Copyright
©The Author(s) 2021.
World J Stem Cells. May 26, 2021; 13(5): 416-438
Published online May 26, 2021. doi: 10.4252/wjsc.v13.i5.416
Published online May 26, 2021. doi: 10.4252/wjsc.v13.i5.416
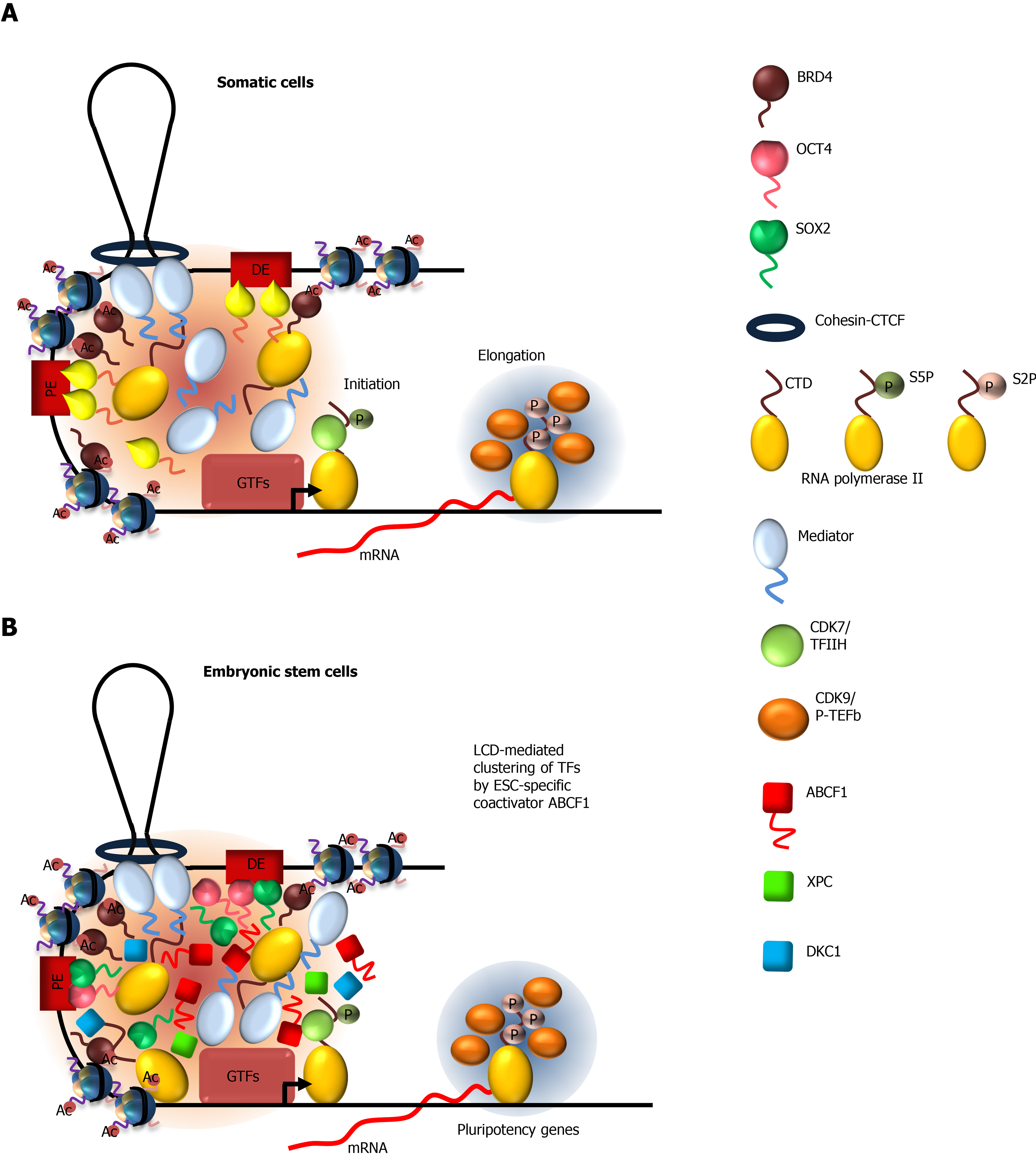
Figure 1 Models depicting the mechanisms by which low-complexity sequence domain-driven interactions between transcription factors and coactivators at gene enhancers contribute to transcriptional activation.
A: Mechanism of transcriptional activation in somatic cells. Low complexity domain (LCD)-mediated interactions between Mediator and TFs, Mediator-bromodomain-containing protein 4 (BRD4), and Mediator-RNA polymerase II (Pol II), as well as binding of BRD4 to acetylated histones, facilitate the formation of a transcription factor-rich compartment at proximal enhancer, and at distal enhancers brought into close proximity through DNA looping by cohesion-CTCF. Increasing local concentration of these factors promotes the formation of the pre-initiation complex composed of general transcription factors and Pol II. Phosphorylation of the C-terminal domain (CTD) at serine 5 (S5P) of Pol II by cyclin-dependent kinase 7/TFIIH disrupts Mediator-Pol II condensates, allowing transcriptional initiation and promoter escape by Pol II. During the early elongation phase of transcription, positive transcription elongation factor b preferentially forms condensates with S5P CTD of Pol II. This results in efficient hyperphosphorylation of the CTD at serine 2 and productive transcription elongation by Pol II; B: Optimal activation of pluripotency genes by stem cell-specific transcription factors octamer-binding transcription factor 4 (OCT4) and SOX2 in ESCs requires cell-specific coactivators ATP-binding cassette subfamily F member 1 (ABCF1), xeroderma pigmentosum, complementation group C (XPC), and dyskerin (DKC1). The LCD of ABCF1 forms selective multivalent interactions with SOX2, XPC, DKC1 and Pol II to promote the assembly of Pol II transcription machinery at pluripotency genes. Activation of OCT4/SOX2-target genes in vivo likely requires both promiscuous LCD-mediated interactions by Mediator and selective LCD-dependent interactions by ABCF1. LCDs are represented by wavy lines. ABCF1: ATP-binding cassette subfamily F member 1; Ac: Acetylated; BRD4: Bromodomain-containing protein 4; CDK: Cyclin-dependent kinase; DE: Distal enhancer; DKC1: Dyskerin; GTFs: General transcription factors; ESC: Embryonic stem cell; General transcription