Published online Jul 15, 2003. doi: 10.3748/wjg.v9.i7.1629
Revised: March 28, 2003
Accepted: April 11, 2003
Published online: July 15, 2003
AIM: To produce the recombinant NS3 protease of hepatitis C virus with enzymatic activity in insect cells.
METHODS: The gene of HCV serine proteinase domain which encodes 181 amino acids was inserted into pFastBacHTc and the recombinant plasmid pFBCNS3N was transformed into DH10Bac competent cells for transposition. After the recombinant bacmids had been determined to be correct by both blue-white colonies and PCR analysis, the isolated bacmid DNAs were transfected into Sf9 insect cells. The bacmids DNA was verified to replicate in insect cells and packaged into baculovirus particles via PCR and electronic microscopic analysis. The insect cells infected with recombinant baculovirus were determined by SDS-PAGE and Western-blot assays. The recombinant protein was soluted in N-lauryl sarcosine sodium (NLS) and purifed by metal-chelated-affinity chromatography, then the antigenicity of recombinant protease was determined by enzyme-linked immunoabsorbant assay and its enzymatic activity was detected.
RESULTS: The HCV NS3 protease domain was expressed in insect cells at high level and it was partially solved in NLS. Totally 0.2 mg recombinant serine proteinase domain with high purity was obtained by metal-chelated-affinity chromatography from 5 × 107 cells, and both antigenicity and specificity of the protein were evaluated to be high when used as antigen to detect hepatitis C patients' sera in indirect ELISA format. In vitro cleavage assay corroborated its enzymatic activity.
CONCLUSION: The recombinant HCV NS3 proteinase expressed by insect cells is a membrane-binding protein with good antigenicity and enzymatic activity.
-
Citation: Hou LH, Du GX, Guan RB, Tong YG, Wang HT.
In vitro assay for HCV serine proteinase expressed in insect cells. World J Gastroenterol 2003; 9(7): 1629-1632 - URL: https://www.wjgnet.com/1007-9327/full/v9/i7/1629.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i7.1629
Hepatitis C virus, the major etiological agent of post-transfusion non-A, non-B hepatitis, is an enveloped virus obtaining a single-stranded RNA genome of approximately 9.5 kb[1-3]. A single polyprotein of 3010-3030 amino acids is translated from this genome in the order of NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. Proteolytic procession of the viral precursor protein is required for replication and is mediated by either viral or host cell proteinases[4-7]. HCV uses a serine proteinase located at the N- terminus of NS3, to process the cleavage of four sites (3/4A, 4A/4B, 4B/5A and 5A/5B). Since the NS3 protein is very important for releasing functional proteins from the polyprotein, it has been currently targeted in the development of drugs and diagnostics[8-11].
In the absence of satisfactory cell culture, protein expression is the primary technological methods for HCV[12-14]. In this report, we described the construction of the recombinant baculovirus, designed to express the NS3 proteinase in insects. The activity of the expressed enzyme has been demonstrated by in vitro assay.
Spodoptera frugiperda Sf9 cells were a gift from Professor Gu Shu-Yan, Institute of Virology, CDC, China. The vector pGEX-3X-NS3N containing the gene serine proteinase located at N-terminus of HCV NS3, was constructed in our lab before[15]. The expression vector pFastBacHTc and E.coli DH10Bac were purchased from Invitrogen Inc.
The primers flanking the vector transfer pFastBacHTc MCS were WB538P (5'-TATTCCGGATTATTCATACC-3') and WHT62P (5'-TGGTATGGCTGATTATGAT-3'). The primers for bacmid were pUC/M13-R (5'-CAGGAAACAGCTATGAC-3') and pUC/M13-F (5'-GTTTTCCCAGTCACGAC-3').
The gene HCV NS3N serine proteinase was excised from pGEX-3X-NS3N with restriction enzyme BamHI and EcoRI, and ligated with vector pFastBacHTc. The recombinant plasmids were transformed into E.coli DH5α. The positive recombinants were identified with PCR and restriction enzyme digestion, then termed as pFBCNS3N.
The recombinant transfer vector pFBCNS3N was transformed into the competent DH10Bac cells. After growth in the shaking incubator at 37 °C for 6 h, the cells were placed on the plates(containing 50 mg/L kanamycin, 7 mg/L gentamicin, 10 mg/L tetracycline, 40 mg/L IPTG and 100 mg/L X-gal) with serial dilution, then incubated at 37 °C for 48 h. The white colonies containing the recombinant bacmid, therefore, were selected for the isolation of recombinant bacmid DNA. The bacmid, termed as bacmid-NS3N, was analyzed by 0.5% agarose gel electrophoresis to confirm the presence of high molecular weight DNA.
