HAND1-SOX17) and phenotypic readout (cKIT distribution) | Cardiomyocyte differentiation | On day 2 | [31] | |
| qPCR | Improved scorecard: 96 specific gene markers | Directed EB differentiation into three germ lineages | At 12 d of differentiation. | [28] | |
| Microarray or RNA-sequencing | TeratoScore: 100 tissue-specific genes representing the three embryonic germ layers and extra- embryonic membranes | Xenograft teratoma formation | A suitable growth period | [22,23,26] | |
| Morphology | Definitive endoderm morphology production | Treated with small molecules | 48 h after induction | [44] | |
| qPCR | SALL3 for ectodermal differentiation | N/A | N/A | [32] | |
| PCR array | Prediction scores for hepatic differentiation based on the expression of the three genes FGF-1, RHOU, and TYMP | N/A | N/A | [33] | |
| Malignant potential | Histology, qPCR, and microarray | TeratoScore: 10 undifferentiated hPSC markers; embryonal carcinoma-like cells with yolk sac elements; undifferentiated hPSC marker and malignancy marker expression | Xenograft teratoma formation | Suitable growth period | [22] |
| TeratoScore: 100 tissue-specific genes; embryonal carcinoma-like cells; undifferentiated hPSC marker and malignancy marker expression | [23] | ||||
| Histology, microarray, karyotype analysis, and whole exome sequencing | Formation of immature teratomas, carcinoma and sarcoma; mutation of cancer-related genes; chromosomal abnormalities | Xenograft teratoma formation; N/A | Suitable growth period; N/A | [40] |
Collectively, the lineage-specific differentiation potential of hPSC lines can be predicted in an early stage using multiple assays, including the teratoma assay, different scorecards calculated by high-throughput sequencing data collected from EBs or monolayer-based differentiated hPSCs, or specific maker expression in undifferentiated hPSCs.
The safety of using hPSCs in clinical application is one of the greatest concerns for both clinicians and patients[34]. Currently, tumorigenicity tests are well-designed, and animal transplantation studies have been used to detect the malignancy potential of hPSC-derived cell products[35,36]. Although residual undifferentiated cells are present among the differentiated cell population, these cells can be ablated by various techniques[37,38]. Additionally, evaluation of hPSCs rather than their derivatives can provide safety-related information. As described above, the teratoma assay is commonly used to measure the pluripotency and differentiation potential of hPSCs[22,39]. It is also feasible to predict the malignancy potential by detecting the immaturity of teratomas and formation of carcinoma or sarcoma in tumors derived from hPSC lines[40]. Furthermore, a combination of histologic examination and “TeratoScore”, which involves computational quantification of gene expression data derived from teratoma tissue, can provide much greater detail for evaluating whether a cell line has the malignant potential[22,23]. It is also important to examine chromo-somal abnormalities by karyotype analysis[40,41] and assess mutations in cancer-related genes[40].
Collectively, multiple screening tools are available for hPSC selection with different features and prediction capabilities, and researchers can choose one or more methods according to the intended applications (Table 1 and Figure 1). However, concordance between two different prediction methods is low[22,23], and even with the same prediction assay, distinct results may be obtained under different differentiation conditions. The lack of concordance of results makes it very difficult for researches to determine the most appropriate method for their purposes. Moreover, each prediction method has limitations, thus largely restricting its wide applications. High-throughput analysis methods such as microarray or RNA-sequencing are informative but generate excessive data, some of which are non-functional. Additionally, iden-tifying objective genes is time-and labor-intensive as well as costly[33]. The platforms used in these assays are not available to most laboratories and require customized downstream analysis, which also restricts their applications. Although teratoma formation reflects the in vivo differentiation capability of hPSCs and gene expression analysis can provide more definitive and quantitative information when combined with histological assessment, these methods are also very time-consuming to be feasible for validating a large number of hPSC lines[1].
