Published online Jul 15, 2003. doi: 10.3748/wjg.v9.i7.1525
Revised: March 28, 2003
Accepted: April 9, 2003
Published online: July 15, 2003
AIM: To prepare and purify TAT-HBV targeted ribonuclease fusion protein, evaluate its transduction activity and investigate its effect on HBV replication in 2.2.15 cells.
METHODS: The prokaryotic expression vector pTAT containing TR gene was used in transforming E.coli BL21 (DE3) LysS and TR was expressed with the induction of IPTG. The TAT-TR fusion protein was purified using Ni-NTA-agrose and PD-10 desalting columns, and analyzed by SDS-PAGE. Transduction efficiency of TAT-TR was detected with immunofluorescence assay and the concentration of HBeAg in the supernatant of the 2.2.15 cells was determined via solid-phase radioimmunoassay (spRIA). MTT assay was used to detect the cytotoxicity of TAT-TR.
RESULTS: The SDS-PAGE showed that the TAT-TR fusion protein was purified successfully, and the purity of TAT-TR was 90%. The visualization of TAT-TR by immunofluorescence assay indicated its high efficiency in transducing 2.2.15 cells. RIA result suggests that TAT-TR could inhibit the replication of HBV effectively, it didn’t affect cell growth and had no cytotoxicity.
CONCLUSION: TAT-TR possesses a significant anti-HBV activity and the preparation of TAT-TR fusion protein has laid the foundation for the use of TR in the therapeutic trial of HBV infection.
- Citation: Ding J, Liu J, Xue CF, Gong WD, Li YH, Zhao Y. Anti-HBV effect of TAT- HBV targeted ribonuclease. World J Gastroenterol 2003; 9(7): 1525-1528
- URL: https://www.wjgnet.com/1007-9327/full/v9/i7/1525.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i7.1525
The introduction of proteins into mammalian cells has been achieved by transfection of expression vectors, microinjection, or infectious virus, etc. Although these approaches have been somewhat successful, the classical manipulation methods are not easily regulated and can be laborious. One approach to resolve these problems is the use of PTD-mediated protein transduction[1,2]. Linked covalently to proteins, peptides, nucleic acids, or as in-frame fusions with full-length proteins, PTD would let them enter any cell type in a receptor and transporter independent fashion[3]. HIV-TAT is a member of protein transduction domains and appears to possess high level of protein-transduction efficiency[4,5]. TAT fusion proteins were shown to transduce into all cells and tissues present in mice[6], including those present across the blood-brain barrier[7,8]. And many, if not most, proteins may be transduced into cells by using this technology. Therefore, TAT PTD may let us address new questions in preclinical research work and even help in the treatment of human disease.
Hepatitis B is a major world-wide health problem[9-13]. Chronic infection is associated with high risk of liver cirrhosis and primary liver carcinoma[14-22]. Currently available therapies are of limited efficiency[23-35]. HBV targeted ribonuclease (TR) gene, a fusion gene of HBVc and hEDN, was constructed by Liu et al[36] in our lab, according to the theory of capsid-targeted viral inactivation (CTVI) which is a promising strategy in anti-virus research. HBVc was used as the target molecule, which was the structure protein of HBV and was indispensable during the packaging of HBV particle. The effector molecule was hEDN, a kind of human ribonuclease that can degrade pgRNA of HBV. Transfection of 2.2.15 cells with the eukaryotic expression vector bearing TR gene suggested that TR inhibited the replication of HBV significantly[37]. Therefore, linking HIV-TAT to TR would provide us a more efficient approach to deliver TR into hepatocytes, and greatly help us to utilize TR in the treatment of HBV infection. Reported here are the purification of TAT-TR fusion protein, the identification of its transduction and the anti-HBV effect on the 2.2.15 cells. To confirm its anti-HBV mechanism, we also prepared and purified TAT-TRmut, TAT-hEDN and TAT-HBVc proteins for use as negative controls.