9 × 105 of Sf9 cells were seeded to each well of a 6-well plate in TNM-FH medium and incubated at 27 °C. Bacmic-NS3N was transfected into Sf9 cells using Lipofectamine reagent (Gibco BRL) after cells attached to the wells. The supernatant was collected as virus stock when cytopathic effect appeared. At the same time, virus particles in the supernatant were precipitated using PEG8000 to extract viral genome for PCR amplification with the primers WB538P, WHT62P and pUC/M13-R, F to confirm the recombinant virus containing the interested gene. The morpha of recombinant baculovirus was observed under transmission electron microscope.
Monolayer insect cells were transfected by the recombinant baculovirus at a multiplicity of infection (MOI) of 10. The cells were harvested after incubation for 72 h at 27 °C, resuspended in lysis buffer (0.5% NP-40, 20 mmol/L Tris-HCl pH7.4, 150 mmol/L NaCl, 100 mg/L PMSF), and disrupted by ultrasonic. The supernatant and precipitation of the mixture were collected for SDS-PAGE and Western blotting. 5 × 107 of Sf9 cells were collected by centrifugation after transfection for 72 h to purify the recombinant serine proteinase. The cell pellet was resuspended in TNG buffer (50 mmol/L Tris-HCl pH7.4, 0.2% NLS (N-Lauryl Sarcosine Sodium), 10% glycerol, 150 mmol/L NaCl, 100 mg/L PMSF), and disrupted by ultrasonic. The interested protein was purified from the supernatant by means of affinity chromatography on Ni-NTA columns (Qiagen).
Fifteen μg recombinant serine proteinase was mixed with 15 μg substrate HCV NS5ab in 100 μL of 25 mmol/L Tris-HCl pH7.4, 10% glycerol, 0.5 mol/L NaCl, 10 mmol/L DTT, 0.5% NP-40. The incubation was carried out at room temperature for different time periods and the result was read through SDS-PAGE.
The identity of recombinant proteins was confirmed by 50 HCV-positive and 10 HCV-negative sera. Microtitre cells were coated by overnight incubation at 4 °C with 100 μL recombinant protein solution (10 mg/L proteins in 0.1 mol/L sodium carbonate buffer, pH9.6). Serum samples diluted at 1:20 in PBSTM(PBS + 0.3% Tween-20 + 3% fat-free milk) were added, and incubated at 37 °C for 1 hour after blocking. The specific binding was revealed using HRP anti-human IgG detection system(Sigma, USA).
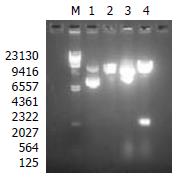
A 540 bp fragment was excised from the plasmid pFBCNS3N with restriction endonuclease enzymes HindIII and EcoRI (Figure 1). The result showed that the construction of recombinant transfer vector was successful.
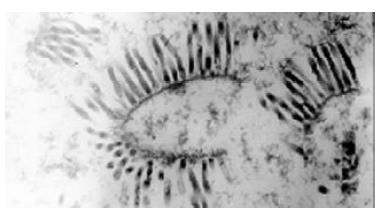
The recombinant baculoviruses were generated after the site-specific transposition of transfer plasmids in E.coli, transfection and packaging in insect cells. Large amounts of baculoviruses were observed in the nuclei of cells under transmission electron microscope (Figure 2). In PCR detection, genome DNA of the recombinant baculoviruses as template, 800 bp and 3000 bp fragments appeared respectively with primers WB538P, WHT62P and pUC/M13-R, F. The result conformed to our previous expectation and revealed HCV serine proteinase gene existed in the genome of recombinant baculoviruses.
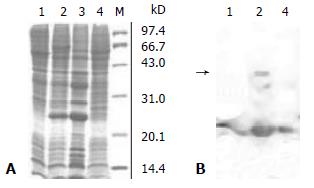
Sf9 cells were transfected with the recombinant baculovirus at a MOI of 10, and serious CPE appeared after 72 h. The total cell extracts were separated by 15% SDS-PAGE and stained with Coomassie blue. A predominant band at 23 kDa (approximately 20% of total proteins) was detected in the cell extracts from bacmic-NS3N infected cells but not in the uninfected cell extracts (Figure 3A). Immunoblot analysis with the HCV-positive sera (Figure 3B) revealed that the 23 kDa protein was clearly detected in cells infected with the recombinant baculovirus.
Though HCV serine proteinase was expressed efficiently in insect cells, a little was laid in the supernatant of the cell lysate. Only by immunoblot analysis was the protein in the supernatant detected. Did the protein's misfolding or binding with endomembrane system in cells lead to unsoluble protein This question was analyzed by changing the composition of the lysis buffer. When the cell lysates were dissolved in TNG buffer containing N-lauryl sarcosine sodium, more interested protein was soluble because TNG as a detergent could dissolve proteins binding with membrane. Therefore, it was presumed that the recombinant HCV serine proteinase binding with membrane resulted in its unsolubility. The HCV serine proteinase was extensively purified from 5 × 107 Sf9 cells and demonstrated a single protein band of 23 kDa (more than 90% of the total proteins by densitometric analysis) by using Ni-NTA columns. The total yield of the purified protein was about 0.2 mg.
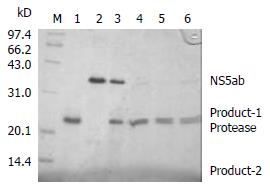
In order to determine whether the baculovirus-expressing cleavage N-terminus of HCV NS3 has a proteinase activity, an In vitro proteolytic assay with HCV NS5ab protein (235 aa) as a substrate containing the cleavage site of HCV NS5a and NS5b was designed. Figure 4 shows that the baculovirus-expressing HCV serine proteinase could cleave the substrate HCV NS5ab (about 35kDa) into bands of 24kDa and 11kDa (Figure 4). This means that the baculovirus-expressing HCV serine proteinase has a high proteolytic activity.
Fifty HCV positive sera and 10 HCV negative sera were evaluated for their reactivities against the purified HCV serine proteinase. A high degree of immunoreactivity was observed in 30 HCV positive sera, showing good antigenicity of HCV serine proteinase.
It has been proved that HCV NS3 protein with 631 amino acids possesses both serine proteinase in its N-terminus and NTP/RNA helicase activity in its C-terminus[16]. Both In vitro translation and transient mammalian cells have shown that NS3 is a serine proteinase and is responsible for proteolytic processing of the non-structural region (NS3, NS4A, NS4B, NS5A and NS5B) of HCV polyprotein. Since the maturation of these non-structural proteins is the prerequisites for replication, and packaging of HCV, NS3 serine proteinase plays an important role in the life cycle of HCV and is an attractive target for antiviral therapy. We expressed only 181 amino acids in HCV NS3 N-terminus because previous experiments had corroborated it with good enzyme activity[17].
Baculovirus-insect cell expression system has two advantages over others: first, in most cases the recombinant protein in insect cells is processed, modified, and targeted to its appropriate cellular location, where it is functionally similar to its authentic counterparts. Second, high levels of heterogenous gene expression, up to 20% of total cellular proteins, are often achieved compared with other eukaryotic expression systems[18-20]. Here we chose the Bac-to-Bac expression system which is based on site-specific transposition of an expression cassette into a baculovirus shuttle vecor (bacmid) propagated in E.coli[21,22]. Colonies containing the recombinant bacmid are white in a background of blue colonies that harbor the unaltered bacmid. Recombinant bacmid DNA can be rapidly isolated from small scale cultures and then is used to transfect insect cells, eliminating the need for multiple rounds of plaque purification.
In our experiment, the interested gene was efficiently expressed in insect cells through the recombinant virus BacNS3N containing HCV NS3 serine proteinase gene, and the identity of the protein was validated by use of immunoblot. But the recombinant protein predominantly lay in the insoluble part, and the solubility could not be improved even by adding 1% NP-40 into the lysate. Overton et al[23] illuminated that HCV NS3 serine proteinase expressed in insect cells was a membrane-associated protein through discontinuous sucrose density gradients and only by adding some detergents did part of the protein gain the enzyme activity. Suzuki et al[24] discovered that HCV serine proteinase from insect cells was soluble and active with 0.5% NLS. Therefore, the TNG buffer including NLS and glycerol was used to dissolve the pellet. The results showed that part of the recombinant protein became soluble and more proteins were soluble when enhancing the percentage of NLS and adding EDTA both. The recombinant protein fused with 6 His tag at its N- terminus could be purified through metal-chelating chromatography. Finally, 0.2 mg purified recombinant protein was obtained from 5 × 107 insect cells. An In vitro detection system with SDS-PAGE and staining was developed with the recombinant HCV NS5ab as a substrate. The system eliminates the need of expensive instrument and reagent, heavy work, as compared with other cleavage systems[25-27] and provides a simple and clear strategy for the screening of inhibitors against the HCV proteinase.
Edited by Ma JY and Wang XL
| 1. | Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Kuo G, Choo QL, Alter HJ, Gitnick GL, Redeker AG, Purcell RH, Miyamura T, Dienstag JL, Alter MJ, Stevens CE. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2495] [Cited by in RCA: 2342] [Article Influence: 65.1] [Reference Citation Analysis (0)] |
| 3. | Alter MJ, Margolis HS, Krawczynski K, Judson FN, Mares A, Alexander WJ, Hu PY, Miller JK, Gerber MA, Sampliner RE. The natural history of community-acquired hepatitis C in the United States. The Sentinel Counties Chronic non-A, non-B Hepatitis Study Team. N Engl J Med. 1992;327:1899-1905. [PubMed] |
| 4. | Okamoto H, Okada S, Sugiyama Y, Kurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697-2704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 323] [Cited by in RCA: 323] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 5. | Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105-1113. [PubMed] |
| 6. | Grakoui A, Wychowski C, Lin C, Feinstone SM, Rice CM. Expression and identification of hepatitis C virus polyprotein cleavage products. J Virol. 1993;67:1385-1395. [PubMed] |
| 7. | Shimotohno K, Tanji Y, Hirowatari Y, Komoda Y, Kato N, Hijikata M. Processing of the hepatitis C virus precursor protein. J Hepatol. 1995;22:87-92. [PubMed] |
| 8. | Yan Y, Li Y, Munshi S, Sardana V, Cole JL, Sardana M, Steinkuehler C, Tomei L, De Francesco R, Kuo LC. Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form. Protein Sci. 1998;7:837-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 190] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 9. | Dymock BW, Jones PS, Wilson FX. Novel approaches to the treatment of hepatitis C virus infection. Antivir Chem Chemother. 2000;11:79-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Llinàs-Brunet M, Bailey M, Fazal G, Ghiro E, Gorys V, Goulet S, Halmos T, Maurice R, Poirier M, Poupart MA. Highly potent and selective peptide-based inhibitors of the hepatitis C virus serine protease: towards smaller inhibitors. Bioorg Med Chem Lett. 2000;10:2267-2270. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 81] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 11. | Bartenschlager R. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepat. 1999;6:165-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 105] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Takehara T, Hayashi N, Miyamoto Y, Yamamoto M, Mita E, Fusamoto H, Kamada T. Expression of the hepatitis C virus genome in rat liver after cationic liposome-mediated in vivo gene transfer. Hepatology. 1995;21:746-751. [PubMed] |
| 13. | Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 450] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 14. | Gale M, Beard MR. Molecular clones of hepatitis C virus: applications to animal models. ILAR J. 2001;42:139-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 15. | Du GX, Hou LH, Guan RB, Tong YG, Wang HT. Establishment of a simple assay in vitro for hepatitis C virus NS3 serine protease based on recombinant substrate and single-chain protease. World J Gastroenterol. 2002;8:1088-1093. [PubMed] |
| 16. | Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832-2843. [PubMed] |
| 17. | Tanji Y, Hijikata M, Hirowatari Y, Shimotohno K. Identification of the domain required for trans-cleavage activity of hepatitis C viral serine proteinase. Gene. 1994;145:215-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Luckow VA. Baculovirus systems for the expression of human gene products. Curr Opin Biotechnol. 1993;4:564-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 73] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Patterson RM, Selkirk JK, Merrick BA. Baculovirus and insect cell gene expression: review of baculovirus biotechnology. Environ Health Perspect. 1995;103:756-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 20. | Marchal I, Jarvis DL, Cacan R, Verbert A. Glycoproteins from insect cells: sialylated or not. Biol Chem. 2001;382:151-159. [PubMed] |
| 21. | Luckow VA, Lee SC, Barry GF, Olins PO. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J Virol. 1993;67:4566-4579. [PubMed] |
| 22. | Joubert AM, Louw AI, Joubert F, Neitz AW. Cloning, nucleotide sequence and expression of the gene encoding factor Xa inhibitor from the salivary glands of the tick, Ornithodoros savignyi. Exp Appl Acarol. 1998;22:603-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Overton H, McMillan D, Gillespie F, Mills J. Recombinant baculovirus-expressed NS3 proteinase of hepatitis C virus shows activity in cell-based and in vitro assays. J Gen Virol. 1995;76:3009-3019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Suzuki T, Sato M, Chieda S, Shoji I, Harada T, Yamakawa Y, Watabe S, Matsuura Y, Miyamura T. In vivo and in vitro trans-cleavage activity of hepatitis C virus serine proteinase expressed by recombinant baculoviruses. J Gen Virol. 1995;76:3021-3029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Shoji I, Suzuki T, Sato M, Aizaki H, Chiba T, Matsuura Y, Miyamura T. Internal processing of hepatitis C virus NS3 protein. Virology. 1999;254:315-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Sali DL, Ingram R, Wendel M, Gupta D, McNemar C, Tsarbopoulos A, Chen JW, Hong Z, Chase R, Risano C. Serine protease of hepatitis C virus expressed in insect cells as the NS3/4A complex. Biochemistry. 1998;37:3392-3401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 27. | Berdichevsky Y, Zemel R, Bachmatov L, Abramovich A, Koren R, Sathiyamoorthy P, Golan-Goldhirsh A, Tur-Kaspa R, Benhar I. A novel high throughput screening assay for HCV NS3 serine protease inhibitors. J Virol Methods. 2003;107:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |