Published online Aug 15, 2003. doi: 10.3748/wjg.v9.i8.1743
Revised: April 24, 2003
Accepted: May 17, 2003
Published online: August 15, 2003
AIM: Point mutation, one of the commonest gene mutations, is the most important molecular pathogenesis of cancer and chronic infection. The commonest methods for detection of point mutation are based on polymerase chain reaction (PCR). These techniques, however, cannot be used in large scale screening since they are neither accurate nor simple. For this reason, this study established a novel method of competitively differentiated PCR (CD-PCR) for screening point mutation in clinical practice.
METHODS: Two competitively differentiated primers for mutant-type and wild-type templates respectively with an identically complemented region in 3’ end except for last 2 base pairs and a different non-complemented region in 5’ end were designed. Thus, competitive amplification might be carried out at a lower annealing temperature at first, and then differentiated amplification at a higher annealing temperature when primers could not combine with initial templates. The amplification was performed in one-tube. The products of CD-PCR were detected using microplate hybridization assay. CD-PCR was evaluated by detecting G1896A variant of hepatitis B virus (HBV) in form of recombinant plasmids and in sera from patients with hepatitis B, and compared with allele-specific PCR (AS-PCR) and competitive AS-PCR.
RESULTS: CD-PCR was successfully established. It could clearly distinguish wild-type and mutant-type plasmid DNA of G1896A variant when the amount of plasmid DNA was between 102-108copies/reaction, while for AS-PCR and competitive AS-PCR, the DNA amount was between 102-104copies/reaction. CD-PCR could detect one copy of G1896A variant among 10-100 copies of wild-type plasmid DNA. The specificity of CD-PCR was higher than those of AS-PCR and competitive AS-PCR in the detection of HBV G1896A variant in sera from patients with hepatitis B. CD-PCR was independent of the amount of HBV DNA in serum. HBV G1896A variant was more often found in HBeAg (-) patients with a lower level of detectable viremia than that with a higher level of detectable viremia (P = 0.0192).
CONCLUSION: CD-PCR is more specific since it is less influenced by the amount of initial templates and the cross amplification between mutant- and wild-type amplified products. It is also simple and time-saving. Thus, CD-PCR might be useful in routine gene typing and point mutation screening. HBV G1896A or other more important mutations have to be routinely detected in patients with a detectable level of viremia after HBeAg/antibody conversion in clinical practice.
- Citation: Peng XM, Chen XJ, Li JG, Gu L, Huang YS, Gao ZL. Novel assay of competitively differentiated polymerase chain reaction for screening point mutation of hepatitis B virus. World J Gastroenterol 2003; 9(8): 1743-1746
- URL: https://www.wjgnet.com/1007-9327/full/v9/i8/1743.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i8.1743
Point mutation is one of the commonest gene mutations not only in human genome but also in genome of pathogens. It is the most important molecular pathogenesis of cancer and chronic infection[1-3]. The commonest method for detecting point mutation is based on polymerase chain reaction (PCR), which can be divided into two types. One type is to analyze the PCR products using direct sequencing, restriction fragment length polymorphism and single strand conformation polymorphism[4-7]. This type of technique is usually accurate, but cannot be used in routine clinical examination since it needs additional manipulation. The other type is allele specific-PCR (AS-PCR), sequence specific-PCR or amplification refractory mutation system[8-13]. This type of techniques is usually simple, but its specificity is influenced by the amount of initial templates of PCR. This deficiency was not so obvious when this type of methods was used to detect point mutation or gene typing of human gene in the past since the initial templates in these samples could be controlled[8,12,13]. Some investigators, however, have made efforts to improve AS-PCR by introducing competitive mechanism, and established the so-called bidirectional AS-PCR and competitive PCR in order to decrease the influence of the amount of initial templates on its specificity[14-19]. AS-PCR has been used to detect point mutation of pathogens recently[10,20-22]. However, the specificity of AS-PCR in detection of pathogen mutations was challenged by uncontrollable amount of initial templates and the requisition of high sensitivity. For these reasons, a novel method of competitively differentiated polymerase chain reaction (CD-PCR) was established in this study and compared with AS-PCR and competitive AS-PCR in the detection of G1896A variant of HBV.
