INTRODUCTION
H pylori is a spiral, Gram-negative bacterium, which chronically infects more than half of population worldwide, and is implicated in gastritis, peptic ulcer and gastric cancer[1-12]. One of its features is high genome diversity[13-15]. Clinical isolates of H pylori from different individuals show enormous variation in their genomic fingerprints[16-18]. Studies based on the two sequenced genomes of H pylori strains (i.e. 26695 and J99) have revealed that among the H pylori genes, 22% are dispensable and about 6% are unique[14,15]. Sequence variation of certain genes or regions, such as the cag pathogenicity island[19,20], vacA[21], babA and babB[22], drug resistant genes[23-26] and restriction-modification genes[27-29], is also remarkable. For a given individual, the genes of the colonized strain even change over the course of colonization[17,30]. Genotyping of the bacterium is important for the epidemiological and pathogenic investigation, and genes that are present in one strain but absent or substantially different in others can be of great interest biologically.
The polymerase chain reaction (PCR)-based randomly amplified polymorphic DNA (RAPD) method has widely applied to the distinction of H pylori isolates due to its sensitivity, efficiency and promptness[16,17]. However, problems with its stability or reproducibility arise. In this study, we obtained RAPD fingerprints of clinical isolates of H pylori under optimized conditions, and then determined the stability of RAPD products by Southern blotting and DNA sequencing techniques.
MATERIALS AND METHODS
H pylori culture
Clinical isolates of H pylori cultured from gastric antra and cardia of 73 patients, 12 with gastric ulcer (GU), 18 with duodenum ulcer (DU), 8 with both GU and DU, and 25 with non-ulcer dyspepsia (NUD), were selected for this study. Each isolate preserved in the brain heart infusion broth (BHI, Gibco, Edinburgh, UK) supplemented with 100 mL/L horse serum and 200 mL/L glycerol was inoculated onto H pylori selective chocolate blood agar containing 40 g/L blood agar base No. 2 (Oxoid, Basingstoke, UK) and 50 mL/L horse blood (Gibco, Edinburgh, UK). Antibiotics (Sigma, St. Louis, MO, USA) were used at the following concentrations: 3 mg/L vancomycin, 5 mg/L trimethoprim, 10 mg/L nalidixic acid and 2 mg/L amphotericin B. The plates were incubated in a microaerobic atmosphere (50 mL/L CO2) in a CO2 incubator (Forma Scientific, Marietta, OH, USA) at 37 °C for up to 5 days.
Extraction of genomic DNA
H pylori cells on the plates were harvested by using a sterile swab, and transferred into an Eppendorf tube containing 1.5 mL TE buffer (10 mmol/L Tris-HCl, pH8.0; 1 mmol/L EDTA, pH8.0). The suspension was centrifuged at 8000 g for 10 min and washed once with TE buffer. The pellet was then suspended in 800 μL TE buffer was incubated with 10 μL of 100 g/L lysozyme (Sigma, St. Louis, MO, USA) at 37 °C for 30 min. The suspension was then lysed with 100 μL of 100 g/L sodium dodecyl sulfate (SDS) by incubating at 37 °C for another 30 min. Then 5 μL of proteinase K (10 g/L) (Boehninger, Mannheim, Germany) was added into the mixture and incubated at 56 °C for 1 h. Afterward, H pylori DNA was purified by extracting twice with equal volume of phenol and once with equal volume of chloroform, followed by centrifugation at 12000 g for 10 min each time. The supernatant was transferred into a new Eppendorf tube, and precipitated with 2 volume of absolute ethanol and 20 μL of 3 M sodium acetate at -20 °C overnight. The DNA preparation was centrifuged at 12000 g for 20 min and washed once with 700 mL/L ethanol. The pellet was then vacuum-dried and suspended in 50 μL distilled water. RNase A was added at the final concentration of 20 mg/L. After incubated at room temperature fo r 30 min, DNA concentration was determined spectrophotometrically.
RAPD for genomic DNA
A universal primer was chosen for the PCR-based RAPD, according to the paper of Akopyanz, et al[31] to fingerprint the genomic DNA. The primer was 5-AAGAGCCCGT-3. The volume of the PCR mixture was 25 μL containing 50 ng of H pylori genomic DNA, 20 pmol primer, 1 unit of Taq DNA polymerase, 0.25 mmol/L dNTPs, 10 mmol/L Tris-HCl, 50 mmol/L KCl, 2 mmol/L MgCl2 and 0.1 g/L gelatin (Promega, Madison WI, USA). PCR was performed with a thermal cycler (Perkin-Elmer 2400, Boston, MA, USA) consisting of an initial step at 94 °C for 5 min. This was followed by 39 cycles of denaturation at 94 °C for 1 min, annealing at 36 °C for 1 min and extension at 72 °C for 1 min. Ten microlitres of the PCR products were electrophoresed in 10 g/L horizontal agarose gel in TBE buffer at 68 V for 2 h. The gels were stained with ethidium bromide (EB, 1 mg/L) and photographed.
DNA fragment cloning and sequencing
At first, two primers were designed: RDF1-CGCAAGCTTCCACACAGAACCAC, and RDF2-CGCGGATCCGGACCTTTACTACAAC. A 23S rDNA fragment (RDF) was amplified according to the gene sequence reported[32]. The PCR mixture was 40 μL in volume containing 4 μL of 10 × polymerase buffer, 2 μL of genomic DNA (20 ng) from the reference strain NCTC11637, 1 μL (2 units) of Taq DNA polymerase, 4 μL of 2.5 mmol/L dNTPs, and 4 μL of 10 mmol/L primers. The sample was subjected to denaturation at 94 °C for 3 min. The PCR program was 30 cycles at 94 °C for 30 s, at 55 °C for 45 s, and at 72 °C for 1 min, followed by an extra extension at 72 °C for 5min. The PCR product (10 μL) was then electrophoresed in a 15 g/L agarose gel. DNA was recovered by using QIAEXÐ agarose gel extract kits (Qiagen, Santa Clarita, CA, USA). The purified PCR product was then cloned into the pT-Adv vector (Clontech, Palo Alto, CA, USA) and transformed into E. coli Top10, which was then inoculated on the selective plates containing 20 µL of 50 g/L ampicillin, 35 µL of 100 mmol/L IPTG and 40 µL of 20 g/L X-gal. White colonies were randomly picked up, suspended in 5 mL of Luria-Bertani medium containing ampicillin (50 mg/L) and cultured at 37 °C overnight. The recombinant plasmid was extracted by using Qiaprep spin miniprep kit (Qiagen, Santa Clarita, CA, USA). In the same way, RDFs from different H pylori isolates were amplified and cloned into the pGEM-T-Easy Vector (Promega, Madison, WI, USA). Then, A 23S rDNA fragment (1050 bp) containing the RDF in the middle was also amplified from different H pylori isolates by using the primers DMSR1-TAAGTTCGCGATAAGGTGTGC and DMSR1-GGTTCCTGCTTAGATGCTTTC. The inserts in the recombinant plasmids and the RDF flanking regions were sequenced by using the Big dye terminator DNA sequencing kit (Perkin-Elmer, Boston, MA, USA) and ABI automated sequencer (GMI, Minnesota, USA).
Southern blotting
Southern blotting was carried out following the protocol provided in the Enhanced chemiluminescence (ECL) direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech, Buckinghamshire, UK). In brief, after agarose gel electrophoresis of the RAPD products, the DNA was submitted to denaturation by immersing the gel (200 cm2) in 200 ml buffer containing 0.5 mol/L NaOH and 1.5 mol/L NaCl for 30 min. Then the gel was neutralized in 200 ml of 0.5 mol/L Tris-HCl and 1.5 mol/L NaCl (pH7.5) for 30 min. Afterward, the DNA was transferred to the Hybond N+ membrane (Amersham Pharmacia Biotech, Buckinghamshire, UK) by using a capillary blotting apparatus. DNA fixation was carried out by exposing the membrane to an UV transilluminator (Vilber Lourmat, Marne la Vallee Cedex 1, France) for 5 min. RDF sequenced and digested by EcoR I from pT-Adv vector was used as a probe. The pre-hybridization and hybridization were performed in a buffer containing 0.5 mol/L NaCl and 50 g/L blocking agent in a hybridization oven at 42 °C for 1 h and 5 h respectively. After stringency washing (twice for 20 min in 6 mol/L urea, 4 g/L SDS and 0.1 × standard saline citrate, SSC; twice for 5 min in 2 × SSC), chemiluminenscence signals were detected by exposing the hyperfilm to the membrane for 5 to 30 min in a dark room.
RESULTS
H pylori gene profiles identified by RAPD
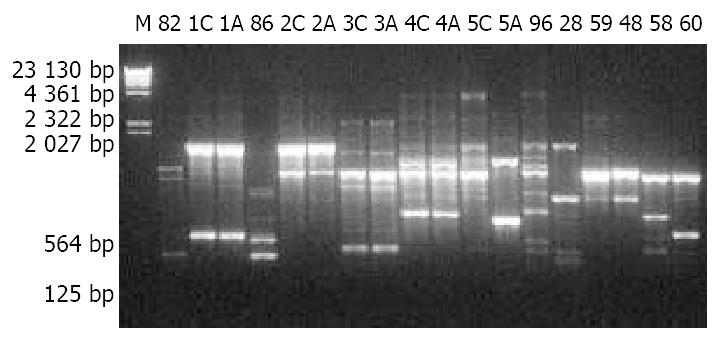
For all H pylori isolates from the 73 individuals, bacterial growth, genomic DNA extraction and RAPD were carried out under the same conditions. RAPD products were analyzed by 10 g/L agarose gel electrophoresis (Figure 1 and Figure 2), which showed that H pylori from gastric antra of different individuals had different RAPD profiles whereas the isolates from antrum and cardia of the same individual had almost identical RAPD profiles with only one exception (lanes 5C and 5A in Figure 1).
Figure 1 10 g/L agarose gel electrophoresis of RAPD products of H pylori isolates.
M: λDNA Hind III markers; 82, 86, 96, 28, 59, 48, 59 and 60: Designations of H pylori isolates; A: Antrum; C: Cardia.
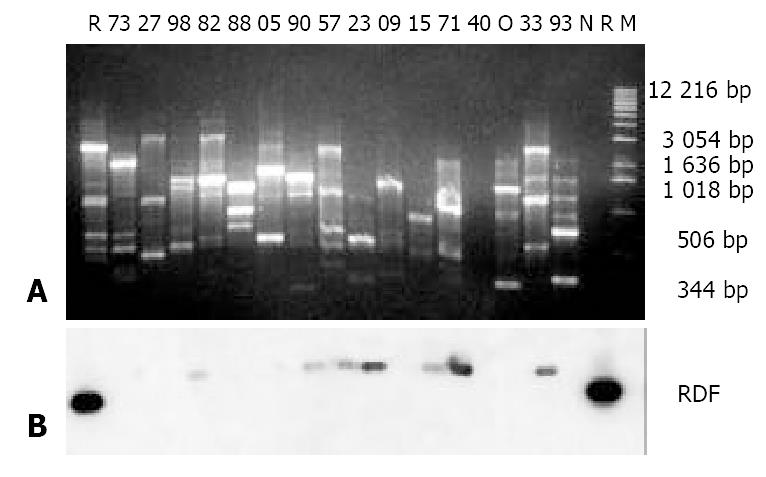
Figure 2 Results of agarose gel electrophoresis of RAPD products (A) and Southern blotting (B).
A: 10 g/L agarose gel elec-trophoresis of RAPD products of H pylori isolates; M: 1 Kb DNA ladder; 73, 27, 98, 82, 88, 05, 90, 57, 23, 09, 15, 71, 40, 33 and 93: Designations of the H pylori isolates; N: H pylori reference strain NCTC11637; R: 1 ng RDF loaded in the well; O: Template blank control; B: Southern blotting results using the RDF as a probe, the dark blots on either side of Picture B corresponding to the 1 ng RDF in the agarose gel in Picture A.
Probe preparation
The sequence of RDF from the reference strain NCTC11637 was 99% identical to that reported in the GenBank. It was a 361 bp fragment of the 23S rDNA (from 2133 to 2493), which is just inside the loop (2134-2684) to form peptidyltransferase center in the ribosome. The fragment was then used as a probe in Southern blotting to investigate if the random PCR primer would pick up the same fragment with the same amount from genomes of different clinical isolates of H pylori.
Southern blotting
Firstly, the sensitivity of Southern blotting, in which the RDF was used as both target and probe, was determined by using the ECL system. The results showed that the sensitive limitation of this system was 4 pg. Then, the probe was used to screen the RAPD products of different H pylori isolates. Parts of the results are shown in Figure 2. H pylori isolates from 27 of the 73 individuals were RDF positive, and the darkness of the blots was quite different from one isolate to another. As the size of the RAPD fragment hybridized with the RDF was bigger (about 400 bp) than that of the RDF, at least one primer binding site would be outside the RDF region.
Sequencing RDFs and their flanking regions
In order to know if there were sequence differences that might facilitate the binding of the random primer, the RDFs and their flanking regions from different H pylori isolates were sequenced. Firstly, the RDFs from 10 different H pylori isolates were cloned for sequencing. The isolates with designated numbers of 15, 22, 38, 58, 90 and 93 were RDF positive and the isolates 20, 41, 84 and 90 were RDF negative in Southern blotting. Blast analysis of the sequenced results exhibited that the RDFs from the 10 isolates were quite similar.
The rates of identity were 98% to 100%. There was no random primer binding siteinside the RDFs. Next, the RDF flanking regions were sequenced by using the PCR products directly. Four isolates were used. Sequences of the upstream regions or the downstream regions of RDFs were also quite similar (98% to 99% identity) between the RDF positive isolates (numbers 38 and 93) and the RDF negative isolates (numbers 84 and 90) in Southern blotting, and there was no random primer binding site inside the upstream regions or the downstream regions.
DISCUSSION
It is known that DNA fingerprints of clinical H pyori isolates are highly diversified[16,17] and the fingerprinting profiles are changeable even for the isolates from different parts of the stomach in a single host[33,34]. The results in this study were basically in according with those obtained in the previous studies. The gene diversity of H pylori is associated with certain diseases[20,35,36], and clinical background and geographic origin of the isolates may be responsible for the changes[37].
It is always considered that the gene of rRNA is relatively conservative, and two copies of 23S rDNA exist in H pylori genome[32,38]. Theoretically, when the RDF probe is used to screen the RAPD products of different clinical isolates, Southern blotting profiles should be the same. However, our results showed that the Southern blotting profiles of 23S rDNA from different H pylori isolates were also polymorphic. Since genomes of different H pylori isolates contain almost identical RDF as well as the flanking regions and the same amount of genomic DNA was used as the templates in this study, instability of using RAPD to amplify this gene fragment at the pg level might be responsible for the results.
The random primer was first proposed for fingerprinting of H pylori by Akopyanz et al[31]. They recommended the primer since it had a relative high CG content and could be used to distinguish different H pylori isolates. In fact, when one employs this primer, there will be numerous potentially amplifiable fragments and only a few of them are visible on agarose gels after EB staining. This depends on the matching extent of the primer to the genomic DNA. For the bright or visible bands, the primer may be completely or mostly matched, but for the light or invisible bands, the primer is probably partially matched.
In conventionally used PCR, the specificity depends mainly on the properties of the primers and the annealing temperature. However, in PCR-based RAPD, the random primer is only 10 mers and the annealing temperature is 36 °C. Therefore, specificity of amplification is relatively low. Since we could not find any primer binding site inside or outside the RDF region in this experiment, we propose that, for amplification of the RDF, the primer be partially matched to the genomic DNA, and thus, the profiles of RAPD and Southern blotting are diversified.
From our experience, RAPD depends highly on the quality and quantity of the template. If we make a comparison between two RAPD profiles of a given H pylori isolate, the concentration, purity and even the integrity of the DNA templates should be considered. The RAPD results will be quite different if the genomic DNA is prepared using two different methods. Sometimes different PCR machines might also give different results. So, when we employed this method, we had tried to treat every sample under the same conditions, and handled the DNA preparation carefully. Even so, RAPD seems to be unstable in amplification of low yield fragments, especially those that do not appear as visible binds in agarose gel electrophoresis.
RAPD has reasonable discriminatory power[39] and is effective for H pylori genotyping grossly on large scale. But stability of this method should be taken into consideration.
Although we found many strain specific RAPD profiles in this study, using the RAPD-based methods to screen H pylori strain specific genes are not recommended. To identify strain specific genes, the PCR sequencing techniques are more reliable and reproducible.