factors; LCD: Low complexity domain; OCT4: Octamer-binding transcription factor 4; P-TEFb: Positive transcription elongation factor b; PE: Proximal enhancer; S2P: Phosphorylated serine 2; S5P: Phosphorylated serine 5; SOX2: SRY-box 2; TF: Transcription factor; XPC: Xeroderma pigmentosum, complementation group C.
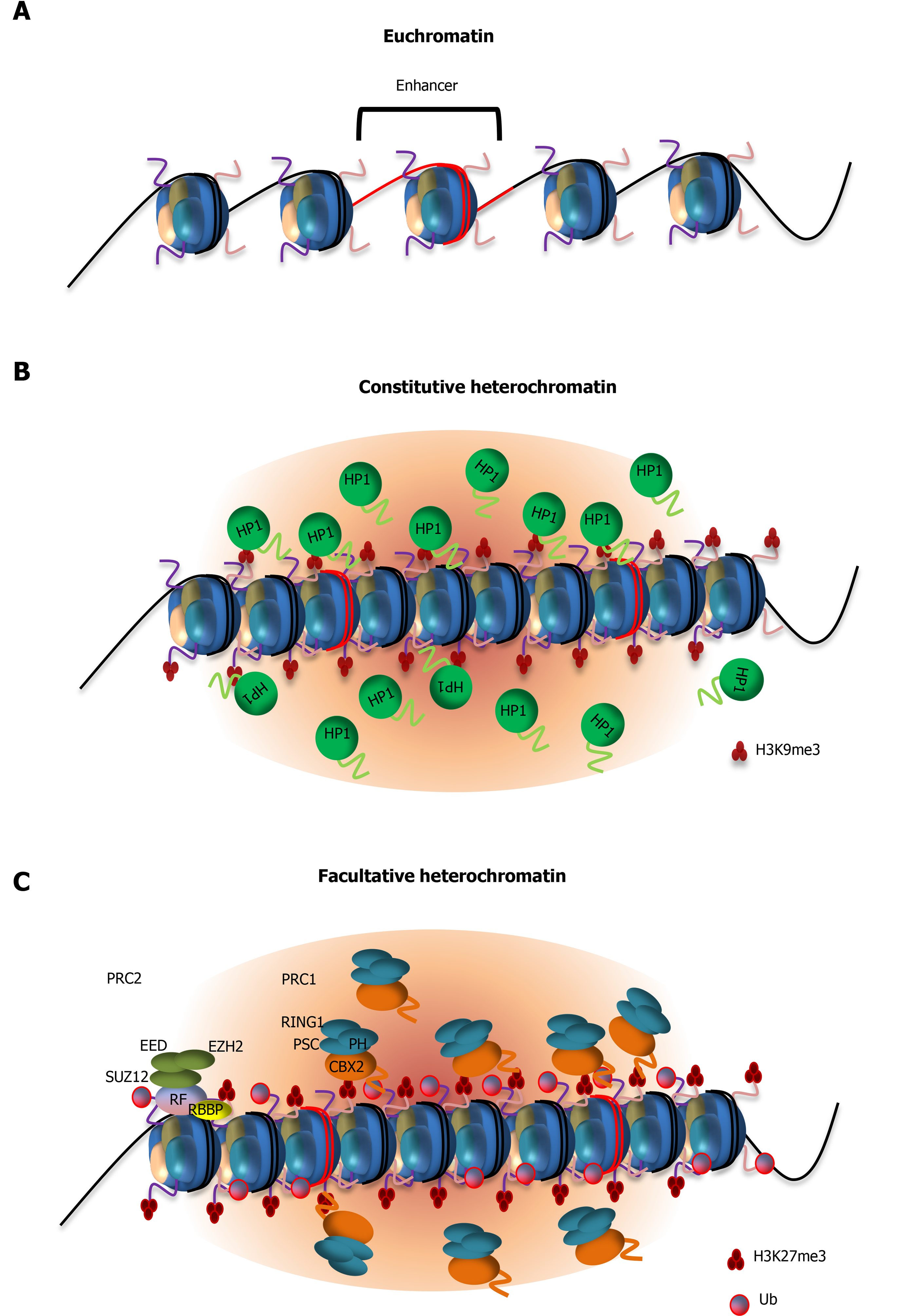
Figure 2 Models showing the role of low complexity domain-mediated protein condensation in gene silencing by heterochromatin formation.
A: Euchromatic regions showing nucleosome-depleted regions containing transcription factor binding sites such as enhancers. Nucleosome-free regions are accessible by transcription factors and thus permissive to gene activation; B: Formation of constitutive heterochromatin requires interactions between heterochromatin protein 1 (HP1) and histone H3 Lysine 9 trimethylation and low complexity domain (LCD)-driven condensation of HP1 with DNA; C: Establishment of facultative heterochromatin is initiated by deposition of histone 3 Lysine 27 trimethylation (H3K27me3) by the histone methyltransferase subunit of polycomb repressive group 2 (PRC2), enhancer of zeste homolog 2. Binding of PRC2 to chromatin is regulated by non-core subunits (e.g., Jumonji and AT-rich interaction domain containing 2, polycomb-likes) which act as recruitment factors[178]. Chromatin compaction is then mediated by the recruitment of PRC1, through recognition of H3K27me3 by its subunit chromobox 2 (CBX2). CBX2 contains an LCD that drives phase separation of PRC1 and is required for proper heterochromatin formation. Subsequent mono-ubiquitination of histone 2A lysine 119 by the ring finger protein 1 subunit of PRC1 is essential for gene silencing. CBX2: Chromobox 2; EZH2: Enhancer of zeste homolog 2; H3K9me3: Histone 3 Lysine 9 trimethylation; H3K27me3: Histone 3 Lysine 27 trimethylation; HP1: Heterochromatin protein 1; JARID2: Jumonji and AT-rich interaction domain containing 2; PCL: Polycomb-like; PRC: Polycomb repressive group; RING1: Ring finger protein 1.
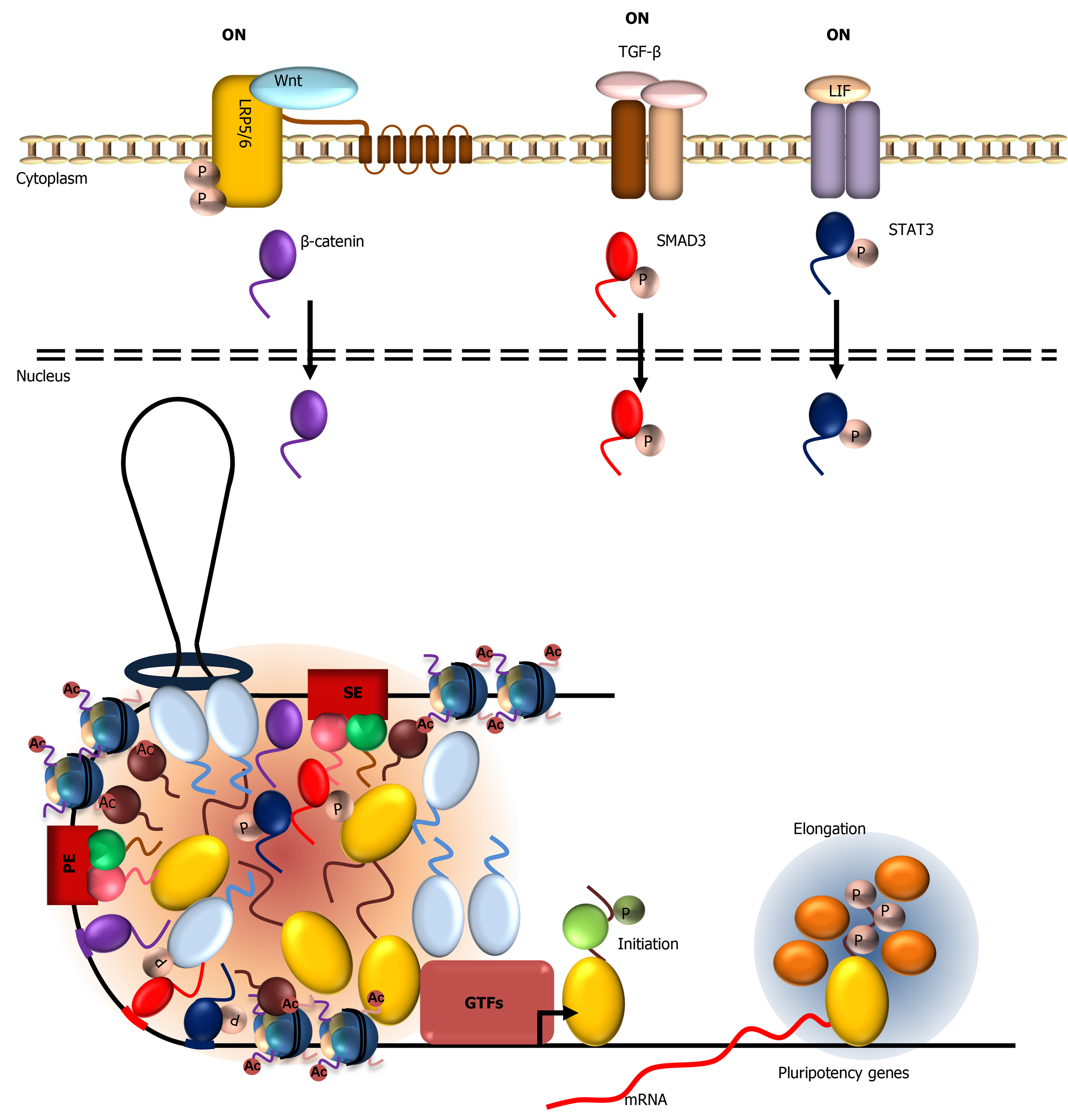
Figure 3 Activation of the wingless-related integration site, transforming growth factor beta, and Janus kinase/signal transducers and activators of transcription signaling pathways leads to nuclear translocation of their respective terminal signaling effectors: β-catenin, SMAD family member 3, and signal transducers and activators of transcription 3.
These low complexity domain (LCD)-containing transcription factors bind their respective signal-responsive elements in pluripotency gene promoters and form LCD-mediated condensates with Mediator to modulate ESC-specific transcriptional activation in a signal-dependent manner. JAK: Janus kinase; STAT: Signal transducers and activators of transcription; SMAD3: SMAD family member 3; TGF-β: Transforming growth factor beta; Wnt: Wingless-related integration site.
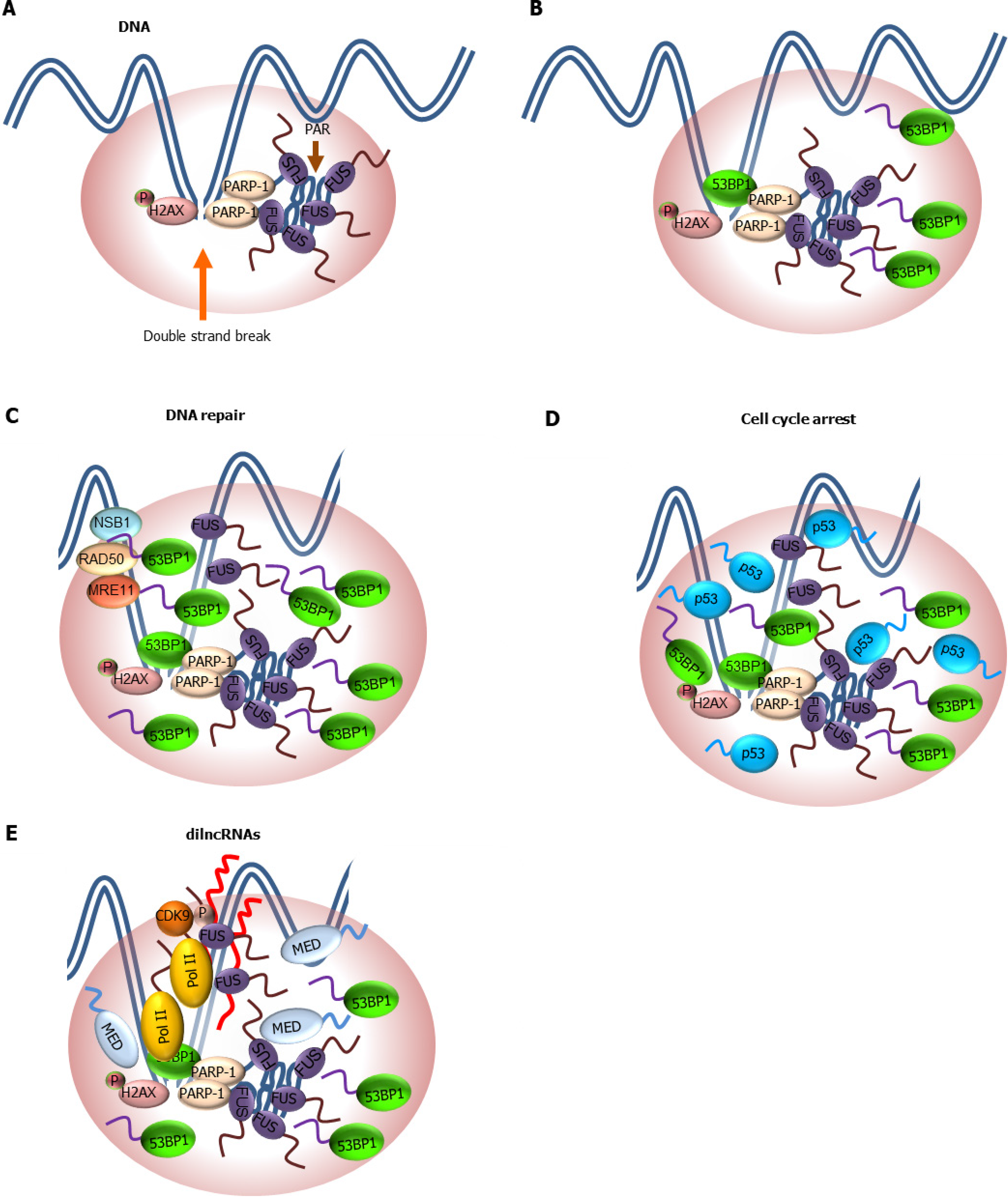
Figure 4 Role of low complexity domain-driven condensate formation in DNA damage response and DNA repair.
A: Double strand break (DSB) triggers phosphorylation of histone variant H2AX (γ-H2AX) and poly-ADP-ribosylation (PARylation) by PARP-1. PARylation facilitates the recruitment of fused in sarcoma (FUS) to DSB site through its RGG domain and formation of FUS condensates driven by its prion-like low complexity domain (LCD); B: γ-H2AX and FUS condensates at DSBs promote the incorporation of a critical downstream effector of DSB repair factor, p53-binding protein 1 (53BP1). However, the Tudor domain but not its disordered region of 53BP1 is required for 53BP1 phase separation; C: Formation of 53BP1/FUS condensates facilitate the recruitment of downstream repair machinery such as meiotic recombination 11 homolog. RAD50 homolog, double strand break repair protein, and Nijmegen breakage syndrome 1 involved in homologous recombination repair of DSBs; D: p53 is incorporated into 53BP1 condensates and activate a p53-dependenent gene expression that results in cell cycle arrest. Disruption of 53BP1 condensates blunts p53-dependent response to DNA damage; E: Assembly of Pol II, mediator and cyclin dependent kinase 9/positive transcription elongation factor b at DSBs leads to transcription of dilncRNAs. dilncRNAs facilitates molecular crowding and phase separation of 53BP1 and other repair factors. It is likely that a network of LCD-mediated protein-protein and protein-nucleic acid interactions drives the formation of repair condensates at DSBs. 53BP1: p53-binding protein 1; dilncRNA: Damage-induced long non-coding RNA; DSB: Double strand break; FUS: Fused in sarcoma; H2AX: Histone variant 2AX; MRE11: Meiotic recombination 11 homolog; NBS1: Nijmegen breakage syndrome 1; PARP-1: Poly-ADP-ribose polymerase-1; RAD50: RAD50 homolog, double strand break repair protein.
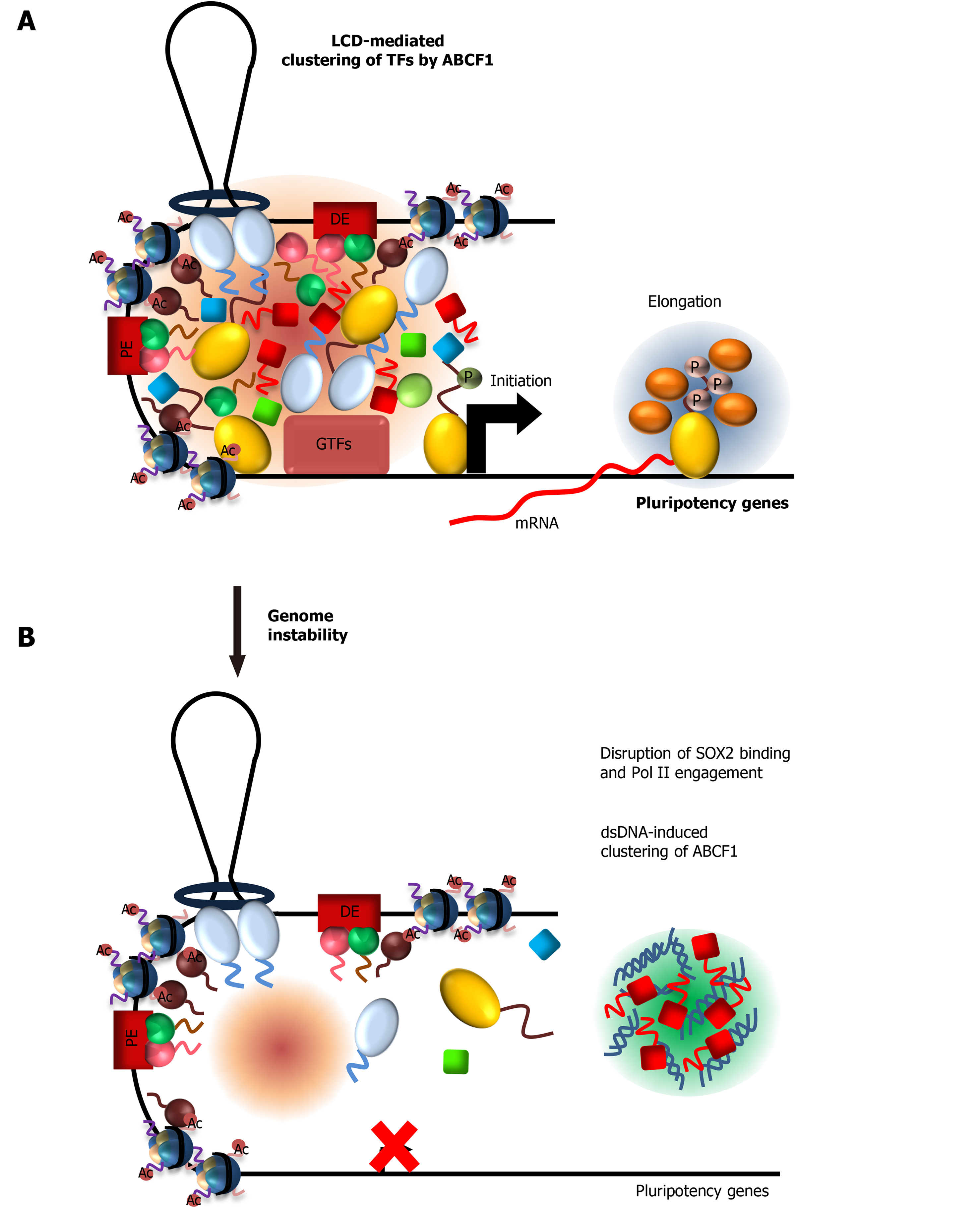
Figure 5 ATP-binding cassette subfamily F member 1 couples stem cell-specific transcription with DNA sensing in Embryonic stem cells.
A: ATP-binding cassette subfamily F member 1 (ABCF1) low complexity domain promotes specific clustering and formation of a hub comprising of sex-determining region Y-box 2 (SOX2), xeroderma pigmentosum, complementation group C (XPC), dyskerin (DKC1), and RNA polymerase II (Pol II) molecules at target gene promoter to stimulate transcription, presumably by increasing local concentration of these factors; B: ABCF1 proteins available for transcription are diverted to bind intracellular double-stranded DNA (dsDNAs) generated from genome instability, due to increased propensity of ABCF1 to form condensates with dsDNAs. Decrease in ABCF1 at gene promoters destabilizes the multivalent interactions between SOX2, XPC, DKC1, and Pol II. This leads to disruption of the protein hub and decrease in gene transcription by Pol II. Downregulation of pluripotency-associated genes promotes differentiation of compromised ESCs and their elimination from the self-renewing population, thereby preserving genome fidelity in ESCs. LCD: Low complexity domain; TF: Transcription factor; dsDNA: Double-stranded DNA; Pol II: RNA Polymerase II; ABCF1: ATP-binding cassette subfamily F member 1; SOX2: Sex-determining region Y-box 2.
- Citation: Vodnala M, Choi EB, Fong YW. Low complexity domains, condensates, and stem cell pluripotency. World J Stem Cells 2021; 13(5): 416-438
- URL: https://www.wjgnet.com/1948-0210/full/v13/i5/416.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i5.416