修回日期: 2004-09-13
接受日期: 2004-09-30
在线出版日期: 2004-11-15
目的: 构建丙型肝炎病毒HCV p7蛋白反式激活相关基因差异表达的差异cDNA, 克隆HCV p7蛋白反式激活相关基因.
方法: 以HCV p7表达质粒pcDNA3.1(-)-p7转染HepG2细胞, 以空载体pcDNA3.1(-)为对照; 制备转染后的细胞裂解液, 从中提取mRNA并合成cDNA, 经Rsa I酶切后将实验组cDNA分成两组, 分别与两种不同的接头衔接, 再与对照组cDNA进行两次消减杂交及两次抑制性PCR, 将产物与T/A载体连接, 构建cDNA消减文库, 并转染大肠杆菌进行文库扩增, 随机挑选克隆PCR后进行测序及同源性分析.
结果: 成功构建人HCV p7蛋白反式激活相关基因差异表达的cDNA. 扩增后得到56个200-1 000 bp插入片段的克隆, 随机挑选其中33个插入片段测序, 并通过生物信息学分析获得其全长基因序列, 结果共获得15种编码基因, 其中1个为未知功能的新基因.
结论: 筛选到的cDNA全长序列, 包括与细胞生长调节、物质代谢、和细胞凋亡密切相关的一些蛋白编码基因.
引文著录: 郭江, 成军, 纪冬, 赵龙凤, 王建军, 刘妍, 黄燕萍. 应用抑制性消减杂交技术克隆和筛选HCVp7蛋白的反式调节基因. 世界华人消化杂志 2004; 12(11): 2590-2593
Revised: September 13, 2004
Accepted: September 30, 2004
Published online: November 15, 2004
AIM: To clone and identify human genes transactivated by hepatitis C virus p7 (HCVp7) protein.
METHODS: Suppression subtractive hybridization (SSH) and bioinformatic techniques were used for screening and cloning of the target genes transactivated by HCVp7 protein. The mRNA was isolated from HepG2 cells transfected with pcDNA3.1(-)-p7 and pcDNA3.1(-) empty vector respectively, and SSH method was employed to analyze the differentially expressed DNA sequence between the two groups. After digestion with restriction enzyme Rsa I, cDNAs of small size were obtained. Then tester cDNA was divided into two groups and ligated to the specific adaptor 1 and 2 respectively. After tester cDNA was hybridized with driver cDNA twice and underwent nested PCR twice, and then the product was subcloned into T/A plasmid vectors to set up the subtractive library. Amplification of the library was carried out after transfected with E. coli strain DH5α. The cDNA was sequenced and analyzed in GenBank with Blast search after PCR.
RESULTS: The subtractive library of genes transactivated by HCVp7 was constructed successfully. The amplified library contained 71 positive clones. Colony PCR showed that 56 clones contained 200-1 000 bp inserts. Sequence analysis was performed in 33 clones randomly, and the full-length sequences were obtained with bioinformatics method. Altogether 15 coding sequences were obtained, including 14 known and 1 unknown.
CONCLUSION: The obtained sequences may be target genes transactivated by HCV p7, and some genes coding proteins get involved in cell cycle regulation, metabolism, and cell apoptosis.
- Citation: Guo J, Cheng J, Ji D, Zhao LF, Wang JJ, Liu Y, Huang YP. Screening and cloning of target genes transactivated by hepatitis C virus p7 protein. Shijie Huaren Xiaohua Zazhi 2004; 12(11): 2590-2593
- URL: https://www.wjgnet.com/1009-3079/full/v12/i11/2590.htm
- DOI: https://dx.doi.org/10.11569/wcjd.v12.i11.2590
丙型肝炎病毒(HCV)是世界范围内慢性肝炎的主要病原之一[1-2], 约有1.7亿人被感染, 常导致严重肝病, 包括肝硬化和肝细胞癌(HCC)[3-5]. HCV的基因组含有一个长的开放阅读框架(ORF), 编码一个约3 010个氨基酸残基(aa)组成的多蛋白, 该多蛋白被细胞和病毒蛋白酶切割产生至少10个病毒基因产物: core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A和NS5B蛋白等[6-8]. p7是由HCV基因组2580-2768核苷酸(nt)之间的基因编码的由63个氨基酸残基(aa)组成的一个小蛋白. 关于其功能研究较少. 我们利用抑制性消减杂交技术构建p7作用于肝母细胞瘤细胞系HepG2细胞的反式调节cDNA消减文库, 筛选差异表达的基因片段, 并应用生物信息学进行分析, 为加深了解p7的功能, 进一步探讨HCV发病及致癌机制提供新的研究方向.
HepG2细胞及感受态大肠杆菌DH5α为本室保存, pcDNA3.1(-)真核表达载体购自Invitrogen公司; FuGENE6转染试剂购自Roche公司, mRNA Purification试剂盒购自Amersham Pharmacia Biotech公司, PCR-Select cDNA Subtraction试剂盒、50×PCR Enzyme Mix、Advantage PCR Cloning试剂盒购自Clontech公司, High Pure PCR Product Purification试剂盒购自Boehringer Mannheim公司, T7、SP6通用引物及pGEM-Teasy载体购自Promega公司. HCV p7真核表达质粒pcDNA3.1(-)-p7由本室构建.用FuGENE6转染试剂将2 μg pcDNA3.1(-)-p7及pcDNA3.1(-)空载体分别转染35 mm平皿HepG2细胞, 48 h后收获细胞. 使用mRNA Purification试剂盒, 直接提取转染了重组表达质粒及空载体的HepG2细胞mRNA, 经琼脂糖凝胶电泳及分光光度计进行定性定量分析.
消减杂交文库的建立采用Clontech公司的PCR-Select cDNA Subtraction Kit, 常规SSH方法按说明书进行. 以转染了重组表达质粒及空载体的HepG2 细胞mRNA为模板逆转录合成双链cDNA(dscDNA), 并分别标记为Tester和Driver, dscDNA经RsaⅠ(一种识别4碱基序列的内切酶)消化, 产生相对较短的平端片段, 纯化酶切产物. 将Tester的dscDNA分为两份, 分别连接试剂盒提供的特殊设计的寡核苷酸接头Adapter 1和Adapter 2, 然后与过量的Driver dscDNA进行杂交; 合并两种杂交产物后再与Driver dscDNA作第2次杂交; 然后将杂交产物做选择性PCR扩增, 使Tester dscDNA中特异性表达或高表达的片段得到特异性扩增. 扩增产物与pGEM-Teasy载体连接, 转化DH5α感受态细菌, 在含氨苄青霉素的LB/X-gal/IPTG培养板上, 37 ℃培养18 h. 共得到71个白色菌落, 以pGEM-Teasy载体多克隆位点两端T7/SP6引物进行菌落PCR扩增, 证明56含有插入片段后(200-1 000 bp), 随机挑选其中个33个克隆增菌, 测序(上海申友公司), 并且应用生物信息学将测得序列GenBank数据库进行同源性分析.
紫外分光检测显示, 转染了真核表达质粒及空载体的HepG2细胞提取mRNA分别为1.58 μg和2.24 μg, A260/A280=2.18.20 g/L琼脂糖凝胶电泳见mRNA为大于0.5 kb清晰慧尾片状条带, 证实mRNA质量完全满足进行消减杂交的要求.
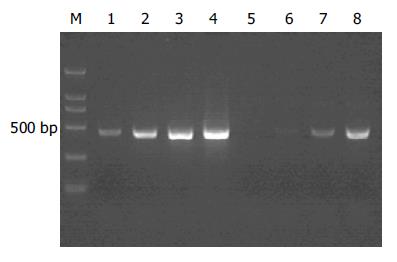
以消减及未消减PCR产物为模板, 用G3PDH引物进行PCR扩增, 分别在18、23、28、33次循环结束时从体系中吸取5 μL进行电泳鉴定. 结果显示: 与未消减组PCR产物相比, 消减组PCR产物中G3PDH基因产物大大减少, 说明所构建的消减文库具有很高的消减效率(图1).
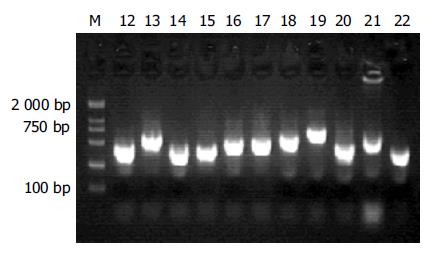
杂交产物经两轮PCR扩增后, 菌落PCR扩增结果显示为200-1 000 bp大小不等的插入片段, 所获得的71个克隆中56含有插入片段, 这些条带可能代表差异表达的基因片段(图2).
挑选33个克隆测序, 与GenBank数据库进行初步比较. 其中32个克隆均与已知基因的部分序列高度同源(95-100%)1个克隆未检索到任何相应的相似序列, 代表了新基因, 命名为HCV p7TP1, GeneBank 注册号为AY596776(表1).
| 同源蛋白名称 | 同源克隆数 | 同源性(%) |
| 核糖体蛋白 | 5 | 98-100 |
| SMT3H2蛋白 | 6 | 95-100 |
| 乌洛托品结合蛋白(FBP3) | 3 | 97-98 |
| 真核翻译启始因子(eIF2A) | 2 | 97-98 |
| 线粒体单倍体 | 3 | 98-99 |
| 二氢二醇脱氢酶 | 3 | 100 |
| MSH5蛋白 | 3 | 99 |
| RNA多聚酶II, TATA | 1 | 95 |
| 盒结合蛋白相关因子(TAF6) | ||
| 内质网硫氧还蛋白(TLP19) | 1 | 96 |
| 环指蛋白 | 1 | 100 |
| 唾液酸糖蛋白受体1(ASGP-R) | 1 | 100 |
| RAD21蛋白 | 1 | 98 |
| T细胞白血病病毒I型结合蛋白1(TAX1BP1) | 1 | 100 |
| 芳香族受体(HOR) | 1 | 98 |
| 新基因序列 | 1 | |
| 克隆总计 | 33 |
HCV p7是一个小的疏水性多蛋白, 编码基因位于结构蛋白和非结构蛋白之间. p7可在黑脂质膜中形成六聚体阳离子通道, 推测其功能与病毒离子孔道蛋白(viroporin)类似, 可使细胞内膜结构不稳定化, 以利于成熟的病毒颗粒释放[9-10], 这一离子通道可被金刚烷胺和长烷基链的亚氨基糖衍生物抑制[11-13]. p7对病毒复制并不是关键的, 但对感染性病毒的产生是必不可少的[14]. p7仍有许多功能没有研究清楚, 对其反式激活作用的研究更少. 为进一步研究p7的功能, 明确p7在丙型肝炎发病机制中的作用, 利用抑制性消减杂交技术[15-17](suppression subtractive hybridization, SSH)筛选并克隆HCV p7反式激活的靶基因, 推测其在体内可能存在功能的线索. SSH方法是近年发展起来的一项新的基因克隆技术, 与传统的方法比较, 具有实验周期短、易操作、可靠性高、假阳性率低等特点, 能有效地分离扩增低丰度特异表达的基因, 可以在较短的时间内获得较理想的实验结果[18-21]. 我们将真核表达载体pcDNA3.1(-)-p7, 转染肝母细胞瘤细胞系HepG2, 并以转染空白载体的相同细胞系作为对照, 以2种转染的细胞系中提取的mRNA为起始材料, 应用SSH方法成功地构建了p7反式激活相关基因差异表达的cDNA消减文库, 挑选33个克隆测序分析, 主要包括两种类型的序列, 即已知功能的基因序列和未知功能的基因序列. 在本次实验中获得1个差异表达的未知序列, 对其基因结构和功能正在研究之中.
在已知功能基因序列中, 主要包括: (1)细胞内结构与细胞生长相关蛋白, 如核糖体L7蛋白、真核翻译起始因子2A(eIF2A)、环指蛋白7、MSH5, 还有线粒体蛋白等; (2)参与细胞内代谢的蛋白基因, 如TLP19、二氢二醇脱氢酶可代谢肝脏的多环芳香烃(DDH), DDH1/2的表达上升与肝癌的大小和疾病进展的相关[22-24]; (3)参与细胞内信号转导的蛋白基因, 如SMT3H2、FBP3、Rad21和TAX1BP1. p7上调TAX1BP基因的表达可能是HCV致癌的机制之一; (4)其他蛋白, 如唾液酸糖蛋白受体(ASGP-R)等.
通过对HCV p7的上述反式激活基因的分析, 我们发现他与体内物质代谢、信号转导、凋亡关系密切[25-26], 在病毒感染后肝细胞恶性变方面有一定的作用, 而且对于HCV病毒颗粒结合和进入肝细胞是重要的. 关于HCV p7在体内与各种活性因子的具体调节机制, 仍需进行详细的实验来研究. HCV p7抑制性消减文库的建立为研究其功能提供了理论依据, 并为HCV致癌机制提供了新的研究方向.
编辑: N/A
| 4. | 刘 妍, 杨 倩, 成 军, 王 建军, 纪 冬, 党 晓燕, 王 春花. 基因表达谱芯片筛选NS5ATP3转染细胞差异表达基因. 世界华人消化杂志. 2004;12:306-310. [DOI] |
| 5. | 李 克, 王 琳, 成 军, 张 玲霞, 段 惠娟, 陆 荫英, 杨 继珍, 刘 妍, 洪 源, 夏 小兵. 酵母双杂交技术筛选克隆HCV核心蛋白结合蛋白基因1. 世界华人消化杂志. 2001;9:1379-1383. [DOI] |
| 6. | Wietzke-Braun P, Braun F, Sattler B, Ramadori G, Ringe B. Initial steroid-free immunosuppression after liver transplantation in recipients with hepatitis C virus related cirrhosis. World J Gastroenterol. 2004;10:2213-2217. [PubMed] |
| 7. | Gong GZ, Jiang YF, He Y, Lai LY, Zhu YH, Su XS. HCV NS5A abrogates p53 protein function by interfering with p53-DNA binding. World J Gastroenterol. 2004;10:2223-2227. [PubMed] |
| 8. | Anand BS, Velez M. Assessment of correlation between serum titers of hepatitis C virus and severity of liver disease. World J Gastroenterol. 2004;10:2409-2411. [PubMed] [DOI] |
| 9. | Griffin SD, Beales LP, Clarke DS, Worsfold O, Evans SD, Jaeger J, Harris MP, Rowlands DJ. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003;535:34-38. [PubMed] [DOI] |
| 10. | Pavlović D, Neville DC, Argaud O, Blumberg B, Dwek RA, Fischer WB, Zitzmann N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc Natl Acad Sci USA. 2003;100:6104-6108. [PubMed] [DOI] |
| 11. | Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3' nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76:5326-5338. [PubMed] [DOI] |
| 12. | Griffin SD, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J Gen Virol. 2004;85:451-461. [PubMed] [DOI] |
| 13. | Branza-Nichita N, Durantel D, Carrouée-Durantel S, Dwek RA, Zitzmann N. Antiviral effect of N-butyldeoxynojirimycin against bovine viral diarrhea virus correlates with misfolding of E2 envelope proteins and impairment of their association into E1-E2 heterodimers. J Virol. 2001;75:3527-3536. [PubMed] [DOI] |
| 14. | Sakai A, Claire MS, Faulk K, Govindarajan S, Emerson SU, Purcell RH, Bukh J. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc Natl Acad Sci USA. 2003;100:11646-11651. [PubMed] [DOI] |
| 15. | Dong XY, Pang XW, Yu ST, Su YR, Wang HC, Yin YH, Wang YD, Chen WF. Identification of genes differentially expressed in human hepatocellular carcinoma by a modified suppression subtractive hybridization method. Int J Cancer. 2004;112:239-248. [PubMed] [DOI] |
| 16. | Galbraith EA, Antonopoulos DA, White BA. Suppressive subtractive hybridization as a tool for identifying genetic diversity in an environmental metagenome: the rumen as a model. Environ Microbiol. 2004;6:928-937. [PubMed] [DOI] |
| 17. | Hou L, Tang JW, Cui XN, Wang B, Song B, Sun L. Construction and selection of subtracted cDNA library of mouse hepatocarcinoma cell lines with different lymphatic metastasis potential. World J Gastroenterol. 2004;10:2318-2322. [PubMed] [DOI] |
| 18. | Sun W, Zhang K, Zhang X, Lei W, Xiao T, Ma J, Guo S, Shao S, Zhang H, Liu Y. Identification of differentially expressed genes in human lung squamous cell carcinoma using suppression subtractive hybridization. Cancer Lett. 2004;212:83-93. [PubMed] [DOI] |
| 20. | Ji W, Wright MB, Cai L, Flament A, Lindpaintner K. Efficacy of SSH PCR in isolating different-tially expressed genes. BMC Genomics. 2002;3:12. [DOI] |
| 22. | Tang DW, Chang KW, Chi CW, Liu TY. Hydroxychavicol modulates benzo[a]pyrene-induced genotoxicity through induction of dihydrodiol dehydrogenase. Toxicol Lett. 2004;152:235-243. [PubMed] [DOI] |
| 23. | Ozeki T, Takahashi Y, Nakayama K, Funayama M, Nagashima K, Kodama T, Kamataki T. Hepatocyte nuclear factor-4 alpha/gamma and hepatocyte nuclear factor-1 alpha as causal factors of interindividual difference in the expression of human dihydrodiol dehydrogenase 4 mRNA in human livers. Pharmacogenetics. 2003;13:49-53. [DOI] |
| 24. | Chen CY, Hsu CP, Hsu NY, Shih CS, Lin TY, Chow KC. Expression of dihydrodiol dehydrogenase in the resected stage I non-small cell lung cancer. Oncol Rep. 2002;9:515-519. [PubMed] [DOI] |
| 25. | Pati D, Zhang N, Plon SE. Linking sister chromatid cohesion and apoptosis: role of Rad21. Mol Cell Biol. 2002;22:8267-8277. [PubMed] [DOI] |