Copyright
©©The Author(s) 2022.
World J Stem Cells. Jan 26, 2022; 14(1): 76-91
Published online Jan 26, 2022. doi: 10.4252/wjsc.v14.i1.76
Published online Jan 26, 2022. doi: 10.4252/wjsc.v14.i1.76
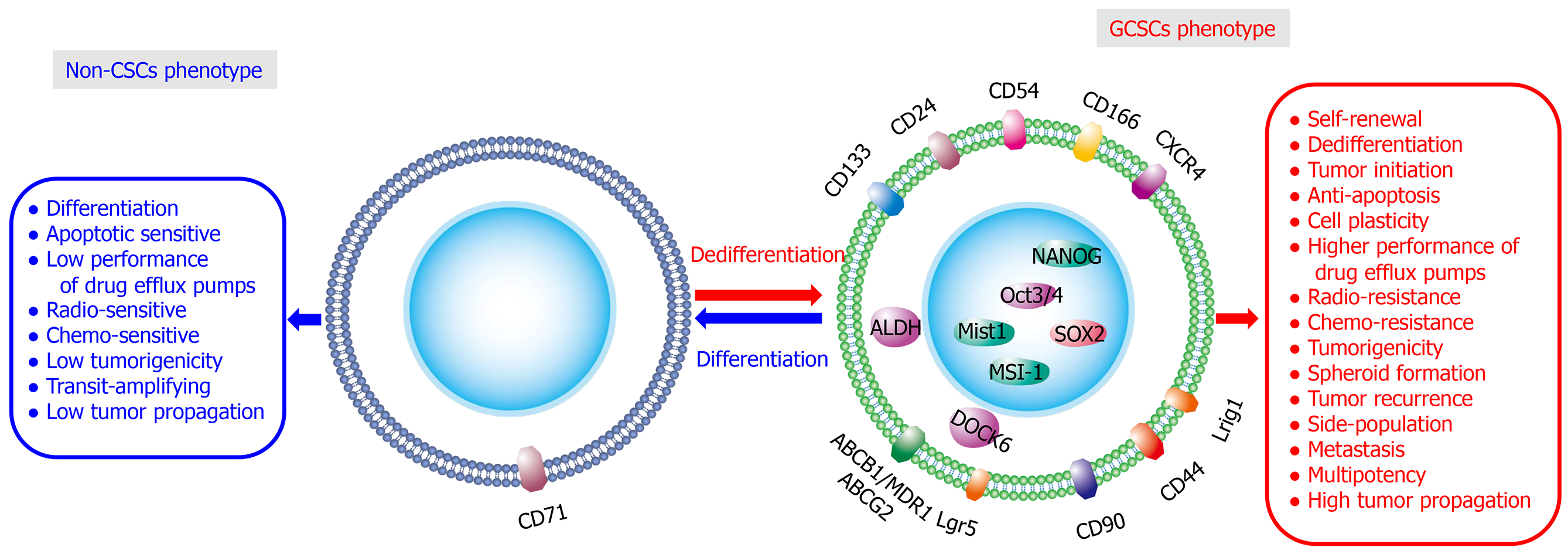
Figure 1 Molecular properties of gastric cancer stem cells.
Main gastric cancer stem cells (GCSCs)-surface markers (such as CD44, CD44v, CD71, CD90, CD133, Lgr5, ALDH1, CXCR4, ABC, and Lrig1) and GCSC-intracellular markers (such as DOCK6, Mist1, MSI-1, NANOG, Oct3/4, and SOX2). GCSCs represent a subpopulation of cancer cells (non-CSCs) existing within heterogeneous tumors implicated in tumor initiation, growth, metastasis, and chemo-/radioresistance, antiapoptosis, cancer recurrence, and metastasis. However, a small number of non-CSCs can dedifferentiate and transform into GCSCs through TME-induced EMT. The newly generated GCSCs from non-CSCs, together with the intrinsic GCSCs, consequently contribute to recurrence and metastasis of cancer. CD44: cluster of differentiation 44; CD44v: CD44v: CD44 splice variant; CD71: cluster of differentiation 71; CD90: cluster of differentiation 90; CD133: cluster of differentiation 133; Lgr5: leucine-rich repeat-containing G-protein coupled receptor 5; ALDH1: aldehyde dehydrogenase 1; CXCR4: C-X-C chemokine receptor type 4; ABC: ATP-binding cassette subfamily; Lrig1: leucine rich repeats and immunoglobulin like domains protein 1; DOCK6: dedicator of cytokinesis 6; Mist1: muscle, intestine and stomach expression 1; MSI-1: musashi RNA binding protein 1; NANOG: nanog homeobox; Oct3/4: octamer-binding transcription factor 3/4; SOX2: sex determining region Y-box 2.
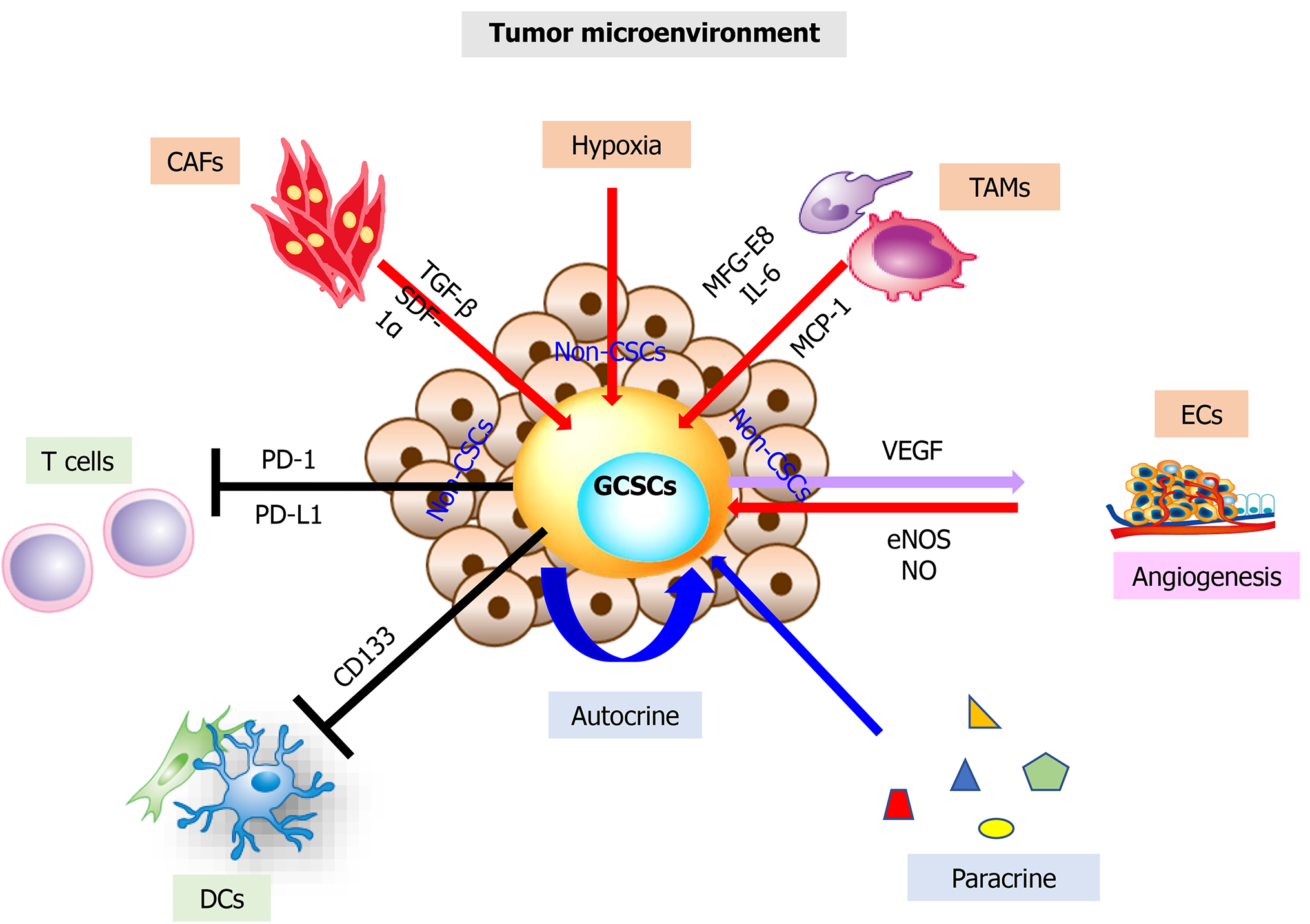
Figure 2 The roles of gastric cancer stem cells in the tumor microenvironment and activated in gastric cancer stem cells.
This figure shows the dynamic regulation of the tumor niche and GCSCs. Cancer cells (non-CSCs) can dedifferentiate by regulating their intracellular signaling pathways, gene expression, and epigenetic modification through the functional connection of the tumor niche to differentiated cancer cells (non-CSCs) to obtain the GCSC phenotype. Stromal cells can support GCSCs development through various kinds of interactions. Tumor-associated macrophages (TAMs), cancer-associated fibroblasts (CAFs), tumor vascular endothelial cells (ECs) and hypoxia not only directly enhance the CSC capabilities of GCSCs by activating the several pathways but also inhibit T cells and dendritic cells (DCs) activity. TGF-β: transforming growth factor-β; SDF-1α: stromal cell-derived factor-1α; PD-1: programmed cell death 1; PD-L1: programmed cell death 1 Ligand 1; CD133: cluster of differentiation 133; MFG-E8: milk fat globule epidermal growth factor 8; NO: nitric oxide; eNOS: endothelial NO·synthase; IL-6: interleukin 6; MCP1: monocyte chemoattractant protein 1; VEGF: vascular endothelial growth factor.
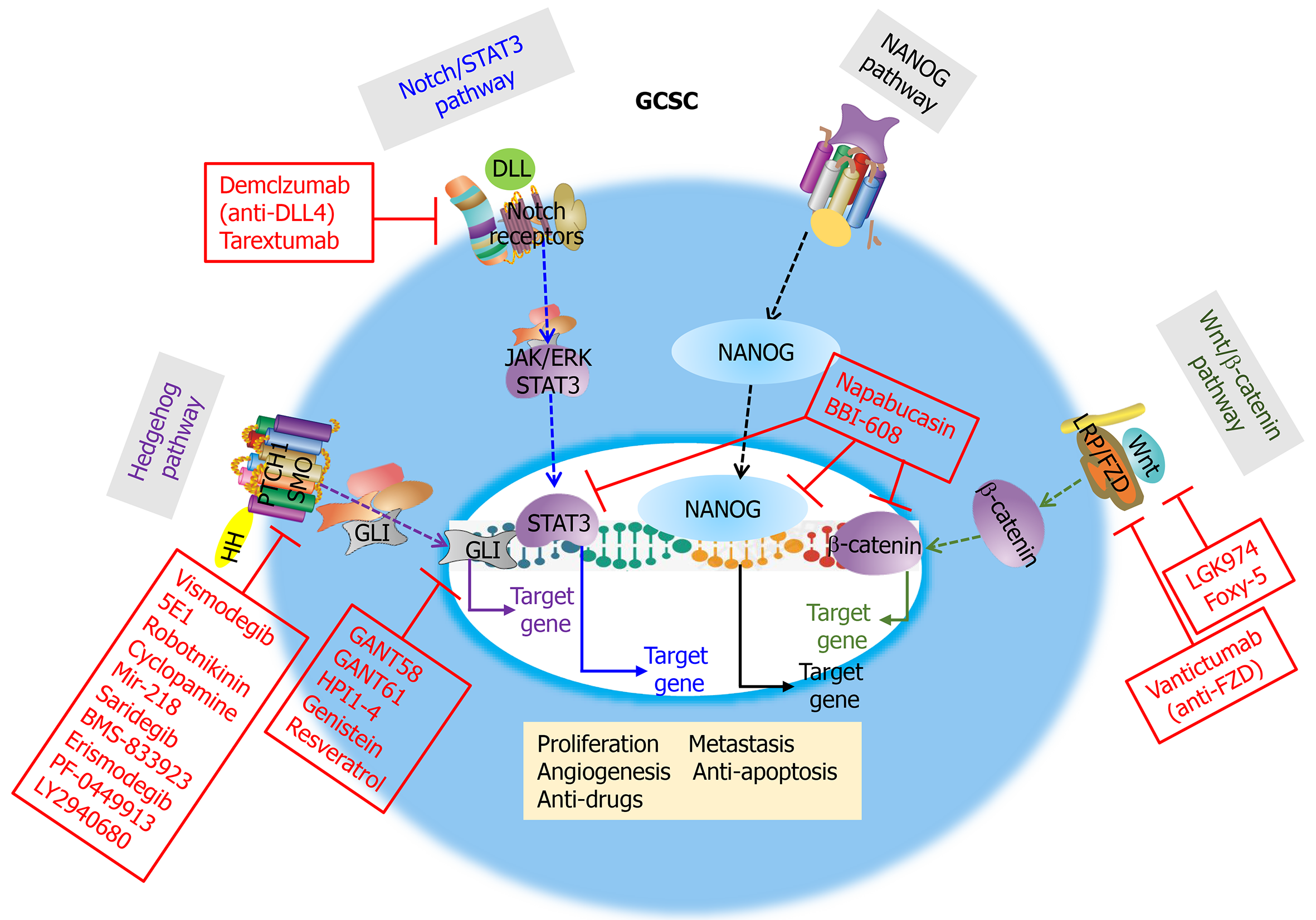
Figure 3 Four signal pathways contribute to stemness properties of cancer stem cells: Hedgehog, Notch/STAT3, NANOG, and Wnt/β-catenin pathways.
Hedgehog pathway: PTCH1-induced inhibition of SMO is reversed by HH binding with PTCH1, leading to the release of the complex of GLI from microtubules, with GLI protein entering the nucleus to transcriptionally activate downstream target genes. Notch/STAT3 pathway: DLL binding-induced Notch activation causes several kinase proteins activation, further converting STAT3 to a transcriptional activator to initiate downstream gene expression. NANOG pathway: Ligand binding-induced NANOG activation causes several kinase proteins activation, further converting STAT3 to a transcriptional activator to initiate downstream gene expression. Wnt/β-catenin pathway: Wnt binds to its receptor, Frizzled to activate LRP protein. The activated LRP protein enhances the phosphorylation of the kinase (a component of the cytoplasmic complex that promotes phosphorylation of β-catenin and its degradation), which inhibits the kinase, further causing the accumulation of free and unphosphorylated β-catenin in the cytoplasm that is then translocated to the nucleus. In the nucleus, β-catenin binds to other transcriptional factors to promote downstream target gene expression.
- Citation: Hsieh HL, Yu MC, Cheng LC, Yeh TS, Tsai MM. Molecular mechanism of therapeutic approaches for human gastric cancer stem cells. World J Stem Cells 2022; 14(1): 76-91
- URL: https://www.wjgnet.com/1948-0210/full/v14/i1/76.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i1.76