Published online Dec 26, 2019. doi: 10.4252/wjsc.v11.i12.1130
Peer-review started: June 19, 2019
First decision: July 31, 2019
Revised: September 23, 2019
Accepted: October 14, 2019
Article in press: October 14, 2019
Published online: December 26, 2019
Processing time: 168 Days and 21 Hours
Cardiovascular disease is the leading cause of death worldwide. Tissue repair after pathological injury in the heart remains a major challenge due to the limited regenerative ability of cardiomyocytes in adults. Stem cell-derived cardiomyocytes provide a promising source for the cell transplantation-based treatment of injured hearts.
To explore the function and mechanisms of miR-301a in regulating cardiomyocyte differentiation of mouse embryonic stem (mES) cells, and provide experimental evidence for applying miR-301a to the cardiomyocyte differentiation induction from stem cells.
mES cells with or without overexpression of miR-301a were applied for all functional assays. The hanging drop technique was applied to form embryoid bodies from mES cells. Cardiac markers including GATA-4, TBX5, MEF2C, and α-actinin were used to determine cardiomyocyte differentiation from mES cells.
High expression of miR-301a was detected in the heart from late embryonic to neonatal mice. Overexpression of miR-301a in mES cells significantly induced the expression of cardiac transcription factors, thereby promoting cardiomyocyte differentiation and beating cardiomyocyte clone formation. PTEN is a target gene of miR-301a in cardiomyocytes. PTEN-regulated PI3K-AKT-mTOR-Stat3 signaling showed involvement in regulating miR-301a-promoted cardiomyocyte differentiation from mES cells.
MiR-301a is capable of promoting embryonic stem cell differentiation to cardiomyocytes.
Core tip: MiR-301a was identified as a miRNA highly enriched in the heart from late embryonic to neonatal mice. Overexpression of miR-301a significantly induced the expression of cardiac transcription factors and promoted cardiomyocyte differentiation of mouse embryonic stem cells. These findings will help improve the efficiency of cardiomyocyte differentiation from stem cells, and strengthen the potential of cell therapeutics to treat heart failure caused by myocardial infarction.
- Citation: Zhen LX, Gu YY, Zhao Q, Zhu HF, Lv JH, Li SJ, Xu Z, Li L, Yu ZR. MiR-301a promotes embryonic stem cell differentiation to cardiomyocytes. World J Stem Cells 2019; 11(12): 1130-1141
- URL: https://www.wjgnet.com/1948-0210/full/v11/i12/1130.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v11.i12.1130
Cardiovascular disease is the leading cause of death worldwide[1]. After myocardial infarction (MI) in adults, the regenerative ability of the differentiated cardiomyocytes is very limited due to the “terminated” cell proliferative ability and the lack of cardiac stem cells[2,3]. Although the cell proliferative potential of cardiomyocytes in adults is occasionally reported[4], this ability cannot produce enough cardiomyocytes for functional recovery of the injured heart. Tissue repair after pathological injury in the heart, including that after MI, is still a major clinical challenge.
MI occurs mostly due to coronary artery disease in which the heart blood flow is blocked, causing damage to the cardiomyocytes. MI may cause heart failure, heart arrhythmia, cardiogenic shock, or cardiac arrest. Although progress has been made in the pharmacologic and device management and gene or cell therapy of heart failure, the mortality in heart failure patients remains significant. All of the current pharmacologic or surgical approaches have limited effects on heart function recovery. Two novel strategies have been suggested to restore the lost cardiomyocytes caused by MI. One is to apply cardiomyocytes differentiated from stem cells or derived from cardiospheres (CDCs)[5,6], and the other is to induce cell cycle reentry in cardio-myocytes[4,7].
Stem cell-based therapy tests in chronic heart failure[8] and preclinical studies applying transplantation of embryonic stem (ES) cell-derived cardiac progenitors in an animal model of MI[9] have suggested strategies to compensate for the lost cardiac cells in damaged hearts. A phase I clinical trial showed that patients treated with CDCs had a reduction in scar mass and an increase in both viable heart mass and systolic wall thickness[6]. As such, stem cells, stem cell-derived cardiomyocytes, and CDCs provide a promising source for cell transplantation-based treatment of injured hearts. In addition, modification of approaches to induce cardiac cell differentiation from stem cells with high efficiency will be crucial to improve the therapeutic effect.
ES cells have been applied to differentiate into cardiomyocytes in the treatment of MI in vivo[9]. During embryogenesis, the heart is derived from mesodermal cells. The mesothelial pericardium forms the outer lining, and the endothelium forms the inner lining and lymphatic and blood vessels of the heart. Myocyte differentiation begins from E7.5 in mouse embryos and day 15 in human embryos. Before birth, cardio-myocytes undergo the hyperplastic to hypertrophic transition. The majority of cardiomyocytes withdraw from the cell cycle and stop proliferation shortly after birth. Here, mouse ES (mES) cells were applied for the induction of cardiomyocyte differentiation in vitro to determine the therapeutic potential of ES cell-based cell transplantation in the treatment of heart failure.
MicroRNAs (miRNAs) have been shown to regulate diverse biological processes, including cell fate decision, organ formation, and stem cell self-renewal and differentiation[10-12]. The aberrant expression of miRNAs in tissues has been closely connected to tissue-related disease. MiRNAs are involved in regulating the development and progression of cancer, cardiovascular disease, and other conditions[11,13-15]. To the best of our knowledge, miR-1 and miR-133 are the most important miRNA families regulating cardiac development and heart function[16,17]. Muscle-specific miR-1 and miR-133a both promote mesoderm formation from ES cells and suppress ectoderm and endoderm fates[18], but later, during further differentiation into cardiac muscle progenitors, these miRNAs show opposing regulatory functions[12,19]. Other miRNAs, including miR-206, miR-708, miR-208a, miR-208b, and miR-499, have also been reported to regulate heart development and heart diseases[20].