Because of these limitations, the methods described above are not sufficient to meet the needs of researchers in routine work. Additionally, hPSC characteristics are not stable during long-term culture, which can be affected by several factors. Further, hPSCs can vary when they are cultured using different commercial products or show different features in different laboratories, even under the same culture conditions. Therefore, a simpler and quicker detection assay with high specificity and sensitivity that can be used to monitor the status of hPSCs at any time is needed. Although most current methods are useful for lineage-direction prediction, fewer targeted markers that are more specific for a given cell type rather than all cell types are urgently needed. Moreover, EBs are ideal candidates for replacing the teratoma assay, and they should be useful for predicting differentiation potential when they are combined with modified detection methods such molecular probes, which could detect targeted markers directly.
Exceptionally, the potential prediction is not a prerequisite when iPSCs from patients with monogenic disease are utilized for disease modeling because the differentiation capability is probably interfered by gene mutation[42]. However, iPSC quality control is still necessary. When using these patient-derived iPSCs for cell therapy, it is also critical to predict their differentiation potential after some special strategies such as gene editing, which can revert their defective capability.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Andrukhov O, Binetruy B, Huang YC, Politi LE, Wang H, Zeng L S-Editor: Ma YJ L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439-452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 852] [Cited by in RCA: 754] [Article Influence: 53.9] [Reference Citation Analysis (0)] |
| 2. | Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1502] [Cited by in RCA: 1912] [Article Influence: 173.8] [Reference Citation Analysis (0)] |
| 3. | Liu C, Oikonomopoulos A, Sayed N, Wu JC. Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development. 2018;145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 190] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 4. | Koyanagi-Aoi M, Ohnuki M, Takahashi K, Okita K, Noma H, Sawamura Y, Teramoto I, Narita M, Sato Y, Ichisaka T, Amano N, Watanabe A, Morizane A, Yamada Y, Sato T, Takahashi J, Yamanaka S. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci USA. 2013;110:20569-20574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 5. | Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, Endo H, Eto K, Toguchida J, Uemoto S, Yamanaka S. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109:12538-12543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 254] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 6. | Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 379] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 7. | Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 646] [Cited by in RCA: 631] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 8. | Sun C, Zhang J, Zheng D, Wang J, Yang H, Zhang X. Transcriptome variations among human embryonic stem cell lines are associated with their differentiation propensity. PLoS One. 2018;13:e0192625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Kim K, Zhao R, Doi A, Ng K, Unternaehrer J, Cahan P, Huo H, Loh YH, Aryee MJ, Lensch MW, Li H, Collins JJ, Feinberg AP, Daley GQ. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117-1119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 432] [Cited by in RCA: 466] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 10. | Dorn I, Klich K, Arauzo-Bravo MJ, Radstaak M, Santourlidis S, Ghanjati F, Radke TF, Psathaki OE, Hargus G, Kramer J, Einhaus M, Kim JB, Kögler G, Wernet P, Schöler HR, Schlenke P, Zaehres H. Erythroid differentiation of human induced pluripotent stem cells is independent of donor cell type of origin. Haematologica. 2015;100:32-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 11. | Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1294] [Cited by in RCA: 1149] [Article Influence: 82.1] [Reference Citation Analysis (0)] |
| 12. | Nishizawa M, Chonabayashi K, Nomura M, Tanaka A, Nakamura M, Inagaki A, Nishikawa M, Takei I, Oishi A, Tanabe K, Ohnuki M, Yokota H, Koyanagi-Aoi M, Okita K, Watanabe A, Takaori-Kondo A, Yamanaka S, Yoshida Y. Epigenetic Variation between Human Induced Pluripotent Stem Cell Lines Is an Indicator of Differentiation Capacity. Cell Stem Cell. 2016;19:341-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 166] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 13. | He W, Kang X, Du H, Song B, Lu Z, Huang Y, Wang D, Sun X, Yu Y, Fan Y. Defining differentially methylated regions specific for the acquisition of pluripotency and maintenance in human pluripotent stem cells via microarray. PLoS One. 2014;9:e108350. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. 2014;10:e1004432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 229] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 15. | Kyttälä A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, Van Handel B, Parkkonen O, Sinisalo J, Jalanko A, Hawkins RD, Woods NB, Otonkoski T, Trokovic R. Genetic Variability Overrides the Impact of Parental Cell Type and Determines iPSC Differentiation Potential. Stem Cell Reports. 2016;6:200-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 195] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 16. | Philonenko ES, Shutova MV, Khomyakova EA, Vassina EM, Lebedeva OS, Kiselev SL, Lagarkova MA. Differentiation of Human Pluripotent Stem Cells into Mesodermal and Ectodermal Derivatives Is Independent of the Type of Isogenic Reprogrammed Somatic Cells. Acta Naturae. 2017;9:68-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 17. | Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, Thykjaer T, Maltby E, Smith K, Abu Dawud R, Jones M, Matin M, Gokhale P, Draper J, Andrews PW. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet. 2005;14:3129-3140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 223] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 18. | Nishihara K, Shiga T, Nakamura E, Akiyama T, Sasaki T, Suzuki S, Ko MSH, Tada N, Okano H, Akamatsu W. Induced Pluripotent Stem Cells Reprogrammed with Three Inhibitors Show Accelerated Differentiation Potentials with High Levels of 2-Cell Stage Marker Expression. Stem Cell Reports. 2019;12:305-318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 19. | Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 790] [Cited by in RCA: 734] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 20. | International Stem Cell Initiative. Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo AB, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K, Fleck RA, Ford A, Gertow K, Gertsenstein M, Gokhale PJ, Hamilton RS, Hampl A, Healy LE, Hovatta O, Hyllner J, Imreh MP, Itskovitz-Eldor J, Jackson J, Johnson JL, Jones M, Kee K, King BL, Knowles BB, Lako M, Lebrin F, Mallon BS, Manning D, Mayshar Y, McKay RD, Michalska AE, Mikkola M, Mileikovsky M, Minger SL, Moore HD, Mummery CL, Nagy A, Nakatsuji N, O'Brien CM, Oh SK, Olsson C, Otonkoski T, Park KY, Passier R, Patel H, Patel M, Pedersen R, Pera MF, Piekarczyk MS, Pera RA, Reubinoff BE, Robins AJ, Rossant J, Rugg-Gunn P, Schulz TC, Semb H, Sherrer ES, Siemen H, Stacey GN, Stojkovic M, Suemori H, Szatkiewicz J, Turetsky T, Tuuri T, van den Brink S, Vintersten K, Vuoristo S, Ward D, Weaver TA, Young LA, Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 793] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 21. | Müller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, Aldenhoff JB, Laurent LC, Loring JF. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315-317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 404] [Cited by in RCA: 352] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 22. | International Stem Cell Initiative. Assessment of established techniques to determine developmental and malignant potential of human pluripotent stem cells. Nat Commun. 2018;9:1925. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 23. | Bouma MJ, van Iterson M, Janssen B, Mummery CL, Salvatori DCF, Freund C. Differentiation-Defective Human Induced Pluripotent Stem Cells Reveal Strengths and Limitations of the Teratoma Assay and In Vitro Pluripotency Assays. Stem Cell Reports. 2017;8:1340-1353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | Andrews PW, Baker D, Benvinisty N, Miranda B, Bruce K, Brüstle O, Choi M, Choi YM, Crook JM, de Sousa PA, Dvorak P, Freund C, Firpo M, Furue MK, Gokhale P, Ha HY, Han E, Haupt S, Healy L, Hei DJ, Hovatta O, Hunt C, Hwang SM, Inamdar MS, Isasi RM, Jaconi M, Jekerle V, Kamthorn P, Kibbey MC, Knezevic I, Knowles BB, Koo SK, Laabi Y, Leopoldo L, Liu P, Lomax GP, Loring JF, Ludwig TE, Montgomery K, Mummery C, Nagy A, Nakamura Y, Nakatsuji N, Oh S, Oh SK, Otonkoski T, Pera M, Peschanski M, Pranke P, Rajala KM, Rao M, Ruttachuk R, Reubinoff B, Ricco L, Rooke H, Sipp D, Stacey GN, Suemori H, Takahashi TA, Takada K, Talib S, Tannenbaum S, Yuan BZ, Zeng F, Zhou Q. Points to consider in the development of seed stocks of pluripotent stem cells for clinical applications: International Stem Cell Banking Initiative (ISCBI). Regen Med. 2015;10:1-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 25. | Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, Rodolfa CT, Dimos JT, Mikkilineni S, MacDermott AB, Woolf CJ, Henderson CE, Wichterle H, Eggan K. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279-286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 410] [Cited by in RCA: 372] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 26. | Avior Y, Biancotti JC, Benvenisty N. TeratoScore: Assessing the Differentiation Potential of Human Pluripotent Stem Cells by Quantitative Expression Analysis of Teratomas. Stem Cell Reports. 2015;4:967-974. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 27. | Zagar L, Mulas F, Garagna S, Zuccotti M, Bellazzi R, Zupan B. Stage prediction of embryonic stem cell differentiation from genome-wide expression data. Bioinformatics. 2011;27:2546-2553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Tsankov AM, Akopian V, Pop R, Chetty S, Gifford CA, Daheron L, Tsankova NM, Meissner A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat Biotechnol. 2015;33:1182-1192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 127] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 29. | Liu LP, Li YM, Guo NN, Li S, Ma X, Zhang YX, Gao Y, Huang JL, Zheng DX, Wang LY, Xu H, Hui L, Zheng YW. Therapeutic Potential of Patient iPSC-Derived iMelanocytes in Autologous Transplantation. Cell Rep. 2019;27:455-466.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 30. | Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J Biosci Bioeng. 2007;103:389-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 358] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 31. | Bargaje R, Trachana K, Shelton MN, McGinnis CS, Zhou JX, Chadick C, Cook S, Cavanaugh C, Huang S, Hood L. Cell population structure prior to bifurcation predicts efficiency of directed differentiation in human induced pluripotent cells. Proc Natl Acad Sci USA. 2017;114:2271-2276. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 32. | Kuroda T, Yasuda S, Sato T, inventors. Evaluation Method of Differentiation Tendency among Undifferentiated Cells, Sall3 Mrna as Evaluation Marker for Differentiation Tendency, and Method of Controlling Differentiation Ability of Undifferentiated Cells. Japan Patent. JP2016073791A. 2016;April 1. |
| 33. | Yanagihara K, Liu Y, Kanie K, Takayama K, Kokunugi M, Hirata M, Fukuda T, Suga M, Nikawa H, Mizuguchi H, Kato R, Furue MK. Prediction of Differentiation Tendency Toward Hepatocytes from Gene Expression in Undifferentiated Human Pluripotent Stem Cells. Stem Cells Dev. 2016;25:1884-1897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 34. | Seki T, Fukuda K. Methods of induced pluripotent stem cells for clinical application. World J Stem Cells. 2015;7:116-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (4)] |
