Published online Jul 15, 2003. doi: 10.3748/wjg.v9.i7.1496
Revised: March 4, 2003
Accepted: March 16, 2003
Published online: July 15, 2003
AIM: To analyze the molecular evolution of different viral genomic regions of HCV in an acute HCV infected patient chronically infected with HIV through a 42-month follow-up.
METHODS: Serum samples of a chronically HIV infected patient that seroconverted to anti HCV antibodies were sequenced, from the event of superinfection through a period of 17 mo and in a late sample (42nd month). Hypervariable genomic regions of HIV (V3 loop of the gp120) and HCV (HVR-1 on the E2 glycoprotein gene) were studied. In order to analyze genomic regions involved in different biological functions and with the cellular immune response, HCV core and NS5A were also chosen to be sequenced. Amplification of the different regions was done by RT-PCR and directly sequenced. Confirmation of sequences was done on reamplified material. Nucleotide sequences of the different time points were aligned with CLUSTAL W 1.5, and the corresponding amino acid ones were deduced.
RESULTS: Hypervariable genomic regions of both viruses (HVR1 and gp120 V3 loop) presented several nonsynonymous changes but, while in the gp120 V3 loop mutations were detected in the sample obtained right after HCV superinfection and maintained throughout, they occurred following a sequential and cumulative pattern in the HVR1. In the NS5A region of HCV, two amino acid changes were detected during the follow-up period, whereas the core region presented several amino acid replacements, once the HCV chronic infection had been established.
CONCLUSION: During the HIV-HCV superinfection, each genomic region analyzed shows a different evolutionary pattern. Most of the nucleotide substitutions observed are non-synonymous and clustered in previously described epitopes, thus suggesting an immune-driven evolutionary process.
- Citation: Flichman D, Kott V, Sookoian S, Campos R. Acute hepatitis C in a chronically HIV-infected patient: Evolution of different viral genomic regions. World J Gastroenterol 2003; 9(7): 1496-1500
- URL: https://www.wjgnet.com/1007-9327/full/v9/i7/1496.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i7.1496
It has been proven that patients with HIV infection are frequently coinfected with other viruses, including that of HCV. Both HIV and HCV share the same route of transmission, and thus, coinfection with these two viruses is rather common among intravenous drug users or transfused patients[1,2]. HCV infection is considered as an HIV opportunistic disease[3].
The clinical impact of HIV-HCV coinfection has been widely studied[4]. However, the viral molecular interaction during the establishment of HCV superinfection is not yet well understood as the acute phase is frequently a subclinical event.
HCV, an RNA virus, shows a marked variability but nucleotide substitutions are unevenly distributed along the entire genome[5]. HCV diversity is the greatest in the envelope proteins E1 and E2, especially in a 27 amino-acid segment at the N-terminus of E2 designated hypervariable region 1 (HVR1). The variation of this region is thought to be related to the maintenance of persistent infection by emerging escape variants[6] and it is considered as the main target for humoral immunity as well as an immunologic decoy[7].
A similar degree of heterogeneity is found within the gp120 V3 loop region of HIV, an RNA virus that also causes persistent infections in the host. This region has been found to elicit neutralizing antibodies as well as cytotoxic and Th-cell responses, properties that are also ascribed to the HVR1[8].
NS5A is an HCV nonstructural protein that is associated with several functions such as being an in vitro transcriptional repressor[9] and tightly associated via the "interferon sensitivity determining region" (ISDR), with the subversion of the IFN activity. ISDR inhibits the cellular double-stranded RNA-activated protein kinase R[10], an effector of the IFN antiviral activity. There is also evidence that the C-terminal domain of NS5A including the ISDR contains transcriptional activity[11], suggesting that NS5A might function as a viral transactivator. The core gene is located in the 5' end of the HCV genome and its primary function is to form the viral nucleocapsid. The core protein has many effects on host-cell signalling pathways that includes the host-cell gene expression[12], apoptosis by interaction with the cytoplasmic tail of the lymphotoxin receptor and with TNF receptor[13,14], transforming activity[15], modulation of lipid metabolism[16,17]. Several studies have shown that the region contains multiple and highly immunogenic humoral[18,19] and cellular [19,20] epitopes.
In a previous report we described the in vivo down regulation of HIV replication in an HIV-infected patient after HCV superinfection[21]. Herein we presented a further study of this patient through the analysis of hypervariable regions of both viruses, and the less variable core and NS5A regions of HCV.
A 16 year-old HIV patient was referred to the Liver Unit because of an acute hepatitis-like illness with the manifestations of jaundice, nausea, abdominal pain, fever, diarrhea, itching and elevated ALT and AST levels. She was in the 30th week of gestation and her prenatal course had been uneventful until this episode. Diagnosis of acute hepatitis C was established by seroconversion of HCV antibodies by an EIA test (HCV EIA 2.0; Ortho Diagnostics) and confirmed with a strip immunoblot assay (RIBA HCV 2.0; Chiron Corporation). HIV and HCV were thought to be acquired by intravenous drug use. Serum HCV and HIV RNAs were detected by RT-PCR using primers for the 5NC and the gag regions, respectively[21] (Table 1).
| Viral region | Nucleotide position | Primer sequence (5'→3') | PCR cycling parameters |
| HIV | 5 min at 94 °C, 40 cycles of | ||
| gag | 30 s at 94°C, 30 s at 55°C and | ||
| OS | 4653-4675 | CCCTACAATCCCCAAAGTCAAGG | 45 s at 72°C and a final |
| OA | 4956-4976 | TACTGCCCCTTCACCTTTCCA | extension of 7 min at 72 °C |
| V3 loop | |||
| OS | 6957 - 6976 | GTACAATGTACACATGGAAT | |
| OA | 7357 - 7376 | GTAGAAAAATTCCCCTCCAC | Idem HIV-gag PCR |
| IS | 7010 - 7029 | TGGCAGTCTAGCAGAAGAAG | |
| IA | 7319 - 7339 | ACAATTTCTGGGTCCCCTCCT | |
| HCV | |||
| 5UT | |||
| OS | 44 - 69 | CCTGTGAGGAACTACTGTCTTCACGC | Idem HIV-gag PCR |
| OA | 321 - 341 | GGTGCACGGTCTACGAGACCT | |
| HVR1 | |||
| OS | 1273-1296 | GCCATATAACGGGTCACCGCATGG | |
| OA | 1681-1704 | TCTCAGGACAGCCTGAAGMGTTGA | Idem HIV-gag PCR |
| IS | 1296-1379 | GCATGGGATATGATGATGAACTGG | |
| IA | 1623-1646 | GGTGTTGAGGCTATCATTGCARTT | |
| core | 5 min at 94 °C, 30 cycles of | ||
| OS | 272 - 291 | CGAAAGGCCTTGTGGTACTG | 1 min at 94 °C, 1 min at 50 °C |
| OA | 1020 - 1037 | CTCGCGRACGCAAGGGAC | and 2 min at 72 °C and a final |
| IS | 281 - 302 | TTGTGGTACTGCCTGATAGGGT | extension of 10 min at 72 °C |
| IA | 956 - 976 | ATACTCGAGTTAGGGCAATCA | |
| NS5A | |||
| OS | 6716 - 6739 | TAGATGGGGTGCGCCTGCAYAGGTT | |
| OA | 7310 - 7332 | GCTTCTTCCGRGGCGGAGGCACWG | Idem HCV core PCR |
| IS | 6734-6753 | TAGGTTTGCGCCCCCCTGMA | |
| IA | 7287-7306 | GGGGACYKTGGAGGTGGAAG |
Thereafter, the patient was followed up for 42 mo and serum samples were taken monthly during the first 17 mo of the analyzed period and in the 42nd month as a late specimen. Serum samples before HCV superinfection were also available. Liver function tests (AST, ALT, γ-glutamyl-transferase, alkaline phosphatase, albumin, gamma globulin, bilirubin and prothrombin time) were routinely performed using automatic standard procedures.
A liver biopsy was obtained at the 15th month of follow-up showing a histological picture of chronic hepatitis (Knodell score 5) without fibrosis.
No antiviral therapy was implemented during the study period because of the patient's refusal. This study followed the ethical standards of the World Medical Association Helsinki Declaration adopted in 1964 and amended in 1996 and was approved by the local Ethics Committee. At all times, written consents were obtained from the woman.
RNA extraction RNA was recovered from 150 mL aliquots as described by Chomczynski and Sacchi[22] using components from a commercially available RNA extraction kit (Trizol, Life Technologies) and resuspended in 20 μL dH2O.
cDNA synthesis and PCRs RNA was used as templates in randomly primed reverse transcription reactions to produce cDNA in the following conditions: 50 mM TrisHCl (pH8.3), 25 mM KCl, 3 mM MgCl2, 0.1 mM DTT, 1.0 mM dNTPs, 18 U RNasin ribonuclease inhibitor (Promega) and 100 U Moloney murine leukemia virus RT (Life Technologies). The reactions were performed for 90 min at 37 °C. After heat inactivation at 95 °C for 5 min and chilling on ice, the cDNA was amplified.
Different genomic viral regions -gp120 V3 loop region of HIV and the HVR1, core and NS5A regions of HCV- were amplified and directly sequenced. The PCR reaction (50 μL) contained 20 mM TrisHCl, 50 mM KCl, 50 pmol of each primer and 1.25 U Taq DNA polymerase (Promega). Nested PCR was performed with 2 μL PCR product as a template, using internal primers under the same conditions as the first round. The first and second round primers and cycling parameters for each region are shown in Table 1 and were made according to Kwok and Higuchi's recommendations[23].
Nucleotide sequencing Each DNA was purified and directly sequenced with a Thermo Sequenase radiolabeled-terminator-cycle sequencing kit (U.S. Biochemical Corporation) using the same cycling parameters of the second PCR round. Confirmatory sequencing was performed on reamplified DNA.
The genomic regions of HCV: HVR1, core and NS5A and the gp120 V3 loop of HIV were sequenced during a 42 month follow-up of a chronically HIV patient acutely infected with HCV. Phylogenetic analysis of core and NS5A grouped the HCV sequences into genotype 1a (data not shown).
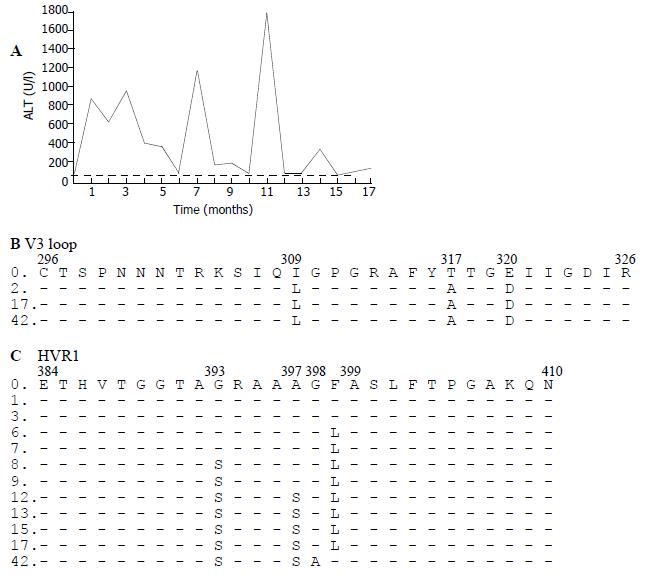
After alignment and comparison of genomic sequences obtained before and after HCV superinfection, the nucleotide sequence of HIV gp120 V3 loop showed three nonsynonymous mutations (I309L, T317A and E320D) that appeared in the sample obtained 2 mo after HCV superinfection and maintained thereafter for the rest of the period (Figure 1).
As far as HCV sequences were concerned, in the HVR1 three nonsynonymous mutations were observed (F399L, G393S and A397S) in the acute phase of the infection, conversely, the mutation dynamics followed a sequential and cumulative pattern, occurring at the 6th, 8th and 12th follow-up months. Each mutation was associated with an ALT flare-up (Figure 1). In the last sample, 42 mo after HCV superinfection, two more amino acid changes were selected (G398A and L399F).
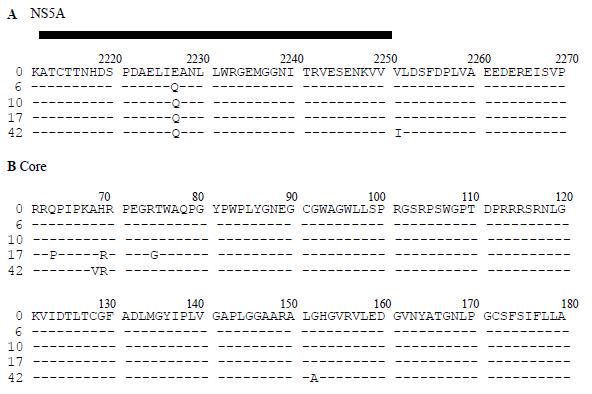
Different patterns of substitutions were observed in the HCV core and NS5A regions. In the core, three nonsynonymous substitutions (Q63P, H69R and R74G) appeared at the 17th month of the follow-up (Figure 2), once chronic hepatitis was fully established. Late on the follow-up, four mutations were selected: P63Q, A68V, G74R and G152A. The amino acids that were resulted from the modifications in positions 63 and 74 were the same as that which was present in the earliest samples.
In the portion of NS5A sequenced (nt 6632-7077), one amino acid substitution was observed in the acute phase (E2227Q) and remained thereafter, and another one was selected at the end of the follow-up (V2251I).
The sequences were submitted to GenBank and assigned accession numbers: AF361170 through AF361176 and AF359345 through AF359354.
In this study, we analyzed the evolution of different viral genomic regions during HCV superinfection of a chronically HIV infected patient.
Most of the nucleotide substitutions fixed during the HCV superinfection process were nonsynonymous and located at described epitopes, suggesting a positive immune selection mechanism driving their molecular evolution.
We found that hypervariable regions of both HCV and HIV had several amino acid modifications during the acute phase of superinfection. However, these changes occurred simultaneously on the gp120 V3 loop whereas they were sequentially selected and showed a cumulative pattern in the HVR1.
The selection of an HIV viral population with three amino acid modifications in the V3 loop region right after HCV superinfection maybe resulted from the overgrowth of markedly different but minor HIV quasispecies from the initial sample.
In contrast, the HVR1 evolved showed a cumulative pattern of mutations during the 17 month period. Each time of sequential incorporation of a nonsynonymous mutation was associated with an ALT flare up and resulted from the selection process, also exerted by the evolving host immune response[24].
It has been proposed that the evolution of hypervariable regions may be caused by the successive accumulation of point substitutions as we have seen in the HVR1-HCV or, alternatively from the selection of a pre-existing minor subpopulations, which was different from the previous one, as observed in V3 loop of HIV[25].
Once the chronic infection was established, two extra amino acid mutations were observed in the HVR1, the first one was at position 398 (G→A), and the second one was at position 399 (L→F). F or L occupied the last position alternatively in most of the HVR sequences published elsewhere[26] and may be considered as a return to a previous state of the quasispecies equilibrium.
The timing of the substitutions observed in the core region, which were late (after 17 mo and 42 mo of infection), implied its involvement in the infection once persistence had been established. Non-synonymous changes clustered in HCV epitopes that stimulated the humoral and cellular response and were previously described[18,20,27], suggesting an active participation of these epitopes in the persistence of HCV infection. Moreover, the sample at the end of the follow-up showed two modifications (aa 63 and 74 of the core protein) that recovered the early amino acid sequences. As far as the HVR sequences concerned, this effect that might be due to the return to a quasispecies equilibrium state favored the outgrowth of the fittest quasispecies for that particular host conditions, as a consequence from the interplaying of virus populations in every situation.
Two substitutions were selected in the NS5A region. One of them occurred at the early phase of the infection whereas the second appeared at the end of the follow-up. The first mutation selected (E2227Q) was located in the ISDR, described as related to the IFN resistance of some HCV genotypes by means of its interactions with the cellular PKR and in a cellular epitope, as previously described[19].
In conclusion, during the HCV superinfection process, each genomic region analyzed presents a different pattern of evolution, and is probably related to their functions in the viral life cycle. The localization of mutations at described epitopes suggests the participation of the host immune system drives the balance of the viral subpopulations towards escape and better fitness ones. In order to study this hypothesis, a quasispecies analysis is worth to be done to address the changes showed during the coinfection by means of the population equilibriums.
Edited by Xu XQ and Wang XL
| 1. | Waldrep TW, Summers KK, Chiliade PA. Coinfection with HIV and HCV: more questions than answers. Pharmacotherapy. 2000;20:1499-1507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 2. | Laskus T, Radkowski M, Piasek A, Nowicki M, Horban A, Cianciara J, Rakela J. Hepatitis C virus in lymphoid cells of patients coinfected with human immunodeficiency virus type 1: evidence of active replication in monocytes/macrophages and lymphocytes. J Infect Dis. 2000;181:442-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 170] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 3. | Centers for Disease Control and Prevention. 1999 USPHS/IDSA guidelines for the prevention of oppotunistic infections in persons infected with human immunodeficiency virus: disease-specific recommendations. USPHS/IDSA Prevention of Opportunistic Infections Working Group: U. S: Public Health Services/Infectious Diseases Society of America. Morbility and Mortality Weekly Report 1999; 1-82. |
| 4. | Rockstroh JK, Woitas RP, Spengler U. Human immunodeficiency virus and hepatitis C virus coinfection. Eur J Med Res. 1998;3:269-277. [PubMed] |
| 5. | Rispeter K, Lu M, Behrens SE, Fumiko C, Yoshida T, Roggendorf M. Hepatitis C virus variability: sequence analysis of an isolate after 10 years of chronic infection. Virus Genes. 2000;21:179-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 6. | Smith DB. Evolution of the hypervariable region of hepatitis C virus. J Viral Hepat. 1999;6 Suppl 1:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Ray SC, Wang YM, Laeyendecker O, Ticehurst JR, Villano SA, Thomas DL. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2938-2946. [PubMed] |
| 8. | Hwang SS, Boyle TJ, Lyerly HK, Cullen BR. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 684] [Cited by in RCA: 696] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 9. | Ghosh AK, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol. 1999;80:1179-1183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 146] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 10. | Gale MJ, Korth MJ, Tang NM, Tan SL, Hopkins DA, Dever TE, Polyak SJ, Gretch DR, Katze MG. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 631] [Cited by in RCA: 608] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 11. | Kato N, Lan KH, Ono-Nita SK, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856-8859. [PubMed] |
| 12. | McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J Viral Hepat. 2000;7:2-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 230] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 13. | Matsumoto M, Hsieh TY, Zhu N, VanArsdale T, Hwang SB, Jeng KS, Gorbalenya AE, Lo SY, Ou JH, Ware CF. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-beta receptor. J Virol. 1997;71:1301-1309. [PubMed] |
| 14. | Zhu N, Khoshnan A, Schneider R, Matsumoto M, Dennert G, Ware C, Lai MM. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J Virol. 1998;72:3691-3697. [PubMed] |
| 15. | Jin DY, Wang HL, Zhou Y, Chun AC, Kibler KV, Hou YD, Kung H, Jeang KT. Hepatitis C virus core protein-induced loss of LZIP function correlates with cellular transformation. EMBO J. 2000;19:729-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 104] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 16. | Barba G, Harper F, Harada T, Kohara M, Goulinet S, Matsuura Y, Eder G, Schaff Z, Chapman MJ, Miyamura T. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc Natl Acad Sci U S A. 1997;94:1200-1205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 499] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 17. | Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, Matsuura Y, Miyamura T, Bréchot C, Barba G. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 138] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Akatsuka T, Donets M, Scaglione L, Ching WM, Shih JW, Di Bisceglie AM, Feinstone SM. B-cell epitopes on the hepatitis C virus nucleocapsid protein determined by human monospecific antibodies. Hepatology. 1993;18:503-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 14] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 19. | Ward S, Lauer G, Isba R, Walker B, Klenerman P. Cellular immune responses against hepatitis C virus: the evidence base 2002. Clin Exp Immunol. 2002;128:195-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Koziel MJ, Dudley D, Afdhal N, Choo QL, Houghton M, Ralston R, Walker BD. Hepatitis C virus (HCV)-specific cytotoxic T lymphocytes recognize epitopes in the core and envelope proteins of HCV. J Virol. 1993;67:7522-7532. [PubMed] |
| 21. | Flichman D, Cello J, Castaño G, Campos R, Sookoian S. In vivo down regulation of HIV replication after hepatitis C superinfection. Medicina (. B Aires). 1999;59:364-366. [PubMed] |
| 22. | Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156-159. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40518] [Cited by in RCA: 39097] [Article Influence: 1028.9] [Reference Citation Analysis (0)] |
| 23. | Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2583] [Cited by in RCA: 2475] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 24. | Hjalmarsson S, Blomberg J, Grillner L, Pipkorn R, Allander T. Sequence evolution and cross-reactive antibody responses to hypervariable region 1 in acute hepatitis C virus infection. J Med Virol. 2001;64:117-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Lu L, Nakano T, Orito E, Mizokami M, Robertson BH. Evaluation of accumulation of hepatitis C virus mutations in a chronically infected chimpanzee: comparison of the core, E1, HVR1, and NS5b regions. J Virol. 2001;75:3004-3009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 26. | Penin F, Combet C, Germanidis G, Frainais PO, Deléage G, Pawlotsky JM. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J Virol. 2001;75:5703-5710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 136] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 27. | Beld M, Penning M, van Putten M, Lukashov V, van den Hoek A, McMorrow M, Goudsmit J. Quantitative antibody responses to structural (Core) and nonstructural (NS3, NS4, and NS5) hepatitis C virus proteins among seroconverting injecting drug users: impact of epitope variation and relationship to detection of HCV RNA in blood. Hepatology. 1999;29:1288-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 53] [Article Influence: 2.0] [Reference Citation Analysis (0)] |