In the current study, we identified miR-301a as a highly enriched miRNA in embryonic and neonatal cardiomyocytes. Although overexpression of miR-301a is frequently observed in diverse tumor types, promoting cell proliferation, invasion, and metastasis of cancer cells[21-23], the functional properties of miR-301a in the heart remain unclear, except one recent report indicating that miR-301a is a novel cardiac regulator of Cofilin-2 in cardiomyocytes[24]. In contrast to its function in tumors, miR-301a may have tissue-specific functions in the heart. Here, we for the first time demonstrated that overexpression of miR-301a significantly induced the expression of cardiac transcription factors in mES cells, thereby promoting cardiomyocyte differentiation and beating cardiomyocyte clone formation. Our findings will be beneficial in the development of an approach with high efficiency to induce stem cell differentiation to cardiomyocytes and strengthen the potential of cell therapeutics for heart failure.
Animal studies were approved by the Institutional Animal Care and Use Committee of the Tongji University School of Medicine. Male C57BL/6J mice were purchased from Silaike Animal Company (Shanghai, China). The hearts were collected from mouse embryos at E11.5, 13.5, 15.5, 17.5, and 19.5 and from neonatal and adult mice and placed into TRIzol for total RNA isolation using a tissue homogenizer.
The murine embryonic stem cell line ES-D3 was originally from ATCC and maintained in “feeder free” culture conditions as described previously[25]. The mES cell culture plates were coated with fetal bovine serum (FBS). The DMEM/F12 medium containing Neurobasal Medium was supplemented with 0.5% N2, 1% B27, 2 mM L-glutamine, 0.055 mmol/L β-mercaptoethanol, 0.05% bovine serum albumin (BSA; Fraction V), 0.1% insulin, 100 U/mL penicillin, 100 μg/mL streptomycin, 3 μmol/L CHIR99021, 0.4 μmol/L PD0325901, and 1000 U/mL LIF. All cells were cultured at 37 °C in a 5% CO2 environment unless stated otherwise.
All primers and miR-301a mimic and negative control oligos were synthesized by GenScript (Nanjing, China). Forward primer sequences for miRNA amplification are as follows: MiR-301a: 5’-CCAGTGCAATAGTATTG-3’; 5S rRNA: 5’-AGTACTTGGATGGGAGACCG-3’. The double-strand miRNA mimic sequence for miR-301a is 5’-CAGU GCAAUAGUAUUGUCAAAGC-3’, and the negative control for the miRNA mimic is 5’-UGGGCGUAUAGACGUGUUACAC-3’. Lipofectamine RNAiMAX (Invitrogen) was applied for oligo transfection, following the manufacturer’s instructions. A final concentration of 50 nM of miRNA mimic or negative control was used. The cells were applied for further assays 24 h after transfection.
Total RNA was extracted with TRIzol reagent (#15596026, Invitrogen, Thermo Fisher Scientific). Then, 500 ng of purified total RNA was applied to prepare the first strand cDNA of miRNA using an M and G miRNA Reverse Transcription Kit (miRGenes, Shanghai, China) following the manufacturer’s instructions. The cDNA was diluted 1:1000 for real-time PCR analysis of miRNAs. For mRNA analysis, a regular approach and random primer were used for reverse transcription. The SYBR Green Master Mix (Applied Biosystem, Thermo Fisher Scientific) and 7900 HT Sequence Detection System (Applied Biosystem, Thermo Fisher Scientific) were used for real-time PCR assays. GAPDH was used for mRNA normalization, and 5S rRNA was used for miRNA normalization. Primer information for all tested genes is shown in detail in Supplemental Table 1.
Cell lysates (50 μg) prepared with RIPA buffer containing protease inhibitor cocktail (Roche Diagnostics) were separated by 10% SDS-PAGE. The proteins were transferred to PVDF membranes. Then, 5% nonfat milk (w/v) was used for the blocking step. The following primary antibodies (1:2000) were used: PTEN (sc-7974, Santa Cruz), p-STAT3 (Tyr705, 9145T, Cell Signaling Technology), p-AKT (Ser473, 4060T, Cell Signaling Technology), p-mTOR (Ser2448, 5536T, Cell Signaling Technology), total STAT3 (4904, Cell Signaling Technology), total AKT (4691, Cell Signaling Technology), total mTOR (2983T, Cell Signaling Technology), MEF2C (sc-365862, Santa Cruz), NKX2.5 (sc-376565, Santa Cruz), GATA4 (sc-25310, Santa Cruz), CTNT (sc-20025, Santa Cruz), α-actinin (Sigma, A7811), and GAPDH (sc-47724, Santa Cruz). HRP-conjugated anti-rabbit IgG (7074S, Cell Signaling Technology) and HRP- conjugated anti-mouse IgG (7076S, Cell Signaling Technology) were used as secondary antibodies (1:3000).
Cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 (Sigma) in 1 × PBS for 10 min at room temperature. Next, 1% BSA was used for blocking for 1 h at room temperature. Then, cells were incubated with the primary antibodies (1:10 to 1:100 dilution), including anti-OCT4 (2750S, Cell Signaling Technology) and anti-NANOG (4903S, Cell Signaling Technology), overnight at 4 °C, and the secondary antibody conjugated to Alexa Fluor 555 (Invitrogen, A21428) was added for 1 h at room temperature. The nucleus was stained with 6-diamidino-2-phenylindole (DAPI, Sigma, D9542) for 30 min at room temperature. All slides were photographed using a fluorescence microscope (Leica, Germany).
The hanging drop technique was applied to form EBs from ES cells. Iscove’s modified Dulbecco’s medium (IMDM, Gibco) containing 20% FBS, 1% penicillin-streptomycin, 2 mM L-glutamine, 0.055 mmol/L β-mercaptoethanol, and 1% MEM nonessential amino acids (Gibco, 11140050) was used for induction of mES cell differentiation. Briefly, a single cell suspension (5 × 104 mES cells/mL) in differentiation medium was split to cell droplets with ~1000 cells in 20 μL for each drop and hung from the bottom of bacterial-grade dishes upside down to culture for 2 d, followed by suspension culture for two more days in 10 mL of differentiation medium still using bacterial-grade dishes on a shaking platform at 40 rpm. This step is for the maturation of EBs. After that, the EBs were moved to 0.1% gelatin-coated plates for adherent culture for an additional 8 d. This step is for cardiomyocyte differentiation.
Alkaline phosphatase activity of the mES clones was examined using a BCIP/NBT alkaline phosphatase color development kit according to the manufacturer’s protocol (Beyotime Institute of Biotechnology, China). The clones were fixed with 70% ethanol and incubated with BCIP/NBT staining work solution for 30 min, followed by washing with ddH2O.