Ni-NTA-agrose was purchased from Qiagen Company. PD-10 desalting columns were purchased from Amersham Pharmacia Biotech. Anti-his mAb was from Santa Cruz Company. Protein molecular mass markers, IPTG and G418, imidazole and MTT were all from Sino-American Biotech. RIA HBVeAg assay kit was purchased from Beimian Dongya Biotechnology Institute. 2.2.15 cells was a kind gift of Prof. Cheng, 302 Hospital of Chinese PLA. hEDN was purified by Li et al[36]. pTAT-HA/TR, TAT-HA/TRmut, pTAT-HA/hEDN and pTAT-HA/HBVc were all prepared in our Lab[38]. PET-30a/TR, PET-30a/TRmut, PET-30a/HBVc and E.coli BL21 (DE3) LysS were maintained in our Lab.
Expression and purification of TAT fusion proteins pTAT-HA/TR, TAT-HA/TRmut, pTAT-HA/hEDN, pTAT-HA/HBVc and pTAT-HA were employed to transform E.coli BL21(DE3) LysS by using CaCl2 perforation. The transformants were separately cultured in 3 mL TB amp (100 μg/L) at 37 °C overnight. 100 μL culture was inoculated into 10 mL fresh TB amp, and incubated for up to 4 h at 37 °C. Then IPTG was added to each tube to a final concentration of 100 μmol/L, and the culture was incubated for an additional 4 h. The induced cells were harvested by centrifugation, and cell lysates were analyzed by 120 g/L SDS-PAGE. The his-tagged fusion proteins were purified by using Ni-NTA-agrose and PD-10 desalting columns according to the manufacturer’s recommendations (Qiagen and Amersham Pharmacia). The purified proteins were analyzed by 120 g/L SDS-PAGE.
Expression and purification of proteins without TAT PTD PET-30a/TR, pET-30a/TRmut, and pET-30a/HBVc transformed E.coli BL21 (DE3) LysS. After the analysis of expression levels, the three proteins were purified in the same way as for TAT fusion proteins.
Culture of 2.2.15 cells Cells were cultured in DMEM containing 150 mL/L fetal bovine serum at 37 °C in 50 mL/L CO2 and 100 mg/L G418.
Identification of TAT fusion protein transduction 2.2.15 cells (2 × 108/L) were plated into 6-well plates with coverships, and allowed to adhere for 24 h. TAT-TR, TAT-TRmut, TAT-hEDN, TAT-HBVc, TR, TRmut, hEDN and HBVc were added into the wells, respectively, at the final concentration of 100 nmol/L. Incubated for 30 min at 37 °C, all cells were immediately washed with sterile PBS (pH8.0), fixed in 20 g/L paraformaldehyde and 1 g/L TritonX-100 diluted in PBS and put on ice for 30 min. Cells were washed three times with cold PBS. Non-specific epitopes were blocked by using 10 g/L BSA for 10 min at 42°C Cells were washed three times with cold PBS, and then incubated with mouse anti-His mAb (1:500) for 15 min at 42 °C. After washing three times in cold PBS, the rabbit anti-mouse IgG labeled with FITC (1:1000) was added to each well and incubated for 10 min at 42 °C. Rinsed with PBS for 1 hour and the coverships were mounted on slides by using 500 mL/L glycerol. The cells were observed by fluorescence microscopy.
Determination of anti-HBV effect of TAT-TR 2.2.15 cells were plated at the density of 2 × 108/L into 12-well plates. TAT-TR, TAT-TRmut, TAT-hEDN and TAT-HBVc were added into the wells, respectively, at the final concentration of 100 nmol/L. 20 μL DMEM was added into wells as mock group. Four parallels were set up for each group. 24 h later, HBVeAg in the supernatant was determined by using spRIA kit as described by the manufacturer.
MTT assay 2.2.15 cells were plated at the density of 2 × 108/L into 96-well plates. After 24 h, TAT-TR, TAT-TRmut, TAT-HEDN, TAT-HBVc were added into (A), (B), (C), (D) groups at the final concentration of 100 nmol/L. 20 μL DMEM was added into well (E). 72 h later, the morphology of cells was observed through inverted microscopy and MTT was applied in each well at the final concentration of 5 g/L. After another 4 h culturing, 150 μL DMSO was added into all wells and the light absorbance at A490 was detected.
All data obtained were processed by SPSS software. P < 0.05 was considered statistically significant.
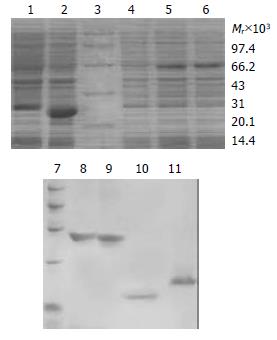
In order to obtain the fusion proteins, pTAT-HA/TR, pTAT-HA/TRmut, pTAT-HA/hEDN and pTAT-HA/HBVc were used to transform E.coli BL21 (DE3) LysS and expressed with the induction of IPTG. The same strain transformed by pTAT-HA was used as negative control. The expression levels were determined by 120 g/L SDS-PAGE. Four predicted new bands could be detected in the lysates of TAT fusion transformants, but not in the control. Then the proteins were purified by using Ni-NTA affinity columns and PD-10 desalting columns (Figure 1). The degrees of purity of the fusion proteins were 90%, 88%, 80% and 85% respectively.
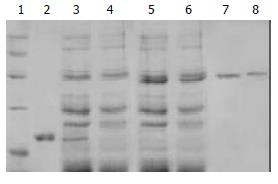
In a similar fashion, pET-30a/TR, pET-30a/TRmut and pET-30a/HBVc were induced to express proteins without TAT PTD by IPTG in BL21 (DE3) LysS, and they produced proteins with predicted molecular masses (Figure 2). Then the proteins were purified by using Ni-NTA affinity columns and PD-10 desalting columns. The degrees of purity of the fusion proteins were 88%, 76% and 81% respectively.
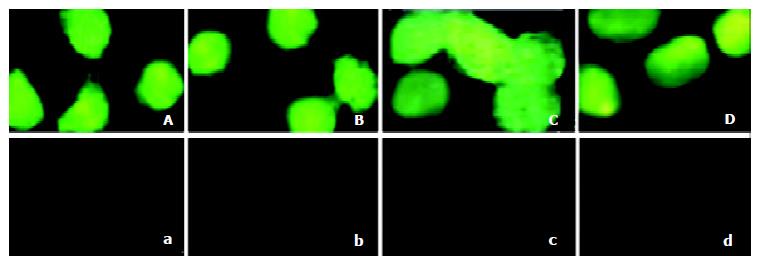
To evaluate the fusion proteins transduction ability in crossing the membrane of 2.2.15 cells. TAT-TR, TAT-TR mut, TAT-hEDN and TAT-HBVc were added in the culture media at the final concentration of 100 nmol/L. TR, TRmut, hEDN and HBVc without TAT PTD were used as negative controls. Under the fluorescence microscope, abundant fluorescence could be seen in cytoplasm of the cells transduced with TAT fusion proteins, but no fluorescence could be found in the control cells (Figure 3). This result clearly suggests that TAT fusion proteins could cross the membrane of 2.2.15 cells with high efficiency.
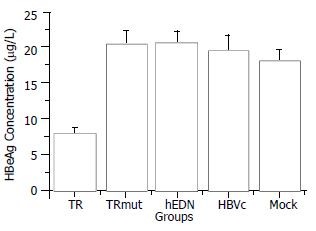
Statistical analysis with SPSS software showed that the mean HBeAg concentration of the TAT-TR group decreased significantly as compared with control groups (The mean difference is significant at the 0.05 level). In addition, there is no significant difference among the mean concentration of control groups (P > 0.05). The HBeAg concentration of TAT-TR group decreased by 60.3% (Figure 4).
After 72 h culture, the morphology of cells was observed under inverted microscope and it was found that there was no discernable difference among the four experiment groups and the DMEM control. MTT assay showed no significant difference among the five groups. Their A490 absorbance values (¯x ± s, n = 4) were 0.4875 ± 0.018, 0.4675 ± 0.022, 0.4690 ± 0.028, 0.4800 ± 0.029 and 0.4855 ± 0.050, respectively, (P > 0.05).
The transduction of proteins into cells was first described in 1988 independently by Green et al[39] and Frankel et al[40]. In 1994, Faweell et al[5] expanded these observations by demonstrating that heterologous proteins chemically cross-linked to a 36 amino acid domain of TAT were able to transduce into cells. Subsequent to the TAT discovery, other transduction domains were identified that resided in the Antennapedia (Antp) protein from Drosophila[41] and HSV VP22 protein from HSV[42]. Although the exact mechanism of protein transduction across the cellular membrane remains unclear, PTD’s widespread application has been realized. TAT-mediated transduction provides several advantages over DNA transfection, the current standard method of intracellular protein expression. Importantly, all eukaryotic cell types tested to date are susceptible to transduction, even osteoclasts, primary cells and peripheral-blood mononuclear cells, which are impervious to DNA transfection and retroviral infection, can be effectively transduced[43,44]. Additionally, as transduction occurs so rapidly (15 min rather than 12 h in serum-free media for transfection), issues of timing can be addressed. The exact intracellular concentration can also be controlled precisely just by varying the amount added to the culture medium. Furthermore, every cell in the population appears to contain a near-identical intracellular protein level[3]. Another dominant advantage of the system allows denatured fusion protein be directly applied without the laborious renature course, and thus it also provides much convenience for protein purification. Once transduced inside the cells, the denatured proteins can be correctly refolded by chaperones[45], and are capable of binding their cognate intracellular targets and performing biochemical functions.
To explore the feasibility of using PTD in the anti-HBV research work and provide an alternative strategy for treatment of hepatitis B in this study, we prepared and purified the TAT-TR fusion protein, and also other control proteins. Transduction of TAT-fusion proteins was detected by immunofluorescence assay. Strong fluorescence appeared in the cytoplasm of the cells applied with four TAT fusion proteins, but not in the control groups. These results showed the high transduction efficiency of TAT fusion proteins. We also investigated TAT-TR’s anti-HBV activity by radioimmunoassay and got an exciting result. As compared with the mock group, concentration of HBeAg in TAT-TR group decreased by 60.3%, which indicated that the purified TAT-TR possessed a potent anti-HBV activity. There was no significant difference between any other group and the mock group, suggesting HBVc or hEDN alone had no effect on HBV replication, and TRmut, a fusion molecule of HBVc with an inactivated mutant hEDN, also had no inhibitory effect. These further confirmed the anti-HBV mechanism of the HBV targeted ribonuclease. While TR was transduced into 2.2.15 cells, it could be packaged into the HBV core particles by HBVc, and then hEDN, as a ribonuclease, could degrade the pgRNA packed in the core particles, thus inhibiting the replication of HBV. We performed MTT assay to reveal whether the TAT fusion proteins were harmful to 2.2.15 cells, and the results indicate that they did not affect the growth of 2.2.15 cells and the purified TAT-TR may be applied in vivo. Therefore, we conclude that the TAT-HBV targeted ribonuclease fusion protein obtained in this study has laid the foundation for using TR in the therapy of HBV infection.
Edited by Lu HM
| 1. | Becker-Hapak M, McAllister SS, Dowdy SF. TAT-mediated protein transduction into mammalian cells. Methods. 2001;24:247-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 287] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 2. | Schwarze SR, Dowdy SF. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol Sci. 2000;21:45-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 346] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 3. | Schwarze SR, Hruska KA, Dowdy SF. Protein transduction: unrestricted delivery into all cells. Trends Cell Biol. 2000;10:290-295. [PubMed] |
| 4. | Vivès E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010-16017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1811] [Cited by in RCA: 1839] [Article Influence: 65.7] [Reference Citation Analysis (0)] |
| 5. | Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci U S A. 1994;91:664-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 942] [Cited by in RCA: 921] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 6. | Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. In vivo protein transduction: delivery of a biologically active protein into the mouse. Science. 1999;285:1569-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1904] [Cited by in RCA: 1912] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 7. | Nagahara H, Vocero-Akbani AM, Snyder EL, Ho A, Latham DG, Lissy NA, Becker-Hapak M, Ezhevsky SA, Dowdy SF. Transduction of full-length TAT fusion proteins into mammalian cells: TAT-p27Kip1 induces cell migration. Nat Med. 1998;4:1449-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 736] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 8. | Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, Wang D, Dowdy SF, Hruska KA. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J Biol Chem. 2000;275:11993-12002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 211] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 9. | Shi H, Wang FS. Host factors in chronicity of hepatitis B virus infection and their significances in clinic. Shijie Huaren Xiaohua Zazhi. 2001;9:66-69. |
| 10. | Befeler AS, Di Bisceglie AM. Hepatitis B. Infect Dis Clin North Am. 2000;14:617-632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 11. | Maddrey WC. Hepatitis B: an important public health issue. J Med Virol. 2000;61:362-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 12. | Lau GK. Hepatitis B infection in China. Clin Liver Dis. 2001;5:361-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Merican I, Guan R, Amarapuka D, Alexander MJ, Chutaputti A, Chien RN, Hasnian SS, Leung N, Lesmana L, Phiet PH. Chronic hepatitis B virus infection in Asian countries. J Gastroenterol Hepatol. 2000;15:1356-1361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 343] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 14. | Guo SP, Wang WL, Zhai YQ, Zhao YL. Expression of nuclear factor-kappa B in hepatocellular carcinoma and its relation with the X protein of hepatitis B virus. World J Gastroenterol. 2001;7:340-344. [PubMed] |
| 15. | Shi DR, Lu L, Wang JH, Dong CL, Cong WT. HBV DNA distibution of hepatitis B virus in pancreas and liver of patients with cirrhosis. Shijie Huaren Xiaohua Zazhi. 2000;8:751-754. |
| 16. | Wu C, Cheng ML, Ding YS, Liu RC, Li J, Wang WL, Hu L. A five -year fellow up survey of risk factor of viral hepatic cirrhosis. Shijie Huaren Xiaohua Zazhi. 2000;8:1365-1367. |
| 17. | Wang HY, Yan RQ, Long JB, Wu QL. Cyclin D1 amplification is associated with HBV DNA intergration and pathology in human hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:98-100. |
| 18. | Fan ZR, Yang DH, Qin HR, Huang CC, Xu C, Qiu QL. Expression of IGF-I, IGF-I receptor mRNA in hepatocellular carcinomas and adjacent nontumor tissue. Shijie Huaren Xiaohua Zazhi. 1999;7:848-850. |
| 19. | Yan JC, Ma Y, Chen WB, Shun XH. Pathological significance of expression of intrahepatic smooth muscle fiber in hepatitis B. Shijie Huaren Xiaohua Zazhi. 2000;8:1242-1246. |
| 20. | Zhai SH, Liu JB, Liu YM, Zhang LL, Du ZH. Expression of HBsAg, HCV-Ag and AFP in liver cirrhosis and hepatocarcinoma. Shijie Huaren Xiaohua Zazhi. 2000;8:524-527. |
| 21. | Feitelson MA. Hepatitis B virus in hepatocarcinogenesis. J Cell Physiol. 1999;181:188-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 22. | Arbuthnot P, Kew M. Hepatitis B virus and hepatocellular carcinoma. Int J Exp Pathol. 2001;82:77-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 203] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 23. | Zhuang L, You J, Tang BZ, Ding SY, Yan KH, Peng D, Zhang YM, Zhang L. Preliminary results of Thymosin-a1 versus interferon-alpha-treatment in patients with HBeAg negative and serum HBV DNA positive chronic hepatitis B. World J Gastroenterol. 2001;7:407-410. [PubMed] |
| 24. | Xie Q, Guo Q, Zhou XQ, Gu RY. Effect of adenine arabinoside monophosphate coupled to lactosaminated human serum albumin on duck hepatitis B virus. Shijie Huaren Xiaohua Zazhi. 1999;7:125-126. |
| 25. | Li J, Tang B. Effect on replication of hepatitis B virus by Chinese traditional medicine. Shijie Huaren Xiaohua Zazhi. 2000;8:945-946. |
| 26. | Zhu Y, Wang YL, Shi L. Clinical analysis of the efficacy of interferon alpha treatment of hepatitis. World J Gastroenterol. 1998;4:85-86. |
| 27. | Xu KC, Wei BH, Yao XX, Zhang WD. Recently therapy for chronic hepatitis B virus by combined traditional Chinese and Western medicine. Shijie Huaren Xiaohua Zazhi. 1999;7:970-974. |
| 28. | Lau GK. Use of immunomodulatory therapy (other than interferon) for the treatment of chronic hepatitis B virus infection. J Gastroenterol Hepatol. 2000;15 Suppl:E46-E52. [PubMed] |
| 29. | Guan R. Interferon monotherapy in chronic hepatitis B. J Gastroenterol Hepatol. 2000;15 Suppl:E34-E40. [PubMed] |
| 30. | Torresi J, Locarnini S. Antiviral chemotherapy for the treatment of hepatitis B virus infections. Gastroenterology. 2000;118:S83-103. [PubMed] |
| 31. | Schiff ER. Lamivudine for hepatitis B in clinical practice. J Med Virol. 2000;61:386-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 32. | Jarvis B, Faulds D. Lamivudine. A review of its therapeutic potential in chronic hepatitis B. Drugs. 1999;58:101-141. [PubMed] |
| 33. | Malik AH, Lee WM. Chronic hepatitis B virus infection: treatment strategies for the next millennium. Ann Intern Med. 2000;132:723-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 109] [Article Influence: 4.4] [Reference Citation Analysis (1)] |
| 34. | Farrell GC. Clinical potential of emerging new agents in hepatitis B. Drugs. 2000;60:701-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 30] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Song YH, Lin JS, Liu NZ, Kong XJ, Xie N, Wang NX, Jin YX, Liang KH. Anti-HBV hairpin ribozyme-mediated cleavage of target RNA in vitro. World J Gastroenterol. 2002;8:91-94. [PubMed] |
| 36. | Li YH, Liu J, Xue CF. Construction of HBV targeted ribonuclease and its expression in 2.2.15 cell line. Xibao Yu Fenzi Mianyixue Zazhi. 2002;18:217-220. |
| 37. | Liu J, Li YH, Xue CF, Ding J, Gong WD, Zhao Y, Huang YX. Targeted ribonuclease can inhibit replication of hepatitis B virus. World J Gastroenterol. 2003;9:295-299. [PubMed] |
| 38. | Ding J, Liu J, Xue CF, Li YH, Gong WD. [Construction and expression of prokaryotic expression vector for pTAT-HBV targeted ribonuclease fusion protein]. Xibao Yu Fenzi Mianyixue Zazhi. 2003;19:49-51. [PubMed] |
| 39. | Green M, Loewenstein PM. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell. 1988;55:1179-1188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1090] [Cited by in RCA: 1126] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 40. | Frankel AD, Pabo CO. Cellular uptake of the tat protein from human immunodeficiency virus. Cell. 1988;55:1189-1193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2021] [Cited by in RCA: 2103] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 41. | Derossi D, Joliot AH, Chassaing G, Prochiantz A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444-10450. [PubMed] |
| 42. | Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 778] [Cited by in RCA: 754] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 43. | Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, Wang D, Dowdy SF, Hruska KA. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J Biol Chem. 2000;275:11993-12002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 211] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 44. | Schutze-Redelmeier MP, Gournier H, Garcia-Pons F, Moussa M, Joliot AH, Volovitch M, Prochiantz A, Lemonnier FA. Introduction of exogenous antigens into the MHC class I processing and presentation pathway by Drosophila antennapedia homeodomain primes cytotoxic T cells in vivo. J Immunol. 1996;157:650-655. [PubMed] |
| 45. | Gottesman S, Wickner S, Maurizi MR. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 408] [Article Influence: 14.6] [Reference Citation Analysis (0)] |