INTRODUCTION
Gastric cancer (GC) is a leading cause of carcinoma morbidity and mortality in China[1]. Meanwhile, the occurrence of colorectal cancer (CRC) has been rising in the past 3 decades. At present, CRC is the fourth cause of death in China[2,3]. It is well known that a multistep process associated with multiple gene abnormalities including inactivation of tumor suppressor genes and activation of oncogenes may be related to the occurrence of these diseases, but the molecular basis of these diseases is still not well understood[4-11]. Recently modern molecular genetic analyses have clarified that multiple chromosome alterations are frequently found in GC and CRC[12-16]. Nonrandom and significant losses of heterozygosity (LOH) at 9p, 3p and 6q have been detected to be more significant by cytogenetic studies, indicating that there may be more than one tumor suppressor gene (TSG) associated with GC and CRC located in these regions[17-24]. Unfortunately, homozygous deletions and point mutations of the known tumor suppressor genes such as p16/MTS1 gene (9p21),VHL gene (3p25-26), PPARγ gene (3p24.2-25) and FHIT gene (3p14.2) have seldom been observed in GC and CRC. These results suggest that these genes might not play important roles in gastric and colorectal tumorigenesis, and several other tumor suppressor genes in these region may contribute to the occurrence and development of GC and CRC[25-33]. NGX6, NAG-7 and BRD7 genes located at 9p21-22, 3p25 and 6q22.1-6q22.33, respectively, are novel tumor related genes cloned by positional candidate cloning strategy. In previous studies, all of them were found to be potential tumor suppressor genes associated with nasopharyngeal carcinoma (NPC)[34-39]. To investigate whether NGX6, NAG-7 and BRD7 genes also play a role in the pathogenesis of gastric and colorectal carcinoma, we analyzed the expression levels of NGX6, NAG-7 and BRD7 genes in 34 human gastric carcinoma tissues, 34 human colorectal carcinoma tissues and their corresponding normal tissues by RT-PCR, dot hybridization and Northern blot analysis.
MATERIALS AND METHODS
Tumor specimens
Fresh surgical specimens of thirty-four gastric carcinomas (GC), thirty-four colorectal carcinomas (CRC) and their corresponding normal tissues were obtained from the Xiang ya Hospital Affiliated to Central South University from January 2000 to July 2000. All tumor specimens were confirmed by pathological diagnosis. Each freshly resected specimen was immediately stored in liquid nitrogen until analysis. Histologically, in the 34 cases of gastric carcinoma, 4 were well-differentiated adenocarcinomas, 22 poorly-differentiated adenocarcinomas, 6 signet ring cell carcinomas and 2 mucoid carcinomas. There were 18 males and 16 females, their age ranged from 30 to 68 years (mean age, 51.7 years). Six cases had lymph node and/or distance metastases. In the 34 cases of colorectal carcinoma, 6 were poorly-differentiated adenocarcinomas, 27 well-differentiated adenocarcinomas and 1 signet ring cell carcinoma. There were 20 males and 14 females, their age ranged from 17 to 72 years (mean age, 47.4 years). Sixteen cases had lymph node and/or distance metastases. No patient had received chemotherapy or radiation therapy before surgery.
RT-PCR
Total RNA was isolated using Trizol reagent (Gibco-BRL, Gaithersburg, MD, USA) according to the protocol provided by the manufacturer. After being treated with DNase-I (Promega), 1-2 μg of total RNA was reversely transcribed into complementary DNA (cDNA) with oligo (dT) using cDNA synthesis kit (Promega). Then 1 μl cDNA product was used as the template to amplify specific fragments in a 25 μl reaction mixture. The PCRs were performed using Taq polymerase and the buffer (Promega) supplied with 0.2 mM dNTPs and 0.2 μM primers. RT-PCR reaction was carried out with an initial denaturation at 95 °C for 5 min, followed by 35 cycles at 94 °C for 50 s, annealing temperature (56 °C) for 50 s, at 72 °C for 60 s, and final extension at 72 °C for 10 min. At the same time, a housekeeping gene, GAPDH1 or GAPDH2 was amplified as an internal control to normalize the relative levels of cDNA, in which primers generated a PCR product of 475 bp or 760 bp. An aliquot (10 μl) of each reaction products was analyzed by 1.0% agarose gel electrophoresis.
Primers corresponding to NGX6, NAG-7 and BRD7 sequences were designed with WWW Primer Picking (Primer 3) and synthesized by TaKaRa. Gene-specific forward and reverse primers for NGX6, NAG-7 and BRD7 genes were designed to produce PCR products of 780 bp, 466 bp and 270 bp, respectively. Primer sequences were as follows: NGX6F1, 5’-GAACGTGGTGGAAATACAGA-3’; NGX6R1, 5’-TTCTACATCTTCTTTGGCCC-3’; NAG-7F1, 5’-ACATCAGCTTGGAGTTATTGA-3’; NAG-7R1,5’-GAAATGTACCACCCTACA-3’; BRD7F1, 5’-TGGAAGCCTCTCACAAGCT-3’; BRD7R1,5’-TGTGTACTAATGCCATGAT-3’; GAPDH1F1, 5’-GTCATCCATGACAACTTTGGTATC-3’; GAPDH1R1, 5’-CTGTAGCCAAATTCGTTGTCATAC-3’; GAPDH2F1, 5’-CGAGATCCCTCCAAAATCAA-3’; GAPDH2R1, 5’-TGCTGTAGCCAAATTCGTTG-3’.
Northern blot analysis
Total RNA was isolated from human gastric and colorectal carcinomas and their corresponding normal tissues by Trizol reagent (Gibco-BRL), and transferred to nylon membrane according to the standard procedure. Hybridization was performed as previously described. 30 μg RNA was separated by electrophoresis through denaturing agarose gels, and blotted onto the nylon membranes (Clontech). RNA was permanently attached to the membrane by UV illumination for 150 s (GS Gene Linker, Bio-Rad, USA), and the membranes were dried in a vacuum at 80 °C for 2 h and sealed in a plastic bag for use. The hybridization probes were obtained by RT-PCR amplification. NGX6, NAG-7 and BRD7 cDNA probes were random-prime labeled with [α-32P]dCTP using primer-a-gene random labeling kit (Promega, USA) following the protocol. Hybridization with RNA blots was carried out at 68 °C overnight in Express Hyb TM hybridization solution (Clontech) in rolling bottles. The membranes were washed twice at room temperature in 2×saline sodium citrate (SSC), 0.05% SDS for 10 min, once at 42 °C in 1×SSC, 0.1% SDS for 15 min and once at 50 °C in 0.1×SSC, 0.1% (w/v) SDS for 30 min, exposure to film (Eastman Kodak, Rochester, NY, USA) for 4 d at -70 °C. After exposure, the blot was again hybridized with a GAPDH probe.
Dot blot analysis
GAPDH and NGX6, NAG-7 and BRD7 cDNA fragments containing open reading frame from cDNA of gastric or colorectal carcinoma samples were obtained by RT-PCR. These cDNAs were reclaimed and purified by using a Kit according to the instruction of its manufacturer (Shanghai Huashun Co.). After alkali dissolution, GAPDH and NGX6, NAG-7 and BRD7 cDNA were blotted onto nylon membranes. cDNA permanently was attached to the membrane by UV illumination for 150 s, and the membranes were dried in a vacuum at 80 °C for 2 h to fix the cDNA. 10 μg total RNA was isolated from 10 cases of human gastric carcinomas, 10 cases of colorectal carcinomas and 10 cases of each corresponding normal tissues respectively, and was reversely-transcribed into cDNA probes with oligo (dT) and [α-32P] dCTP using cDNA synthesis kit after treated with DNase I and RNasin at 37 °C for 1 h to remove contaminated DNA (1 μg total RNA of each case was used). Then the four cDNA probes were hybridized with GAPDH and NGX6, NAG-7 and BRD7 cDNA blots respectively as Northern hybridization described above.
Statistical analysis
Chi-square test was used for the comparison between two groups. A P value less than 0.05 was considered statistically significant.
RESULTS
Expression of NGX6, NAG-7 and BRD7 genes in gastric and colorectal cancer tissues by RT-PCR
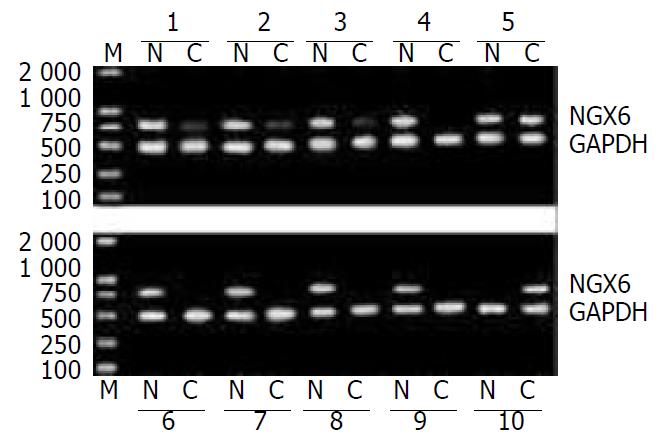
In 34 pairs of CRC and corresponding normal tissues, 25 (73.5%) showed decreased or absent expression of NGX6 in cancer samples compared with their corresponding normal tissues. The expression of NGX6 in colorectal carcinoma was significantly lower than that in normal tissues (χ2 = 15.06, P < 0.005). Representative cases of NGX6 expression detected by RT-PCR are shown in Figure 1. The down-regulation rate of NGX6 in the patients with lymph-node and/or distance metastases was 93.8% (15/16), significantly higher than that in those without lymph-node or distance metastases (55.6%, 10/18) (P < 0.05). The down-regulation rates of NGX6 in well differentiated and poorly differentiated adenocarcinomas were 46.2% and 85.7%, respectively. There was no apparent relevance between NGX6 down-expression and pathologic type of colorectal carcinomas (P > 0.05).
Figure 1 Expression of NGX6 in colorectal carcinoma and its corresponding normal tissues was examined by RT-PCR.
The RT-PCR products with NGX6 primers produced 780 bp frag-ments and GAPDH primers produced 466 bp fragments. Lane M, DL-2000 marker; Lane N, normal tissues; Lane C, colorectal carcinoma tissues. This figure showed the representative re-sults from several individual patients.
In 34 pairs of GC and their corresponding normal tissue specimens, NGX6 expression was detected in 20 GC tissues (58.8%) and 24 corresponding normal tissues (70.6%) as an expected PCR product of 780 bp in length. The expression level of NGX6 did not display any difference between gastric carcinoma and its corresponding normal tissues (χ2 = 1.03, P > 0.05).
NAG-7 expression was detected in 88.2% (30/34) GC tissues and 82.3% (28/34) corresponding normal tissues as well as in 76.2% (26/34) CRC tissues and 76.2% (26/34) corresponding normal tissues as an expected PCR product of 466 bp in length. BRD7 expression was detected in all samples as an expected PCR product of 270 bp in length. The expression level of NAG-7 and BRD7 did not display any significant difference between cancer samples and normal tissues (P > 0.05).
Expression of NGX6, NAG-7 and BRD7 genes in gastric and colorectal carcinoma tissues by Northern blot analysis and dot hybridization
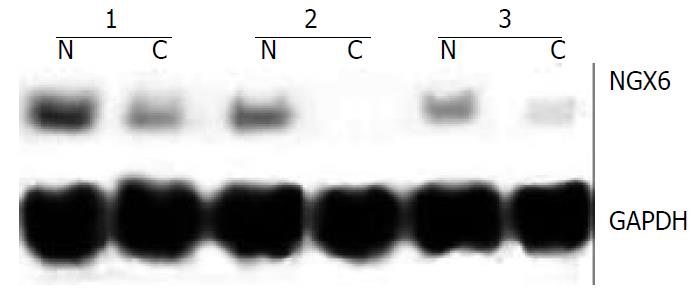
Northern blot analysis and dot hybridization were performed to further investigate the expression of NGX6, NAG-7 and BRD7. Northern blot analysis showed that NGX6 was expressed as one single transcript of 2.4 kb, corresponding well with the size of the cloned cDNA. NGX6 was strongly expressed in its corresponding normal colorectal epithelial tissues, whereas faint signals or no signals were found in colorectal carcinoma tissues. Representative cases of NGX6 expression detected by Northern hybridization are shown in Figure 2. However, no signals were discerned in gastric carcinoma and its corresponding normal epithelial tissues. NAG-7 and BRD7 mRNA was expressed at the same level in the cancer samples and normal tissues. The results of dot hybridization were consistent with those of Northern analysis. Both dot hybridization and Northern analysis confirmed the results of RT-PCR. The expression of NGX6 was significantly down-regulated in colorectal carcinoma tissues.
Figure 2 Northern blot was used to detect the expression abun-dance of NGX6 gene in human colorectal carcinoma and its adjacent normal tissues.
The expression level of NGX6 was down-regulated in colorectal carcinoma tissues. C, human colorectal carcinoma tissue; N, adjacent normal colorectal tissue.
DISCUSSION
NGX6, NAG-7 and BRD7 genes located at 9p21-22, 3p25 and 6q22.1-6q22.33 respectively are novel tumor related genes that were cloned by positional candidate cloning strategy. The cDNA sequence of NGX6, NAG-7 and BRD7 genes compared with Genbank and EMBO database using the BLAST program has verified that all of them are unique genes with no homology to any previously reported human genes, and their GenBank accession numbers are AF188239, AF086709 and AF179285, respectively. The NGX6 gene encodes a transmembrane protein of 338 amino acids containing four transmembrane regions, an EGF-like domain signature and a number of potential phosphorylation sites for protein kinase C (PKC), casein kinase II, tyrosine kinase and several N-myristylation sites. The NAG-7 gene encodes a transmembrane protein of 94 amino acids including one PKC phosphorylation site and one myristylation site. The BRD7 gene encodes a protein of 508 amino acids including a bromodomain and several important phosphorylation sites. In our previous study, their mRNA expression levels in nasopharyngeal carcinoma (NPC) cells were all significantly lower than those in normal nasopharyngeal epithelium. The proliferation rate of NPC cells was slower after transfection with these genes. The growth of xenografts was also inhibited after the transfected cells were injected into nude mice in vivo. These results suggest that the downregulation of these genes might play important roles in the occurrence and development of NPC[34-39].
Allelic imbalance or loss of heterozygosity (LOH) studies have been used to identify regions on chromosomes that may contain putative tumor suppressor genes. Deletions of chromosome 9p, 3p, and 6q regions have been observed at a high frequency in many types of sporadic tumors, including gastric and colorectal cancers[17-24]. NGX6, NAG-7 and BRD7 genes located at 9p21-22, 3p25 and 6q22.1-6q22.33 respectively, are the frequent sites of LOH in GC and CRC. We were interested in investigating if expression of NGX6, NAG-7 and BRD7 genes was altered in GC and CRC. In this study, RT-PCR, Northern blot and dot hybridization were used to detect the expression abundance of the three genes in gastric and colorectal carcinoma tissues, as well as their corresponding normal tissues. The results of RT-PCR showed that the down-regulation rate of NGX6 in colorectal carcinoma tissues with lymph-node or distant metastases was significantly higher than that both in its corresponding normal tissues (P < 0.005) and in carcinomas without lymph-node or distant metastases (P < 0.05). Dot hybridization and Northern analysis confirmed the results of RT-PCR. However, no significant correlation was found between the expression of NGX6 and pathologic type of colorectal carcinomas. The results suggest that down-regulation of NGX6 might be closely associated with the development, progression and metastasis of CRC. It is reasonable to predict that the NGX6 gene plays an important role in suppressing CRC tumorigenesis; losses of its function may contribute to the development of CRC. Deletion of this gene may occur as a later event associated with tumor progress, and may contribute to the metastatic potential of CRC. So, it is speculated that NGX6 gene located on chromosomes 9p21-22 may be a candidate tumor suppressor gene and a putative metastasis suppressor gene associated with CRC. The possible mechanism of this gene suppressing CRC tumorigenesis remains unclear. We speculated that the possible mechanism might be as follows. Firstly, NGX6 gene may be involved in cell cycle regulation and play an important role in tumor growth suppression. Our previous studies showed that significant accumulation of cells in the G0-G1 fraction was observed in HNE1 cell transfected with NGX6 gene by using flow cytometric analysis, and several genes related to cell cycle and transcription regulation were differentially expressed in NGX6 overexpressed nasopharyngeal carcinoma cells by cDNA array assay[34,35,40]. Combined with our data presented here, it is reasonable to speculate that deletion or down-regulation of NGX6 gene in colorectal cancer may contribute to tumor cell proliferation and tumor metastasis. Secondly, NGX6 gene encodes a putative transmembrance protein including an important structural feature-EGF-like domain signature. In our study, NGX6 gene was significantly down-regulated in CRC, indicating that NGX6 protein might function as an inhibited membrane receptor or a negative growth factor in the initiation and progression of CRC. However, many previous studies showed that the proteins with EGF-like domains such as EGF, TGF-a were overexpressed in tumor tissues and might play a role in promoting tumor growth and metastasis[41-43]. Recently, Williams et al[44] identified two novel mucin genes, which encoded two putative transmembrane mucins with EGF-like domains and were commonly downregulated in colorectal carcinoma. This detection further confirmed that the transmembrance protein with EGF-like domain could act as a suppressive membrane receptor or a negative growth factor. Finally, NGX6 gene may exert functions by up-regulating and down-regulating some proteins. Recently, Li et al[45] found that seven proteins were up-regulated and seven proteins were down-regulated in NGX6 transfected cells using high-resolution two-dimensional electrophoresis. These proteins included Fas, zinc-finger protein (ZNF), RAB, and Ah receptor-interacting protein (AIP), which may affect the signaling pathway, alter the cell metabolism, and inhibit cell proliferation, induce cell apoptosis and prevent tumor invasion. Therefore, all these protein roles on cells indicate that NGX6 gene has the capacity to suppress both NPC and CRC tumorigenesis.
In this study, we did not find the existence of up- or down-regulated NGX6 in 34 gastric carcinoma specimens and their corresponding normal tissues by RT-PCR. The expression of NAG-7 and BRD7 genes did not display any significant difference between gastric and colorectal carcinomas and their corresponding normal tissues. The results of RT-PCR were also confirmed by Northern blot analysis and dot hybridization. This seems to suggest that NAG-7 and BRD7 genes play no roles in the pathogenesis of gastric and colorectal carcinomas, and NGX6 gene may not contribute to the occurrence and development of gastric carcinoma.
In summary, the results of this study have shown that NGX6 is down-regulated in colorectal carcinoma, and the down-regulation of this gene has a close correlation with lymph node or distant metastasis of CRC, suggesting that NGX6 plays an important role in tumor suppression. However the mechanism of this gene is still unclear. Further studies are needed to better understand the function of this novel gene and its role in CRC tumorigenesis in vivo.