Data are presented as the mean ± SE. A standard two-tailed Student’s t-test with SPSS 21.0 software was used for statistical analyses, in which P < 0.05 was considered significant.
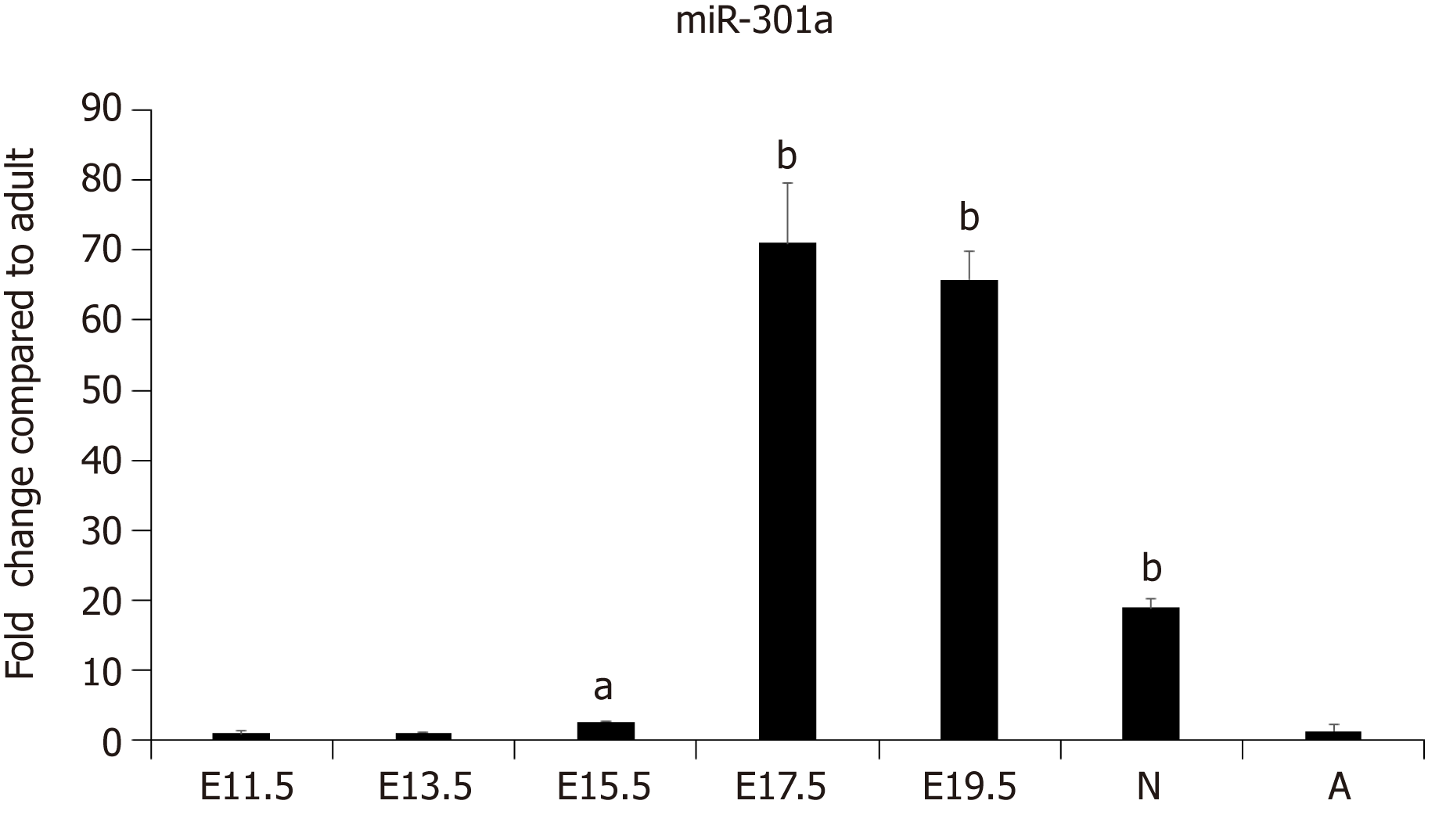
Our previous work demonstrated the enrichment of a subset of miRNAs, including miR-708 and miR-301a, in the cardiomyocytes of neonatal rodents[26]. To further determine the expression pattern and function of miR-301a during heart development, we collected mouse embryo hearts at days 11.5, 13.5, 15.5, 17.5, and 19.5, as well as hearts from 3-day-old neonatal and 6-wk-old adult mice. The expression analysis of miR-301a indicated much higher levels in the hearts of late-stage embryos and 3-day-old postnatal mice than adult hearts. In particular, the levels in the hearts from E17.5 to newborn mice were over 10-fold higher than those in adult hearts (Figure 1), suggesting the potential function of miR-301a in regulating heart development, cardiomyocyte differentiation, and cardiomyocyte proliferation.
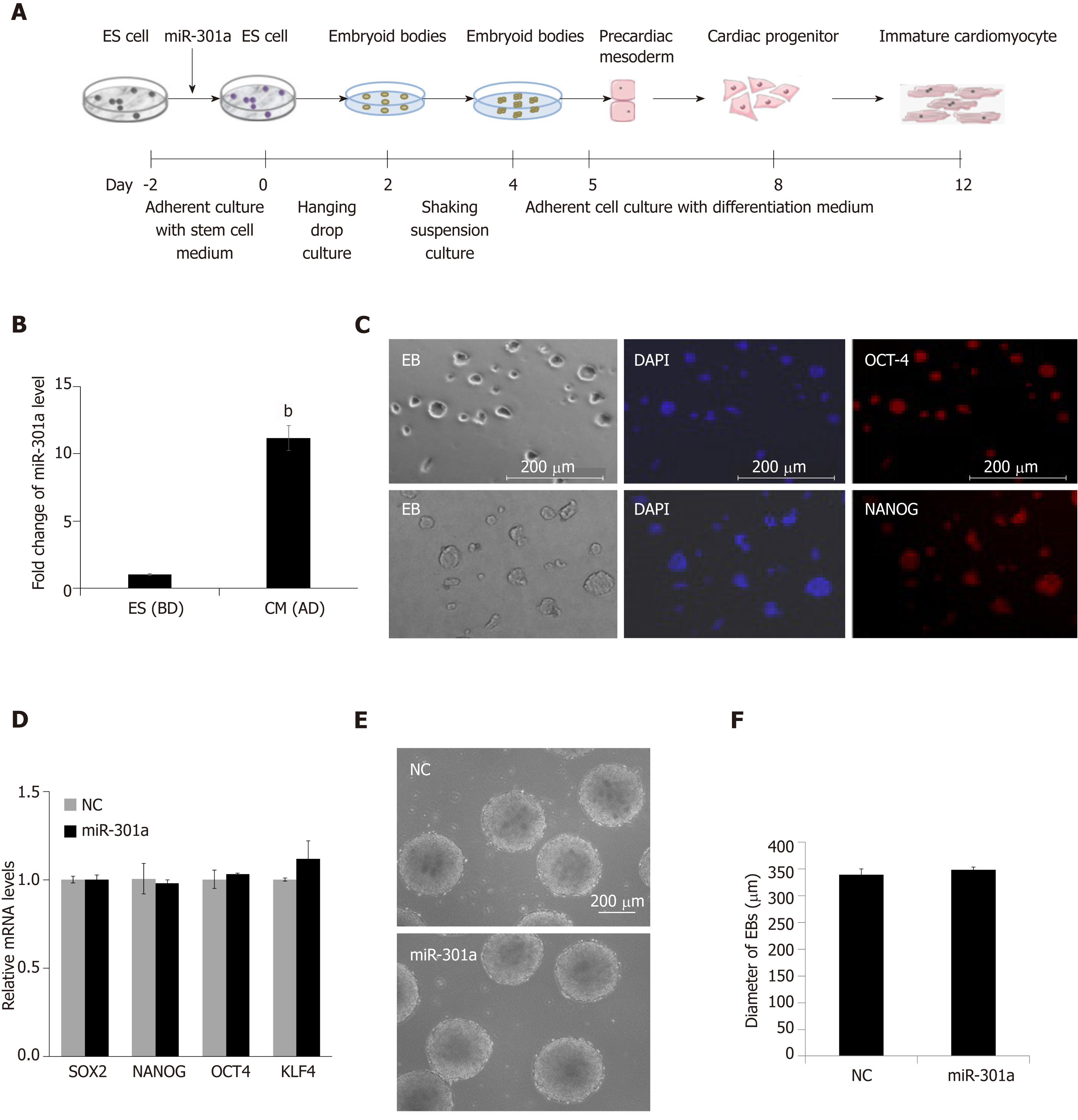
To explore the role of miR-301a during heart development, we established a cell-culturing system using the hanging drop technique to induce mES cell differentiation into cardiomyocytes in vitro (Figure 2A), which can partly mimic the development of the embryonic heart in vivo. We compared the expression of miR-301a in mES cells before differentiation and mES cell-derived cardiomyocytes at day 12 after differentiation, demonstrating a ~10-fold upregulation of miR-301a in differentiated cardiomyocytes compared to mES cells (Figure 2B). To further determine the expression of miR-301a at different stages of EBs during cardiac differentiation, we performed a quantitative analysis of miR-301a in EBs at days 4, 8, and 12. As shown in Supplemental Figure S1, a ~5-fold increase in miR-301a was observed in EBs at both days 8 and 12 compared to day 4.
As shown in Figure 2A, spheroid structured EBs were formed from day 2 to day 4 at the beginning of mES cell differentiation. EB formation is often considered to initiate differentiation toward the three germ lineages. Embryonic organoids derived from EB culture show remarkable parallels to embryonic development. Immunofluorescence staining with the stem cell markers OCT4 and NANOG was applied to ES clones, confirming the stemness maintenance at the early stage (day 2 under adherent culture) of ES cell differentiation (Figure 2C). To determine the regulatory effect of miR-301a on stem properties and EB formation, we performed alkaline phosphatase staining of mES clones and did not find a difference in stemness between the two groups with or without overexpression of miR-301a (Supplemental Figure S2).
MiR-301a-overexpressing mES cells were cultured in parallel with a negative control (NC) under differentiation conditions using the hanging-droplet method to develop EBs (Figure 2A). A quantitative analysis of gene expression of stem cell markers, including SOX2, OCT4, NANOG, and KLF4, was applied to EBs at day 2 and did not show a significant difference between the NC and miR-301a groups (Figure 2D). Subsequent analysis of EBs at day 4 did not show any changes in morphology, diameter, or amount between the NC and miR-301a groups, as shown in Figure 2E and 2F. These observations suggest the very limited effect of miR-301a on the self-renewal and stemness of mES cells during the early period of EB formation.
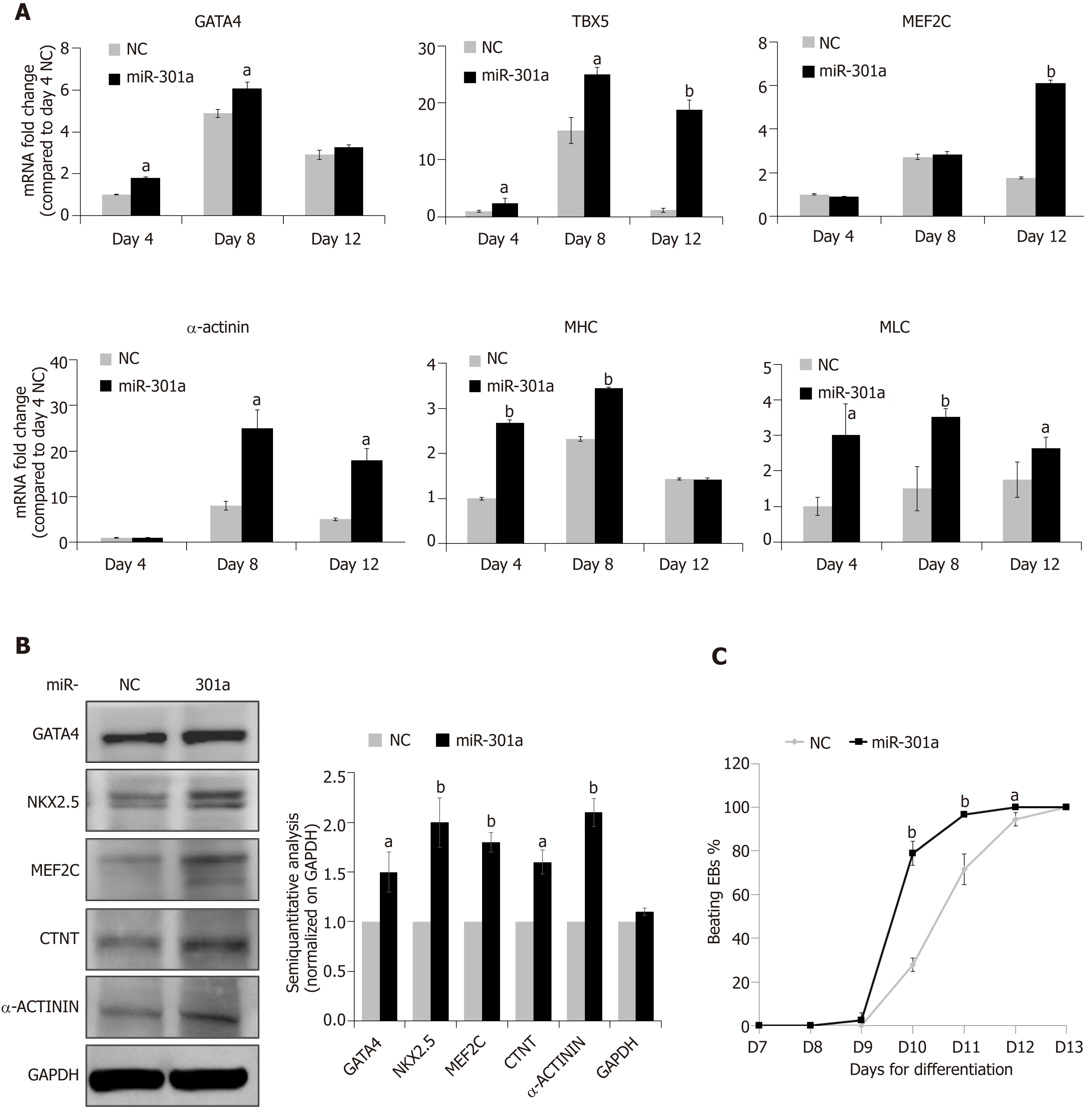
MiR-301a overexpression in mES cells does not regulate stemness, but can it control cardiac differentiation? To address this question, we assessed several cardiac-specific markers and cardiac-specific transcription factors at day 4 (EB formation, corresponding to E7.5 mouse embryos), day 8 (cardiac differentiation, corresponding to E10.5-E16.5 mouse embryos), and day 12 (formation of immature cardiomyocytes, corresponding to E17.5 and thereafter mouse embryos) after mES cell differentiation following the procedure in Figure 2A. As shown in Figure 3A, mid-stage cardiac markers, including GATA-4, TBX5 and MEF2C, and late-stage cardiac markers, including α-actinin, α-sarcomeric myosin heavy chain (α-MHC), and myosin light chain (MLC), showed higher levels at the stages of cardiac differentiation (days 8 and 12) compared to the stage of early EB formation (day 4) (fold changes are indicated in Figure 3A, days 8 and 12 vs day 4 in the NC group). Meanwhile, compared to the control, miR-301a overexpression induced the expression of these cardiac markers during mES differentiation (Figure 3A). Proteomic analysis further demonstrated the increased expression of the cardiac markers by miR-301a, including GATA-4, MEF2C, NKX2.5, CTNT, and α-actinin, at day 12 after mES cell differentiation (Figure 3B). A semiquantitative analysis (cardiac markers normalized on GAPDH) of the Western blot results clearly showed the upregulation of these cardiac markers by miR-301a (Figure 3B). Does this kind of regulation occur in mES cells? To address this question, a gene expression analysis of these cardiac markers in mES cells with or without overexpression of miR-301a was performed. Meanwhile, EBs undergoing differentiation at day 8 were used as positive controls. As shown in Supplemental Figure S3, very low or undetectable levels of these cardiac markers were detected in mES cells. Their expression in mES cells was not affected by miR-301a.
During cardiac differentiation from mES cells, beating of the cardiac clones was observed from day 9 to day 12 and is shown in Supplemental Videos 1-5. Quantitative analysis of the percentage of beating EBs at different time points indicated a greater number in the miR-301a group compared to the control group (Figure 3C). Furthermore, the beating EBs were first observed in the miR-301a group at day 9, one day earlier than that in the control group. At day 11, more than 90% of EBs were beating in the miR-301a group, while 75% were beating in the control group (Figure 3C). Taken together, these findings demonstrated the promotion of cardiac differentiation from mES cells by overexpression of miR-301a.
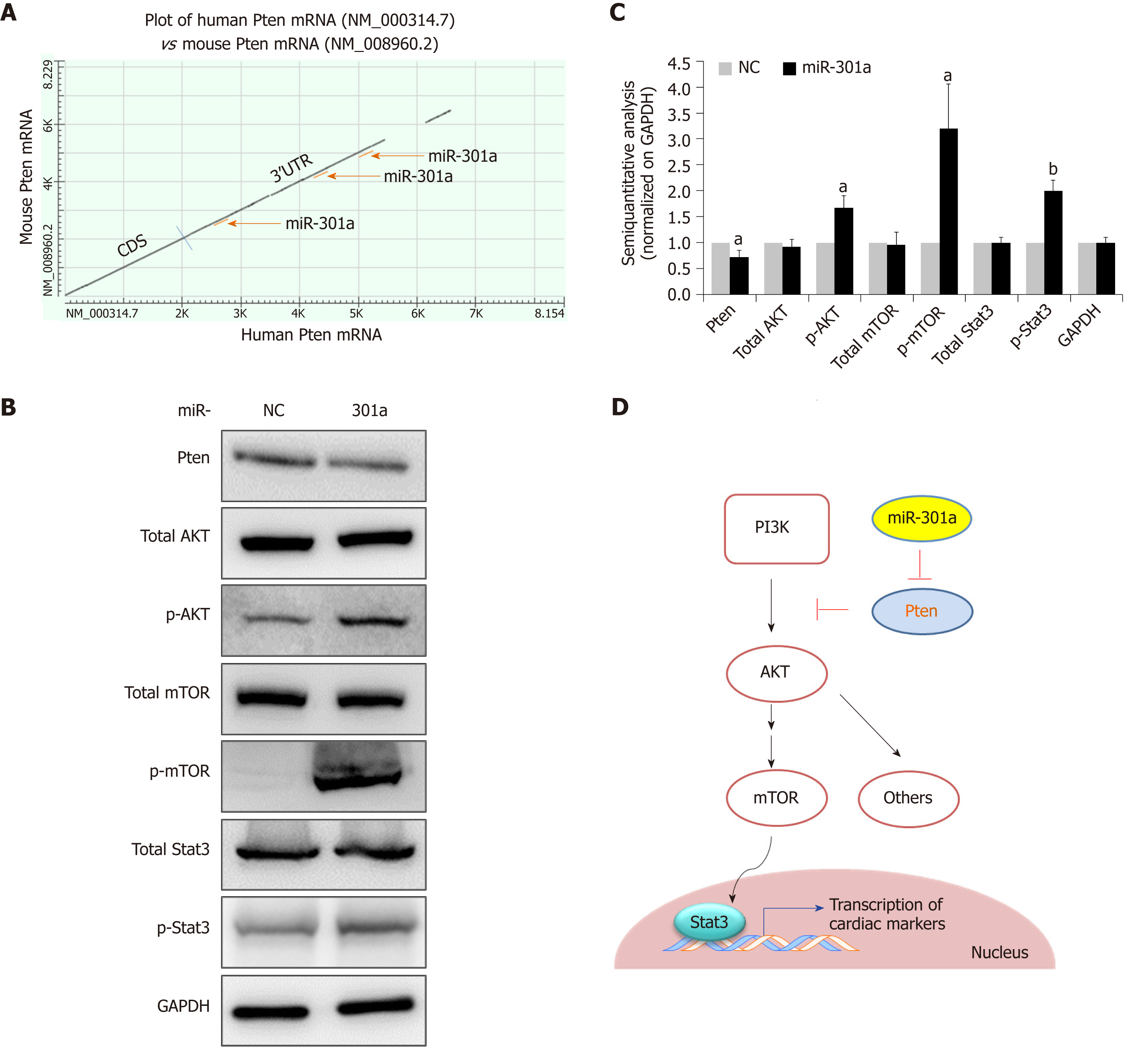
We next determined the mechanism by which miR-301a induces cardiac differentiation from mES cells. Both bioinformatics analysis and literature[27] suggested PTEN as a potential target gene of miR-301a in cardiomyocytes. As shown in Figure 4A, three binding sites to miR-301a are predicted in the highly conserved 3’ untranslated region of human and rodent PTEN mRNAs. Considering the function of PTEN in regulating cell survival and cell differentiation, the downstream PI3K-AKT and mTOR-Stat3 signaling pathways were further analyzed during mES cell differentiation. As shown in Figure 4B, activation of the PI3K-AKT signaling pathway was accompanied by inhibition of PTEN by miR-301a overexpression. A semi-quantitative analysis (normalized on GAPDH) clearly indicated the upregulation of p-AKT, p-mTOR, and p-Stat3 in the miR-301a-mES-derived cardiac cells at day 12 after cell differentiation, while total AKT, total mTOR, and total STAT3 did not show differences between the two groups (Figure 4C). These findings suggest that the activation of PI3K-AKT-mTOR-Stat3 signaling by miR-301a may contribute to the promoted cardiac differentiation shown in Figure 3.
The cardiovascular system is the first system developed in an embryo, and the heart is the first functional organ developed during embryogenesis. The earliest heart precursor cells are detectable in an embryo as early as E6.5 in mice and days 18-19 following fertilization in humans. Cardiac cell differentiation from the precursor cell population is initiated and guided by transcription factors, such as NKX2.5, GATA4, GATA6, and TBX5. Thereafter, signaling pathways, including fibroblast growth factors and bone morphogen proteins, modulate the function of these transcription factors and induce early progenitor cells to further differentiate into mature ventricular or atrial cardiomyocytes[28].
Noncoding genomes play important roles during mES cell differentiation and cell fate determination. A group of miRNAs, including miR-1, miR-133, miR-208a, miR-208b, and miR-499, have been demonstrated to regulate heart development, cardiovascular diseases, and cardiac remodeling[13,16-20]. MiR-1 is enriched in embryonic cardiac cells and mES cell-derived cardiomyocytes, and it promotes not only mesoderm formation from mES cells but also further differentiation into cardiac muscle progenitors[12,16]. In this study, we observed enrichment of miR-301a in the hearts from embryos and neonatal mice. Functional assays demonstrated the induction of cardiac differentiation from mES cells by miR-301a overexpression. Our results contribute to knowledge of the noncoding genome in regulating stem cell differentiation to cardiomyocytes. We demonstrated the potential of miR-301a as a novel target miRNA candidate to induce cardiomyocyte differentiation.
ES cell differentiation, cell fate determination, and organoid morphogenesis are closely related to studies of developmental biology and mammalian embryogenesis. ES cells are derived from the early blastocyst stage of embryo development. Because of the similarities between embryogenesis and ES cell differentiation, the process of EB formation and ES cell differentiation can partly resemble the development of embryonic organoids during embryogenesis. The formation of EBs is the typical first step for ES cell differentiation, from which differentiation begins, and three types of tissues are formed, including mesodermal tissues (muscle, bone, connective tissue, etc.), ectodermal tissues (nervous system, hair, eyes, etc.), and endodermal tissues (epithelium, gastrointestinal tract, etc.). The induced cardiomyocyte differentiation method from EBs has been well developed[9,25].
Stem cell-derived cardiomyocytes represent a good source of cells for studying early cardiac development as well as cell-based therapies in postnatal pathologies. The protocol for cardiomyocyte differentiation was first developed for ES cells[29] and then adapted to iPS cells[30]. In the past decade, cardiomyocyte differentiation protocols have been modified and improved to be more reliable and more efficient, which has contributed to preclinical trials of stem cell-derived cell transplantation therapeutics to treat heart diseases, including heart failure. Herein, we demonstrated that the small RNA molecule miR-301a can induce the expression of cardiac transcription factors and promote cardiomyocyte differentiation from mES cells. As such, miR-301a showed potential to be applied towards the modification of the current cardiomyocyte differentiation protocols.
The enrichment of miR-301a in active cardiomyocytes was originally determined from our miRNA screening study in the hearts of neonatal rodents[26], which was recently confirmed by Rangrez et al[24], who detected much higher expression of miR-301a in isolated cardiomyocytes than in fibroblasts. That study also found that miR-301a negatively regulates SRF signaling through inhibiting the expression of the target gene Cofilin-2 in cardiomyocytes, suggesting the therapeutic potential of miR-301a in the treatment of cardiac disorders caused by the deregulation of Cofilin-2[24]. Here, we first showed that miR-301a has a high level in the hearts of late-stage embryos, while it is low in undifferentiated ES cells and cardiomyocytes in early-stage embryos. Subsequent functional assays demonstrated the induction of cardiomyocyte differentiation from mES cells by miR-301a, suggesting a cell-type specific function for this miRNA. Our findings add to knowledge of miR-301a in the treatment of heart disease by ES cell-based strategies. Notably, the development of a stem cell-specific gene expression system or a cardiomyocyte-targeted local delivery system for miR-301a will be required given the possible oncogenic side effects of miR-301a. As discussed above, the upregulation of miR-301a has been reported in multiple tumor types[21-23].
The activation of mTOR-STAT3 signaling by the PI3K-AKT pathway has been validated in cardiomyocytes[31,32]. STAT3 is essential for cardiomyocyte differentiation, directly promoting the expression of cardiac markers, including TBX5, NKX2.5, and GATA4[33]. PTEN dephosphorylates PIP3 [phosphatidylinositol (3,4,5)-trisphosphate] to from PIP2 [phosphatidylinositol (4,5)-bisphosphate], thereby inhibiting the PI3K/AKT pathway and regulating cell proliferation and cell differentiation in cardiomyocytes[34]. PTEN is a key regulator of the PI3K/AKT pathway. PTEN/ PI3K/AKT signaling-mediated miRNA regulation of ES cell differentiation to cardiomyocytes has been reported for miR-1[35]. Target interactions between miR-301a and PTEN have been demonstrated in cervical cancer[36] and pancreatic cancer[37]. In the current study, we demonstrated that miR-301a activates the PI3K-AKT-mTOR-STAT3 signaling pathway, promoting cardiomyocyte differentiation from mES cells, which was mediated by the target interaction between miR-301a and PTEN. These findings further demonstrated the importance of miRNAs and AKT signaling in regulating mES cell differentiation to cardiomyocytes.
In conclusion, we demonstrated that miR-301a promotes transcriptional activation of the cardiomyocyte-driving genes during mES cell differentiation to cardiomyocytes, and PTEN is a target gene of miR-301a in cardiomyocytes. PTEN-regulated PI3K-AKT-mTOR-STAT3 signaling is involved in regulating miR-301a-promoted cardiomyocyte differentiation from mES cells (Figure 4D). As discussed above, application of a heart-specific local delivery system for miR-301a administration will be required to avoid potential side effects of miR-301a. These findings will shed light on the induction of stem cell-derived cardiomyocyte differentiation and strengthen the potential of miR-301a in cell therapeutics in the treatment of heart disease.
After myocardial infarction (MI) in adults, the regenerative ability of the differentiated cardiomyocytes is very limited due to the “terminated” cell proliferative ability and the lack of cardiac stem cells. Tissue repair after pathological injury in the heart, including that after MI, is still a major clinical challenge. Two novel strategies have been suggested to restore the lost cardiomyocytes caused by MI. One is to apply cardiomyocytes differentiated from stem cells or derived from cardiospheres, and the other is to induce cell cycle reentry in cardiomyocytes.
Stem cells and stem cell-derived cardiomyocytes have been demonstrated to be a promising source for cell transplantation-based treatment of injured hearts. Modification of approaches to induce cardiac cell differentiation from stem cells with high efficiency will be crucial to improve the therapeutic effect.
To explore the function of miR-301a in regulating cardiomyocyte differentiation of mouse embryonic stem cells, and provide experimental evidence for applying miR-301a to the cardiomyocyte differentiation induction from stem cells.
The hanging drop technique was applied to form embryoid bodies from mouse embryonic stem cells with or without overexpression of miR-301a. Cardiac markers including GATA-4, TBX5, and MEF2C, and α-actinin were used to determine cardiomyocyte differentiation from mouse embryonic stem cells.
MiR-301a was identified as a miRNA highly enriched in the heart from late embryonic to neonatal mice. Overexpression of miR-301a in mouse embryonic stem cells significantly induced the expression of cardiac transcription factors, thereby promoting cardiomyocyte differentiation and beating cardiomyocyte clone formation. PTEN was demonstrated to be a target gene of miR-301a in cardiomyocytes. PTEN-regulated AKT-mTOR-Stat3 signaling was involved in regulation of miR-301a-induced cardiomyocyte differentiation.
MiR-301a is capable of promoting embryonic stem cell differentiation to cardiomyocytes. As such, miR-301a has potential as a novel target gene to induce cardiomyocyte differentiation.
These findings will be beneficial in development of an approach with high efficiency to induce stem cell differentiation to cardiomyocytes, and strengthen the potential of cell therapeutics to treat heart failure caused by myocardial infarction.
Manuscript source: Unsolicited manuscript
Specialty type: Cell and tissue engineering
Country of origin: Japan
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Li SC, Miloso M, Scarfì S, Zheng YW S-Editor: Zhang L L-Editor:Wang TQ E-Editor: Xing YX
| 1. | Joseph P, Leong D, McKee M, Anand SS, Schwalm JD, Teo K, Mente A, Yusuf S. Reducing the Global Burden of Cardiovascular Disease, Part 1: The Epidemiology and Risk Factors. Circ Res. 2017;121:677-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 687] [Article Influence: 85.9] [Reference Citation Analysis (0)] |
| 2. | Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2692] [Cited by in RCA: 2438] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 3. | Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737-1746. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 610] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 4. | Wang WE, Li L, Xia X, Fu W, Liao Q, Lan C, Yang D, Chen H, Yue R, Zeng C, Zhou L, Zhou B, Duan DD, Chen X, Houser SR, Zeng C. Dedifferentiation, Proliferation, and Redifferentiation of Adult Mammalian Cardiomyocytes After Ischemic Injury. Circulation. 2017;136:834-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 165] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 5. | Liu YW, Chen B, Yang X, Fugate JA, Kalucki FA, Futakuchi-Tsuchida A, Couture L, Vogel KW, Astley CA, Baldessari A, Ogle J, Don CW, Steinberg ZL, Seslar SP, Tuck SA, Tsuchida H, Naumova AV, Dupras SK, Lyu MS, Lee J, Hailey DW, Reinecke H, Pabon L, Fryer BH, MacLellan WR, Thies RS, Murry CE. Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol. 2018;36:597-605. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 345] [Cited by in RCA: 444] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 6. | Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, Berman D, Czer LS, Marbán L, Mendizabal A, Johnston PV, Russell SD, Schuleri KH, Lardo AC, Gerstenblith G, Marbán E. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1099] [Cited by in RCA: 1057] [Article Influence: 81.3] [Reference Citation Analysis (1)] |
| 7. | Borden A, Kurian J, Nickoloff E, Yang Y, Troupes CD, Ibetti J, Lucchese AM, Gao E, Mohsin S, Koch WJ, Houser SR, Kishore R, Khan M. Transient Introduction of miR-294 in the Heart Promotes Cardiomyocyte Cell Cycle Reentry After Injury. Circ Res. 2019;125:14-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 84] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 8. | Menasché P, Vanneaux V. Stem cells for the treatment of heart failure. Curr Res Transl Med. 2016;64:97-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 9. | Menasché P, Vanneaux V, Fabreguettes JR, Bel A, Tosca L, Garcia S, Bellamy V, Farouz Y, Pouly J, Damour O, Périer MC, Desnos M, Hagège A, Agbulut O, Bruneval P, Tachdjian G, Trouvin JH, Larghero J. Towards a clinical use of human embryonic stem cell-derived cardiac progenitors: a translational experience. Eur Heart J. 2015;36:743-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 10. | Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006;2:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 267] [Cited by in RCA: 282] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 11. | Tili E, Michaille JJ, Gandhi V, Plunkett W, Sampath D, Calin GA. miRNAs and their potential for use against cancer and other diseases. Future Oncol. 2007;3:521-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 12. | Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219-229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 495] [Cited by in RCA: 460] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 13. | Colpaert RMW, Calore M. MicroRNAs in Cardiac Diseases. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 14. | Landrier JF, Derghal A, Mounien L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 153] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 15. | Zhao L, Wang Z. MicroRNAs: Game Changers in the Regulation of α-Synuclein in Parkinson's Disease. Parkinsons Dis. 2019;2019:1743183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Lu TY, Lin B, Li Y, Arora A, Han L, Cui C, Coronnello C, Sheng Y, Benos PV, Yang L. Overexpression of microRNA-1 promotes cardiomyocyte commitment from human cardiovascular progenitors via suppressing WNT and FGF signaling pathways. J Mol Cell Cardiol. 2013;63:146-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242-3254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 627] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 18. | Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 19. | Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721-8726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 533] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 20. | Bernardo BC, Ooi JY, Lin RC, McMullen JR. miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart. Future Med Chem. 2015;7:1771-1792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 186] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 21. | Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu Y, Gu Q, Zhu Z, Liu B. Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer. J Gastroenterol. 2013;48:1023-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Chen Z, Chen LY, Dai HY, Wang P, Gao S, Wang K. miR-301a promotes pancreatic cancer cell proliferation by directly inhibiting Bim expression. J Cell Biochem. 2012;113:3229-3235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 23. | Hu H, Zhang Q, Chen W, Wu T, Liu S, Li X, Luo B, Zhang T, Yan G, Lu H, Lu Z. MicoRNA-301a Promotes Pancreatic cancer Invasion and Metastasis through the JAK/STAT3 Signaling Pathway by Targeting SOCS5. Carcinogenesis. 2019;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 24. | Rangrez AY, Hoppe P, Kuhn C, Zille E, Frank J, Frey N, Frank D. MicroRNA miR-301a is a novel cardiac regulator of Cofilin-2. PLoS One. 2017;12:e0183901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519-523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2986] [Cited by in RCA: 2649] [Article Influence: 155.8] [Reference Citation Analysis (0)] |
| 26. | Deng S, Zhao Q, Zhen L, Zhang C, Liu C, Wang G, Zhang L, Bao L, Lu Y, Meng L, Lv J, Yu P, Lin X, Zhang Y, Chen YH, Fan H, Cho WC, Liu Z, Yu Z. Neonatal Heart-Enriched miR-708 Promotes Proliferation and Stress Resistance of Cardiomyocytes in Rodents. Theranostics. 2017;7:1953-1965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 27. | Ma F, Zhang J, Zhong L, Wang L, Liu Y, Wang Y, Peng L, Guo B. Upregulated microRNA-301a in breast cancer promotes tumor metastasis by targeting PTEN and activating Wnt/β-catenin signaling. Gene. 2014;535:191-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 28. | Nakajima Y, Imanaka-Yoshida K. New insights into the developmental mechanisms of coronary vessels and epicardium. Int Rev Cell Mol Biol. 2013;303:263-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 541] [Cited by in RCA: 514] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 30. | Batalov I, Feinberg AW. Differentiation of Cardiomyocytes from Human Pluripotent Stem Cells Using Monolayer Culture. Biomark Insights. 2015;10:71-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 31. | Ge W, Ren J. mTOR-STAT3-notch signalling contributes to ALDH2-induced protection against cardiac contractile dysfunction and autophagy under alcoholism. J Cell Mol Med. 2012;16:616-626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 32. | Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549-564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 356] [Article Influence: 32.4] [Reference Citation Analysis (0)] |
| 33. | Snyder M, Huang XY, Zhang JJ. Stat3 directly controls the expression of Tbx5, Nkx2.5, and GATA4 and is essential for cardiomyocyte differentiation of P19CL6 cells. J Biol Chem. 2010;285:23639-23646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Oudit GY, Penninger JM. Cardiac regulation by phosphoinositide 3-kinases and PTEN. Cardiovasc Res. 2009;82:250-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 184] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 35. | Glass C, Singla DK. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. Am J Physiol Heart Circ Physiol. 2011;301:H2038-H2049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 36. | Peng LN, Shi WT, Feng HR, Wei CY, Yin QN. Effect of miR-301a/PTEN pathway on the proliferation and apoptosis of cervical cancer. Innate Immun. 2019;25:217-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 37. | Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, Liu B, Su L, Qiu Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kγ to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018;78:4586-4598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 530] [Article Influence: 75.7] [Reference Citation Analysis (0)] |