Copyright
©2013 Baishideng Publishing Group Co.
World J Stem Cells. Oct 26, 2013; 5(4): 98-105
Published online Oct 26, 2013. doi: 10.4252/wjsc.v5.i4.98
Published online Oct 26, 2013. doi: 10.4252/wjsc.v5.i4.98
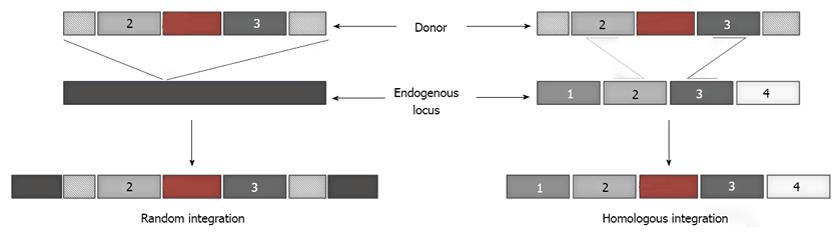
Figure 1 Random and homologous integration.
Introduction of a foreign gene into mammalian cells can either result in its random integration into the endogenous chromosomal DNA, or site-specific integration at the desired location dictated by the homology between the donor DNA and the endogenous target locus.
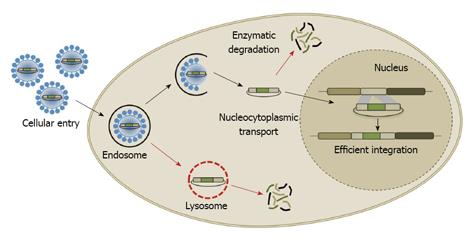
Figure 2 Barriers to gene targeting.
The different barriers that gene delivery vectors must overcome for successful gene targeting include cell binding and internalization, intracellular trafficking and endosomal escape, translocation through the nuclear envelope, and efficient site-specific integration.
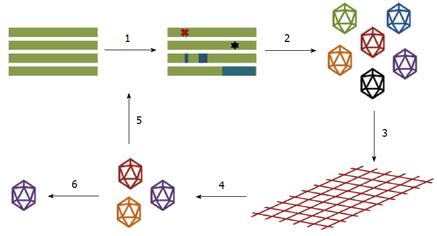
Figure 3 Schematic of steps in the directed evolution of adeno-associated virus.
The wild-type adeno-associated virus (AAV) cap genes are mutated to create large genetic libraries (1) and the mutant cap genes are packaged to generate libraries of AAV particles (2), with each AAV composed of a variant capsid surrounding a viral genome encoding that capsid. The resulting AAV libraries can be subjected to appropriate selective pressures (3) to isolate vectors with modified capsids that facilitate the AAV variants to efficiently surmount these pressures (4). Examples include isolation of AAV variants with the ability to evade neutralizing antibodies, altered receptor binding and cell tropism, and enhanced gene delivery. Successful AAV variants are amplified and recovered (6), or can be subjected to additional rounds of mutagenesis and selection (5). Adapted from Maheshri et al[33] and Bartel et al[35].
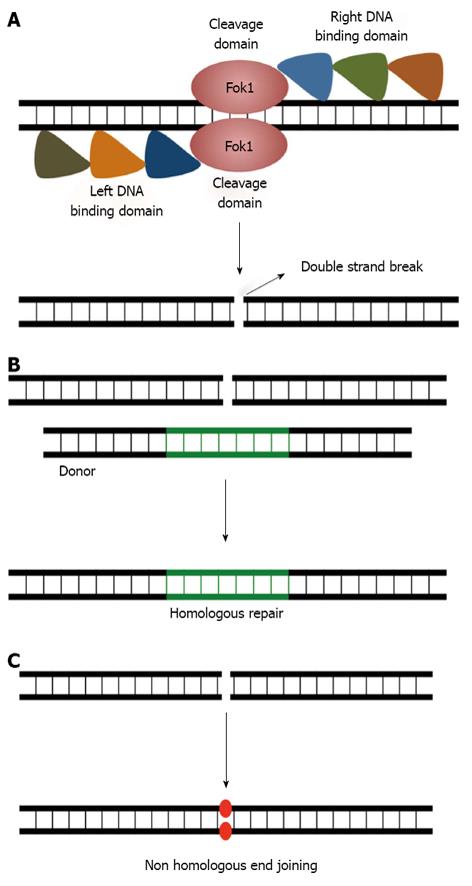
Figure 4 Gene editing using Zinc finger nucleases.
A: Schematic showing a zinc finger nuclease (ZFN) dimer bound to its target locus to introduce a site-specific double-stranded break (DSB) in the chromosome, each ZFN monomer consists of three to six zinc finger domains fused to the DNA-cleavage domain of the FokI endonuclease. B: The DNA DSB can be repaired via either the homologous recombination (HR), or C: the non-homologous end-joining pathway. HR requires a homologous DNA template to accurately repair DSBs in the chromosome, and ensures high fidelity DNA repair. In contrast, non-homologous end joining does not rely on sequence homology between the DNA ends for ligation and can be error-prone.
- Citation: Ramamoorthi K, Curtis D, Asuri P. Advances in homology directed genetic engineering of human pluripotent and adult stem cells. World J Stem Cells 2013; 5(4): 98-105
- URL: https://www.wjgnet.com/1948-0210/full/v5/i4/98.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v5.i4.98