| 35. | Kawamata S, Kanemura H, Sakai N, Takahashi M, Go MJ. Design of a Tumorigenicity Test for Induced Pluripotent Stem Cell (iPSC)-Derived Cell Products. J Clin Med. 2015;4:159-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 36. | Kanemura H, Go MJ, Shikamura M, Nishishita N, Sakai N, Kamao H, Mandai M, Morinaga C, Takahashi M, Kawamata S. Tumorigenicity studies of induced pluripotent stem cell (iPSC)-derived retinal pigment epithelium (RPE) for the treatment of age-related macular degeneration. PLoS One. 2014;9:e85336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 37. | Itakura G, Kawabata S, Ando M, Nishiyama Y, Sugai K, Ozaki M, Iida T, Ookubo T, Kojima K, Kashiwagi R, Yasutake K, Nakauchi H, Miyoshi H, Nagoshi N, Kohyama J, Iwanami A, Matsumoto M, Nakamura M, Okano H. Fail-Safe System against Potential Tumorigenicity after Transplantation of iPSC Derivatives. Stem Cell Reports. 2017;8:673-684. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 38. | Zarogoulidis P, Darwiche K, Sakkas A, Yarmus L, Huang H, Li Q, Freitag L, Zarogoulidis K, Malecki M. Suicide Gene Therapy for Cancer - Current Strategies. J Genet Syndr Gene Ther. 2013;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 39. | Bulic-Jakus F, Katusic Bojanac A, Juric-Lekic G, Vlahovic M, Sincic N. Teratoma: from spontaneous tumors to the pluripotency/malignancy assay. Wiley Interdiscip Rev Dev Biol. 2016;5:186-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 40. | Yasuda S, Kusakawa S, Kuroda T, Miura T, Tano K, Takada N, Matsuyama S, Matsuyama A, Nasu M, Umezawa A, Hayakawa T, Tsutsumi H, Sato Y. Tumorigenicity-associated characteristics of human iPS cell lines. PLoS One. 2018;13:e0205022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 41. | International Stem Cell Initiative. Amps K, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, Baker J, Baker D, Munoz MB, Beil S, Benvenisty N, Ben-Yosef D, Biancotti JC, Bosman A, Brena RM, Brison D, Caisander G, Camarasa MV, Chen J, Chiao E, Choi YM, Choo AB, Collins D, Colman A, Crook JM, Daley GQ, Dalton A, De Sousa PA, Denning C, Downie J, Dvorak P, Montgomery KD, Feki A, Ford A, Fox V, Fraga AM, Frumkin T, Ge L, Gokhale PJ, Golan-Lev T, Gourabi H, Gropp M, Lu G, Hampl A, Harron K, Healy L, Herath W, Holm F, Hovatta O, Hyllner J, Inamdar MS, Irwanto AK, Ishii T, Jaconi M, Jin Y, Kimber S, Kiselev S, Knowles BB, Kopper O, Kukharenko V, Kuliev A, Lagarkova MA, Laird PW, Lako M, Laslett AL, Lavon N, Lee DR, Lee JE, Li C, Lim LS, Ludwig TE, Ma Y, Maltby E, Mateizel I, Mayshar Y, Mileikovsky M, Minger SL, Miyazaki T, Moon SY, Moore H, Mummery C, Nagy A, Nakatsuji N, Narwani K, Oh SK, Oh SK, Olson C, Otonkoski T, Pan F, Park IH, Pells S, Pera MF, Pereira LV, Qi O, Raj GS, Reubinoff B, Robins A, Robson P, Rossant J, Salekdeh GH, Schulz TC, Sermon K, Sheik Mohamed J, Shen H, Sherrer E, Sidhu K, Sivarajah S, Skottman H, Spits C, Stacey GN, Strehl R, Strelchenko N, Suemori H, Sun B, Suuronen R, Takahashi K, Tuuri T, Venu P, Verlinsky Y, Ward-van Oostwaard D, Weisenberger DJ, Wu Y, Yamanaka S, Young L, Zhou Q. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol. 2011;29:1132-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 489] [Cited by in RCA: 438] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 42. | Shalom-Feuerstein R, Serror L, Aberdam E, Müller FJ, van Bokhoven H, Wiman KG, Zhou H, Aberdam D, Petit I. Impaired epithelial differentiation of induced pluripotent stem cells from ectodermal dysplasia-related patients is rescued by the small compound APR-246/PRIMA-1MET. Proc Natl Acad Sci USA. 2013;110:2152-2156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 43. | Yamamoto T, Takenaka C, Yoda Y, Oshima Y, Kagawa K, Miyajima H, Sasaki T, Kawamata S. Differentiation potential of Pluripotent Stem Cells correlates to the level of CHD7. Sci Rep. 2018;8:241. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 44. | Siller R, Naumovska E, Mathapati S, Lycke M, Greenhough S, Sullivan GJ. Development of a rapid screen for the endodermal differentiation potential of human pluripotent stem cell lines. Sci Rep. 2016;6:37178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |