Published online Aug 15, 2003. doi: 10.3748/wjg.v9.i8.1844
Revised: January 15, 2003
Accepted: February 24, 2003
Published online: August 15, 2003
AIM: To establish transgenic mice expressing tamoxifen-inducible Cre-ERt recombinase specifically in the liver and to provide an efficient animal model for studying gene function in the liver and creating various mouse models mimicking human diseases.
METHODS: Alb-Cre-ERt transgenic mice were produced by microinjecting the construct with Cre-ERt fusion gene of DNA fragments into fertilized eggs derived from inbred C57BL/6 strain. Transgenic mice were identified by using PCR and Southern blotting. Expression of Cre-ERt fusion gene was analyzed in the liver, kidney, brain and lung from F1 generation transgenic mice at 8 weeks of age by reverse transcription (RT)-PCR.
RESULTS: Four hundred and fourteen fertilized eggs of C57 BL/6 mice were microinjected with recombinant Alb-Cre-ERt DNA fragments, and 312 survival eggs injected were transferred to the oviducts of 12 pseudopregnant recipient mice, 6 of 12 recipient mice became pregnant and gave birth to 44 offsprings. Of the 44 offsprings, two males and one female carried the hybrid Cre-ERt fusion gene. Three mice were determined as founders, and were back crossed to set up F1 generations with other inbred C57BL/6 mice. Transmission of Cre-ERt fusion gene in F1 offspring followed Mendelian rules. The expression of Cre-ERt mRNA was detected only in the liver of F1 offspring from two of three founder mice.
CONCLUSION: Transgenic mice expressing tamoxifen-inducible Cre-ERt recombinase under control of the liver-specific promoter are preliminary established.
- Citation: Zhu HZ, Chen JQ, Cheng GX, Xue JL. Generation and characterization of transgenic mice expressing tamoxifen-inducible cre-fusion protein specifically in mouse liver. World J Gastroenterol 2003; 9(8): 1844-1847
- URL: https://www.wjgnet.com/1007-9327/full/v9/i8/1844.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i8.1844
The liver plays a central role in regulation of carbohydrate, lipid, and urea metabolism as well as in production of most plasma proteins and detoxification of exogenous chemicals. The ability to create defined genetic modifications in vitro and in vivo provides a promising way to understand relevant functions of numerous hepatic genes in health and disease. Ligand-dependent chimeric Cre recombinases are powerful tools to induce specific DNA rearragements in cultured cells and mice[1-3]. To introduce defined genetic modifications in a temporally controlled manner in the liver, an alternative approach is to establish transgenic mice expressing tamoxifen-inducible Cre recombinase under control of the liver specific promoter. To this end, we constructed the fusion gene of Cre and the ligand binding domain (LBD) of a mutated human estrogen receptor (ER) that recognizes anti-estrogen 4-hydroxytamoxifen (4-OHT), Cre-ERt , under control of a liver-specific regulatory sequence from the mouse albumin gene. By using microinjection technology, two transgenic mice lines expressing Cre-ERt only in the liver were finally obtained.
Restriction enzymes and other reagents Enzymes EcoRI,
SmaI, SacII HindIII, BamHI, Bgl I, T4 DNA ligase, dNTP, Taq DNA polymerase, MMLV reverse transcriptase, etc, were purchased from NEB, Sangon and Sigma companies, respectively.
Mice C57BL/6, ICR mice (clean animal) were purchased from Laboratory Animal Center, Shanghai, China.
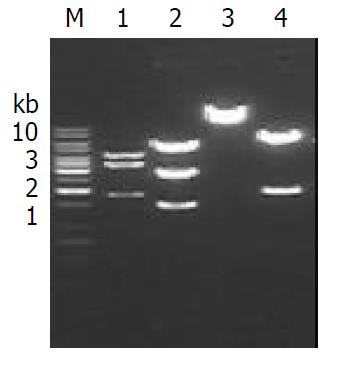
Construction of recombinant plasmid Construction of the Alb-Cre-ERt (Figure 1) was performed according to Sambrook et al[4]. In brief, a 0.6 Kb SmaI-SacII fragment containing CMV promoter from plasmid pCMV-Cre-ERt (kindly provided by Prof. Chambon P) was replaced with a 2.3 Kb SacII-EcoRV fragment containing albumin enhancer and promoter (alb e/p) from p2335A (kindly provided by Prof. Palmiter). Identity of the constructs was confirmed by restriction enzyme mapping and DNA sequence analysis.
Production of transgenic mice Transgenic mice were generated by standard methods[5]. In brief, the transgene construct was linearized using Bgl l. The fragment was gel-purified with QIAGEN Kit. After diluted to 1 μg/ml final con centration in microin jection buffer, DNA was microinjected into the male pronucleus of fertilized C57BL/ 6 mice oocytes. Then injected eggs were transplanted into the oviduct of pseudopregnant mice. Operated mice got pregnant and gave birth. Selected mice were bred with wild-type animals to establish transgenic lines that were maintained at heterozygosity.
Detection of transgenic integration Genomic DNA was extraction from transgenic mice tails (about 1-1.5 cm) at 4 weeks of age. Total DNA was extracted with the method described by Sambrook[4].
PCR assay The primers were designed on Cre gene sequence and synthesized by Cybersyn Biotechnology Company. The sequence of primers for PCR was as follows: the 5’ primer for Cre was 5’-ATCCGAAAAGAAAACGTTGA-3’ and the 3’ primer was 5’-ATCCAGGTTACGGATATAGT-3’. The PCR reaction condition contained 2 µl of 10×PCR Buffer, 0.4 µl of 25 mM Mg2+, 1 µl of 15 mM dNTP, 1 µl of 10 pmol each of primer, 2u of Taq DNA polymerase, 1 µg sample DNA. A 20 µl total reaction volume was obtained by adding sterile water, the reaction procedure of PCR was: beginning at 95 °C for 5 min, then 30 cycles at 94 °C denature for 30 s, annealing at 58 °C for 40 s and extension at 72 °C for 40 s, final extension at 72 °C for 10 min. 1.8% agarose gel was prepared and 10 µl PCR products was loaded in the gel, then electrophoresed at 80 voltage for 40 min, the result of electrophoresis was observed.
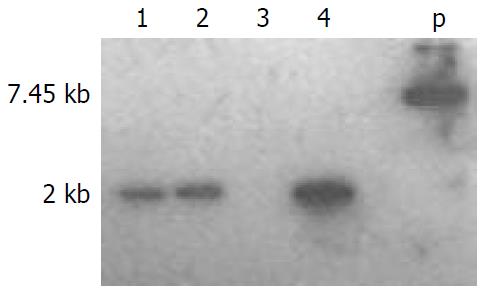
Southern blot analysis Southern-blotting analysis was performed with the following standard procedures[4]: 10 µg tail DNA was digested with EcoRI and separated by electrophoresis on a 0.6% agarose gel, and transferred onto a nylon membrane (Boehringer Mannheim). The nylon membrane was prehybridized for 5 h at 42 °C and hybridized overnight with a probe from a 0.7 kb BamHI-XhoI Cre fragment that was a random primer labeled using α-32P dCTP (Amersham Pharmacia Biotech Inc). Bands were detected by X-ray film after 6 days of exposure.
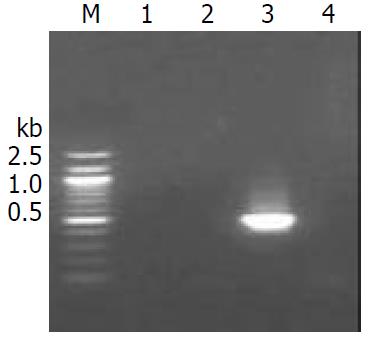
Detection of Cre-ERt mRNA synthesis in mice The level of Cre-ERt mRNA was estimated by RT-PCR. Total RNA was isolated from F1 generation of 4 weeks old mouse tissues (liver, kidney, brain and lung) by Sambrooks method[4]. The upstream primer was 5’-CAGAACCTGAAGATGTTCG-3’ and downstream was 5’ -GGATCATCAGCTACACCAG-3’, resulting in a 505 bp fragment of the Cre-ERt cDNA. First stranded cDNA was synthesized as follows: The reverse transcriptase reactions contained 1 µg RNA, 5 µl of 10×PCR Buffer, 3 µl of 15 mM dNTP, 1 µl of RNasin (80 u/µl), 2 µl 10 pmol upstream primer, 1 µl MMLV(200 u). A 50 µl total reaction volume was obtained by adding sterile DEPC treated water. Reaction conditions were at 48 °C for 45 min for reverse transcription. The products of reverse transcriptase reactions were used directly in PCR reactions, adding 2 u Taq DNA polymerase, 3 µl of 10 pmol upstream/downstream primers, respectively. PCR denaturation was at 96 °C for 2 min, then for 35 cycles of reaction (denaturation at 94 °C for 30 s, annealing at 62 °C for 40 s, extension at 72 °C for 40 s, and a final extension at 72 °C for 7 min).
The SmaI-SacII (blunted) fragment containing Cre-ERt of plasmid pCMV-Cre-ERt was isolated; the SacII (blunted)-EcoRV fragment containing albumin enhancer and promoter (alb e/p) of p2335A were isolated, respectively. The two individual fragments were ligated to generate palb-Cre-ERt. The identity of palb-Cre-ERt was confirmed by restriction enzyme mapping (Figure 2) and DNA sequence analysis (data not shown), indicating that the albumin enhancer and promoter were correctly inserted and linked.
Recombinant Cre-ERt DNA fragment was microinjected into the male pronucleus of the 414 fertilized eggs of C57 BL/6 mice, and 312 survival eggs injected were transferred to the oviducts of 12 pseudopregnant recipient mice, 6 of 12 recipient mice became pregnant and gave birth to 44 offspring mice. The survial rate and birth rate of zygotes were 75% (312/414) and 14% (44/312), respectively. DNA extracted from tails of 44 offspring mice was screened by PCR assay and Southern blot analysis (Figure 3). The results showed that three mice carried the Cre-ERt gene. Three mice(two male, one female) were determined as founders. Total integration rate and efficiency of transgene were 6% (3/44) and 0.7% (3/414), respectively.
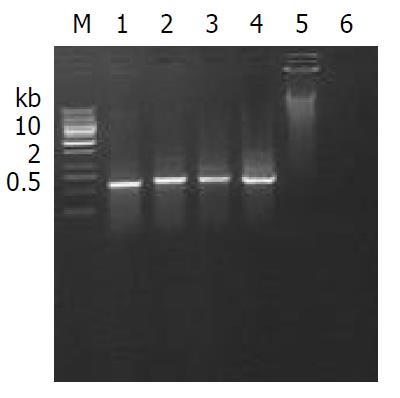
The founders were back crossed to set up F1 offsprings with other inbred C57BL/6 mice or transgenic littermates, respectively. All offsprings were screened by PCR (Figure 4), indicating that the integrated Cre-ERt gene was stably transmitted to subsequent generations (Table 1).
| Founder line | Sex | No. of littermates | No. of offsprings | No. of transgenic mice | Ratio (%) |
| 1 | M | 3 | 12 | 5 | 41.6 |
| 2 | M | 4 | 18 | 8 | 44.4 |
| 3 | F | 2 | 9 | 5 | 55.5 |
We performed RT-PCR to examine whether Cre-ERt transgene expressed only in liver tissues of F1 mice of transgenic mice lineages. The RT-PCR product (505 bp) was detected only in the liver of two of three transgenic mouse lineage (Figure 5).
By using transgenic mice carrying Cre recombinase under the control of a liver-specific promoter, the spatial window of gene modification can be strictly fixed to the liver. More recently, the generation and characterization of transgenic mice carrying Cre recombinase under the control of constitutive liver-specific promoters have been described[5-8]. These investigators used the constructs in which either mouse albumin regulation elements and α-fetoprotein enhancers or promoters and upstream enhancers of rat albumin gene were used to drive the Cre recombinase expression. These lines, AlfpCre, AlbCre, and Albumin-cre, allowed precise targeting of gene modifications in the liver. Nevertheless, the time of recombination remained strictly dependent on the promoter activity in these transgenic mice. In AlfpCre mice, Cre was shown to be constitutively active shortly after the appearance of the liver bud. Using the AlfpCre mice, mutagenesis of hepatic genes that serves essential functions during development could result in embryonic lethality, thus precluding analysis of their putative functions at subsequent stages. As for the temporal control of Cre expression in the liver, it was possible by using adenoviruses carrying the Cre transgene[9-11]. However, the immune response led to elimination of infected cells and could have deleterious effects on the general health status of mice[12]. An alternative approach for the temporal regulation of Cre activity is to create chimeric Cre fused to mutated ligand-binding domains of steroid hormone receptors which are insensitive to the endogenous ligands but still responsive to synthetic drugs. At present, tamoxifen-inducible Cre recombination can be achieved in many tissues of Cre-ERt transgenic adult mice and in developing mouse embryo as well as in cultured cells[13-18]. By combining the use of liver-specific promoters driving the expression of Cre and a ligand-activated Cre, gene modifications could be achieved in the liver at selected time points. Tannour-Louet et al[19] established transgenic mice expressing tamoxifen-inducible Cre-mer recombinase under the control of the transthyretin promoter (TTR-Cre ind), the result showed a high recombination efficiency in the fetal and adult liver after treatment with 4-OHT. Imai et al.[20] established transgenic mice expressing tamoxifen-inducible Cre-ERt recombinase under control of the human 1-antitrypsin promoter, the result showed a low recombination efficiency, making it useful only to produce genetic chimeras which are not suitable for hepatic gene inactivation. In this study, the promoter and upstream enhancer of the mouse albumin gene were used to drive tamoxifen-inducible Cre-ERt recombinase expression for producing transgenic mice. After injecting the construct into oocytes, two lines expressing Cre-ERt in the liver, but not in other tested organs, were finally obtained. The results demonstrated that the promoter and upstream enhancer of the mouse albumin gene were liver-specific expression.
When the transgene results from random integration of a DNA fragment injected into a fertilized egg‘s pronucleus, its level of expression in the resulting transgenic animal varied and depended upon certain factors, such as integration site, transgene cope number, genetic background of mouse, promoter/enhancer of fusion gene, etc. In this study, we only got two transgenic mice lines expressing Cre-ERt in the liver from three founder mice. The result was consistent with that of Imai et al[20] in which three of 20 transgenic founder animals expressed Cre-ERt in the liver. These results indicate that not every transgenic founder mouse expresses Cre recombinase effectively, the chromosomal integration site may affect the expression pattern of transgenes.
Taken together, we have produced transgenic mice expressing Cre-ERt gene specific in the liver. This work will certainly permit a better understanding of hepatic gene functions and establish various mouse models mimicking human diseases.
We thank Prof. Chambon P of Universite Louis Pasteur for providing plasmid pCMV Cre-ERt, Prof. Palmiter of University of Washington for plasmid p2335A, Prof. Orkin SH of Howard Hughes Medical Institute for Rosa26 reporter mice. This work was also supported by the Postdoctoral Foundation of China.
Edited by Xu XQ and Wang XL
| 1. | Rossant J, McMahon A. "Cre"-ating mouse mutants-a meeting review on conditional mouse genetics. Genes Dev. 1999;13:142-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 88] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 2. | Le Y, Sauer B. Conditional gene knockout using Cre recombinase. Mol Biotechnol. 2001;17:269-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 3. | Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 286] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 4. | Sambrook J, Fritsch EF, Maniatis T. "Molecular Cloning: A Labo-ratory Manual," 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. 1989;9.31-9.61. |
| 5. | Hogan B, Costantini f, Lacy E. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbour, Cold Spring Harbor Laboratory, New York. 1980;116-188. |
| 6. | Kellendonk C, Opherk C, Anlag K, Schütz G, Tronche F. Hepatocyte-specific expression of Cre recombinase. Genesis. 2000;26:151-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 7. | Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 8. | Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA. 1999;96:7324-7329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1007] [Cited by in RCA: 997] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 9. | Wang Y, Krushel LA, Edelman GM. Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc Natl Acad Sci USA. 1996;93:3932-3936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 121] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 10. | Rohlmann A, Gotthardt M, Willnow TE, Hammer RE, Herz J. Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nat Biotechnol. 1996;14:1562-1565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 112] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 11. | Okuyama T, Fujino M, Li XK, Funeshima N, Kosuga M, Saito I, Suzuki S, Yamada M. Efficient Fas-ligand gene expression in rodent liver after intravenous injection of a recombinant adenovirus by the use of a Cre-mediated switching system. Gene Ther. 1998;5:1047-1053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Akagi K, Sandig V, Vooijs M, Van der Valk M, Giovannini M, Strauss M, Berns A. Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res. 1997;25:1766-1773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 196] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 13. | Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1033] [Cited by in RCA: 1092] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 14. | Zheng B, Zhang Z, Black CM, de Crombrugghe B, Denton CP. Ligand-dependent genetic recombination in fibroblasts : a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol. 2002;160:1609-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 180] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 15. | Vallier L, Mancip J, Markossian S, Lukaszewicz A, Dehay C, Metzger D, Chambon P, Samarut J, Savatier P. An efficient system for conditional gene expression in embryonic stem cells and in their in vitro and in vivo differentiated derivatives. Proc Natl Acad Sci USA. 2001;98:2467-2472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 36] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch JL, Chambon P, Metzger D. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc Natl Acad Sci USA. 1997;94:14559-14563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 182] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 17. | Li M, Indra AK, Warot X, Brocard J, Messaddeq N, Kato S, Metzger D, Chambon P. Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature. 2000;407:633-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 246] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 18. | Utomo AR, Nikitin AY, Lee WH. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat Biotechnol. 1999;17:1091-1096. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 156] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 19. | Tannour-Louet M, Porteu A, Vaulont S, Kahn A, Vasseur-Cognet M. A tamoxifen-inducible chimeric Cre recombinase specifically effective in the fetal and adult mouse liver. Hepatology. 2002;35:1072-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 2.4] [Reference Citation Analysis (0)] |