One hundred serum samples with (hepatitis B surface antigen) HBsAg(+), anti-(hepatitis B e antigen) HBe(+) and anti-(hepatitis B core antigen) HBc(+), 60 serum samples with HBsAg(+), HBeAg(+) and anti-HBc(+) and 40 serum samples without HBV serum markers were collected. The serum markers were demonstrated by enzyme-linked immunoabsorbent assay. Recombinant plasmid pG1896A was constructed as described before[23]. TZ19U-HBV that contained double copies of HBV DNA (adw) was a gift from Professor Huang Zhimin (Zhongshan University, Guangzhou, China) and was used as wild-type DNA control. T4 DNA ligase and pfu DNA polymerase were purchased from Promega (USA). DNA gel extraction kits and plasmid isolation kits were purchased from Qiagen (Germang). Anti-digoxigenin and anti-fluorescein labeled with horseradish peroxidase were purchased from Roche (USA). Primers shown in Table 1 were designed with the Omega 2 software and synthesized in Bioasia Biological Engineering Company (Shanghai, China).
| Denomination | Sequence (5’→3’) |
| BIO-PCP | BIO-GAGACTCTAAGGCTTCTCGATACAGAGCTGAGG |
| PCA | GCAGTATGGTGAGGTGAGCAATGCTCAG |
| DIG-PCMd | DIG-CTCACGCTACATTGTGTGCCTTGGGTGGCTTCA |
| DIG-PCMc | DIG-TGTGCCTTGGGTGGCTTCA |
| FLU-PCWd | FLU-GTCCGTAGTCTCGTTGTGCCTTGGGTGGCTTGG |
| FLU-PCWc | FLU-TGTGCCTTGGGTGGCTTGG |
| PCS | CCACCGTGAACGCCCATCAG |
| PCSc | CCCGAATTCCACCGTGAACGCCCATCAG |
| PCAc | CCCAAGCTTGCAGTATGGTGAGGTGAGCAATG |
Principles of CD-PCR Two competitively differentiated primers (CDP) with different labels in 5’ end were designed. CDP had a complemented region of 17-20 bp long with penultimate mismatch and a 3’ terminus matching the mutant or the normal bases of the templates. The annealing temperature of this part was about 52-54 °C. There was also a non-complemented region of 14 bp long, which was different from each other in 5’ end. The total annealing temperature of CDP was about 75-80 °C. PCR was performed in one tube. Competitive amplification was performed first when CDP competed for templates under lower annealing temperature. After certain amounts of products of each primer were obtained, differentiated amplification was then performed under higher annealing temperature when CDP could only use its own products as templates. This amplification model would keep the products ratio of mutant and wild templates and allow the templates to be fully amplified.
G1896A variant detected by CD-PCR Wild-type and G1896A mutant-type plasmids were used for the optimization of CD-PCR. A total volume of 30 μl was used in the final conditions of PCR reaction. The reaction mixture consisted of 10 mmol/L Tris-HCl, pH 8.5,50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.2 mmol/L dNTPs, 2U pfu DNA polymerase, 20 pmol FLU-PCWd, 20 pmol DIG-PCMd, 20 pmol PCA and 5 μl plasmid or extracted DNA. The cycling conditions were as follows: 3 cycles (first set) at 94 °C for 40 s, at 52 °C for 40 s and at 72 °C for 90 s followed by 35 cycles (second set) at 94 °C for 40 s, and at 72 °C for 90 s. The PCR products were then hybridized with solidified biotin-labeled probe PCP in two wells of microplate. The color reaction was obtained after captured PCR products reacted with anti-DIG or anti-FLU respectively, which were conjugated with horseradish peroxidase.
G1896A variant detected by competitive AS-PCR Except for the replacement of primer FLU-PCWd and DIG-PCMd with primer FLU-PCWc and DIG-PCMc, the components of competitive AS-PCR were similar to CD-PCR. The cycling conditions were as follows: 25 cycles (first set) at 94 °C for 40 s, at 60 °C for 40 s, and at 72 °C for 90 s followed by 10 cycles (second set) at 94 °C for 40 s, at 52 °C for 40 s, and at 72 °C for 90 s. The PCR products were then detected similar to CD-PCR.
G1896A variant detected by AS-PCR Except for the absence of FLU-PCWc, the components and the cycling conditions were similar to competitive CD-PCR. The PCR products were visualized under an ultraviolet transilluminator.
Quantification of serum HBV DNA by fluorescent quantitive PCR The HBV DNA level of 200 sera was detected using fluorescent quantitive PCR following the instruction (Taqmen, Roche).
DNA sequencing A fragment of HBV precore in positive or negative sera by all three methods and in sera that were positive for AS-PCR and competitive AS-PCR, and negative for CD-PCR was analyzed using DNA sequencing after it was amplified using primers PCSc and PCAc and cloned into plasmid pUC19.
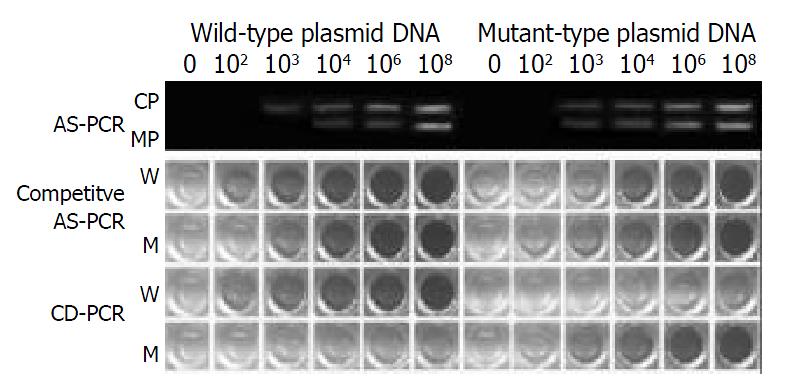
The typical results of HBV G1896A variant detected by means of AS-PCR, competitive AS-PCR and CD-PCR are shown in Figure 1. To detect G1896A variant, the sensitivity of AS-PCR was 103 copies/reaction, and there was non-specific amplification when the amount of wild-type initial templates was higher than 104 copies/reaction. For competitive AS-PCR, the wild-type and mutant-type plasmid DNA could be clearly distinguished when the initial templates were 102-104 copies/ reaction. CD-PCR could clearly distinguish them in the range of 102-108 copies/reaction. There was some degree of non-specific reaction when the amount of initial templates was higher than 104 copies/reaction, but the difference of color intensities between wild-type and mutant-type product wells was obvious. CD-PCR could detect one variant G1896A copy among 100 wild-type copies of DNA when the initial templates were 102-105 copies/reaction, but one out of 10 copies when the initial templates were 105-108 copies/reaction.
Among 200 sera, HBV DNA was positive in 104 cases using AS-PCR, in 119 cases using competitive AS-PCR and in 122 cases using CD-PCR. The sensitivity for HBV DNA detection by CD-PCR was similar to that of competitive AS-PCR, but higher than that of AS-PCR (χ2 = 5.47, P = 0.0193). The results of G1896A variant in sera are shown in Table 2. Among the 104 cases with positive HBV DNA detected by all the three methods, there were 33 positive cases, and 31 negative cases of G1896A variant detected by both CD-PCR and AS-PCR, and 40 cases that were positive by AS-PCR but not by CD-PCR.
HBV G1896A mutation was confirmed by DNA sequencing in 3 positive sera by all the three methods, and excluded in 3 negative sera by all the three methods and in 4 sera that were positive by AS-PCR and competitive AS-PCR, but negative by CD-PCR.
The relationship between G1896A variant detected by AS-PCR, competitive AS-PCR and CD-PCR and HBV DNA level in the sera is shown in Table 3. More G1896A variants were found in patients with positive HBeAg by AS-PCR and competitive AS-CR. These could be false positives. These false-positive findings occurred more often in patients with a higher level of HBV DNA in serum. CD-PCR seemed more specific since only 4 patients were found to be infected with both wild-type HBV and G1896A variants in HBeAg positive cases. By CD-PCR, HBV G1896A variant was more often found in HBeAg (-) patients with a lower level of viremia (P = 0.0192).
AS-PCR has been considered as a simplest and quickest method for the detection of known point mutations[5,10-19]. False positive results, however, seem inevitable since the amplification of wild-type templates by allele-specific or sequence-specific primers for mutant-type templates can not be completely prevented. Once non-specific products occur, they will be amplified subsequently. Thus the ratio of specific and non-specific products will decrease, and become even smaller as the plateau of PCR reaches at the late stage. It is understandable that false band will occur when a large amount of initial templates or too many cycles are used in AS-PCR. To improve the specificity of AS-PCR, the specificity of primers was designed by introducing penultimate mismatch[24], and the cycles of specific amplification were reduced by introducing basic amplification[6,25]. The application of competitive mechanism is another important step to improve AS-PCR.
Competitive AS-PCR and bidirectional or tetra-primer AS-PCR are common assays that have a competitive mechanism[6,14-19,25]. Bidirectional or tetra-primer AS-PCR is in fact a competition for PCR system, while competitive AS-PCR is the competition for both templates and PCR system. However, these PCRs can not exclude the non-specific amplification from initial templates in each cycle. For competitive AS-PCR, it is possible to reduce the influence of initial templates, but a new problem will occur, which is that the initial products will become the templates for non-specific amplification in subsequent cycles. This might be the underlying mechanism that the competitive AS-PCR seemed worse than AS-PCR in this study. Therefore, to reduce the influence of the initial templates and products would be a very important measure to improve AS-PCR assay on specificity of amplification
For CD-PCR, competitive amplification was performed first, and followed by differentiated amplification, which was different from competitive AS-PCR. This protocol would allow allele- or sequence-specific amplification to begin at a low level of templates. Thus, the influence of initial templates could be controlled to a minimal level. After a few cycles, differentiated amplification was carried out under higher annealing temperature at which primers could not combine with initiated templates and the initial products could not be used as templates for non-specific amplification in subsequent cycles under higher annealing temperature either. These might be the mechanisms that CD-PCR had much better performance than that of AS-PCR and competitive AS-PCR in this study. The molecular weights of wild-type and mutant-type products were the same, therefore, products of CD-PCR could not be distinguished by electrophoresis. It was convenient and time-saving when microplate hybridization assay was used to demonstrate the products in this study. Automatic detection was possible too when CD-PCR was adapted to real-time fluorescent PCR assay as used in AS-PCR or competitive AS-CR[26,27]. CD-PCR could also be used by combining with other high resolution techniques[28,29]. It could be used in gene typing for organ transplantation or gene polymorphism in addition to the detection of gene point mutations as AS-PCR[30,31].
Detectable viremia was found in sera of some patients with hepatitis B after HBeAg/antibody conversion[32-34]. This type of viremia resulted in G1896A mutation in 60.4% out of such patients, especially in patients with a much lower level of HBV DNA. This result is conformable to the fact that the replication ability of G1896A variant has decreased[35], and suggests that G1896A or other more important mutations have to be detected in patients with a detectable level of viremia after HBeAg/ antibody conversion in clinical practice.
Edited by Yuan HT and Wang XL
| 1. | Bressac B, Kew M, Wands J, Ozturk M. Selective G to T mutations of p53 gene in hepatocellular carcinoma from southern Africa. Nature. 1991;350:429-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 972] [Cited by in RCA: 872] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 2. | Steinberg JL, Yeo W, Zhong S, Chan JY, Tam JS, Chan PK, Leung NW, Johnson PJ. Hepatitis B virus reactivation in patients undergoing cytotoxic chemotherapy for solid tumours: precore/core mutations may play an important role. J Med Virol. 2000;60:249-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 3. | Yoo BC, Park JW, Kim HJ, Lee DH, Cha YJ, Park SM. Precore and core promoter mutations of hepatitis B virus and hepatitis B e antigen-negative chronic hepatitis B in Korea. J Hepatol. 2003;38:98-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 78] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 4. | Chen RY, Edwards R, Shaw T, Colledge D, Delaney WE, Isom H, Bowden S, Desmond P, Locarnini SA. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology. 2003;37:27-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 96] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Peng XM, Peng WW, Yao JL. Codon 249 mutations of p53 gene in development of hepatocellular carcinoma. World J Gastroenterol. 1998;4:125-127. [PubMed] |
| 6. | Peng XM, Yao CL, Chen XJ, Peng WW, Gao ZL. Codon 249 mutations of p53 gene in non-neoplastic liver tissues. World J Gastroenterol. 1999;5:324-326. [PubMed] |
| 7. | Ding Y, Le XP, Zhang QX, Du P. Methylation and mutation analysis of p16 gene in gastric cancer. World J Gastroenterol. 2003;9:423-426. [PubMed] |
| 8. | Suzuki F, Suzuki Y, Tsubota A, Akuta N, Someya T, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. Mutations of polymerase, precore and core promoter gene in hepatitis B virus during 5-year lamivudine therapy. J Hepatol. 2002;37:824-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Moses JH, Greville WD, Downes J, McClenahan W, Kennedy A, Dunckley H. A new HLA-A*02 allele, A*0234, detected by polymerase chain reaction using sequence-specific primers (PCR-SSP). Tissue Antigens. 2000;55:175-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 10. | Ma CL, Fang DX, Chen HB, Li FQ, Jin HY, Li SQ, Tan WG. A mutation specific polymerase chain reaction for detecting hepatitis B virus genome mutations at nt551. World J Gastroenterol. 2003;9:509-512. [PubMed] |
| 11. | Hodgson DR, Foy CA, Partridge M, Pateromichelakis S, Gibson NJ. Development of a facile fluorescent assay for the detection of 80 mutations within the p53 gene. Mol Med. 2002;8:227-237. [PubMed] |
| 12. | Stoehr R, Knuechel R, Boecker J, Blaszyk H, Schmitt R, Filbeck T, Hofstaedter F, Hartmann A. Histologic-genetic mapping by allele-specific PCR reveals intraurothelial spread of p53 mutant tumor clones. Lab Invest. 2002;82:1553-1561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Kirby GM, Batist G, Fotouhi-Ardakani N, Nakazawa H, Yamasaki H, Kew M, Cameron RG, Alaoui-Jamali MA. Allele-specific PCR analysis of p53 codon 249 AGT transversion in liver tissues from patients with viral hepatitis. Int J Cancer. 1996;68:21-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 14. | Waterfall CM, Eisenthal R, Cobb BD. Kinetic characterisation of primer mismatches in allele-specific PCR: a quantitative assessment. Biochem Biophys Res Commun. 2002;299:715-722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 15. | Horvath AD, Kirov SA, Karaulanov EE, Ganev VS. Detection of apoB-100 R3500Q mutation by competitive allele-specific polymerase chain reaction. J Clin Lab Anal. 2001;15:256-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 16. | Sasvari-Szekely M, Gerstner A, Ronai Z, Staub M, Guttman A. Rapid genotyping of factor V Leiden mutation using single-tube bidirectional allele-specific amplification and automated ultrathin-layer agarose gel electrophoresis. Electrophoresis. 2000;21:816-821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 17. | Hamajima N, Saito T, Matsuo K, Kozaki K, Takahashi T, Tajima K. Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Jpn J Cancer Res. 2000;91:865-868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 164] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 18. | Hamajima N, Saito T, Matsuo K, Tajima K. Competitive amplification and unspecific amplification in polymerase chain reaction with confronting two-pair primers. J Mol Diagn. 2002;4:103-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | McKinzie PB, Parsons BL. Detection of rare K-ras codon 12 mutations using allele-specific competitive blocker PCR. Mutat Res. 2002;517:209-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Gramegna M, Lampertico P, Lobbiani A, Colucci G. Detection of the hepatitis B virus major pre-core mutation by the amplification refractory mutation system technique. Res Virol. 1993;144:307-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 21. | Nainan OV, Khristova ML, Byun K, Xia G, Taylor PE, Stevens CE, Margolis HS. Genetic variation of hepatitis B surface antigen coding region among infants with chronic hepatitis B virus infection. J Med Virol. 2002;68:319-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 22. | Kinoshita M, Seno T, Fukui T, Shin S, Tsubota A, Kumada H. A detection method for point mutation in the precore region of human hepatitis B virus (HBV)-DNA using mutation-site-specific assay. Clin Chim Acta. 1994;228:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 23. | Peng XM, Gu L, Chen XJ, Huang YS, Gao ZL. The Defect and Improvement of Allele-specific PCR during Detection of G1896A Variant of Hepatitis B Virus. J Practical Med. 2003;19:467-469. |
| 24. | Fishbein WN, Davis JI, Foellmer JW, Nieves S, Merezhinskaya N. A Competitive Allele-specific Oligomers Polymerase Chain Reaction Assay for the cis Double Mutation in AMPD1 That Is the Major Cause of Myo-adenylate Deaminase Deficiency. Mol Diagn. 1997;2:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 25. | Hersberger M, Marti-Jaun J, Hänseler E, Speck RF. Rapid detection of the CCR2-V64I, CCR5-A59029G and SDF1-G801A polymorphisms by tetra-primer PCR. Clin Biochem. 2002;35:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Glaab WE, Skopek TR. A novel assay for allelic discrimination that combines the fluorogenic 5' nuclease polymerase chain reaction (TaqMan) and mismatch amplification mutation assay. Mutat Res. 1999;430:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Suemizu H, Ohnishi Y, Maruyama C, Tamaoki N. Two-color allele-specific polymerase chain reaction (PCR-SSP) assay of the leptin receptor gene (Leprdb) for genotyping mouse diabetes mutation. Exp Anim. 2001;50:435-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 28. | Tian H, Brody LC, Fan S, Huang Z, Landers JP. Capillary and microchip electrophoresis for rapid detection of known mutations by combining allele-specific DNA amplification with heteroduplex analysis. Clin Chem. 2001;47:173-185. [PubMed] |
| 29. | McClay JL, Sugden K, Koch HG, Higuchi S, Craig IW. High-throughput single-nucleotide polymorphism genotyping by fluorescent competitive allele-specific polymerase chain reaction (SNiPTag). Anal Biochem. 2002;301:200-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Donohoe GG, Laaksonen M, Pulkki K, Rönnemaa T, Kairisto V. Rapid single-tube screening of the C282Y hemochromatosis mutation by real-time multiplex allele-specific PCR without fluorescent probes. Clin Chem. 2000;46:1540-1547. [PubMed] |
| 31. | See D, Kanazin V, Talbert H, Blake T. Electrophoretic detection of single-nucleotide polymorphisms. Biotechniques. 2000;28:710-714, 716. [PubMed] |
| 32. | Seo Y, Yoon S, Nakaji M, Yano Y, Nagano H, Ninomiya T, Hayashi Y, Kasuga M. Hepatitis B virus DNA in anti-HBe-positive asymptomatic carriers. Intervirology. 2003;46:43-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 33. | Yotsuyanagi H, Hino K, Tomita E, Toyoda J, Yasuda K, Iino S. Precore and core promoter mutations, hepatitis B virus DNA levels and progressive liver injury in chronic hepatitis B. J Hepatol. 2002;37:355-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 34. | Yuen MF, Sablon E, Yuan HJ, Hui CK, Wong DK, Doutreloigne J, Wong BC, Chan AO, Lai CL. Relationship between the development of precore and core promoter mutations and hepatitis B e antigen seroconversion in patients with chronic hepatitis B virus. J Infect Dis. 2002;186:1335-1338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 69] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Karino Y, Toyota J, Sato T, Ohmura T, Yamazaki K, Suga T, Nakamura K, Sugawara M, Matsushima T, Hino K. Early mutation of precore (A1896) region prior to core promoter region mutation leads to decrease of HBV replication and remission of hepatic inflammation. Dig Dis Sci. 2000;45:2207-2213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |