INTRODUCTION
It has been well known in recent years that dioxin-like chemicals (DLCs) such as 2, 3, 7, 8- tetrachlorodibenzo-p-dioxin (TCDD) can produce a wide variety of species- and tissue- specific toxic and biological effects, such as epidermal lesion, wasting syndrome, birth defect, hepatotoxicity, lethality, alteration in endocrine homeostasis, tumor promotion, myelotoxicity, immunotoxicity and induction of numerous enzymes (e.g. cytochrome P4501A1)[1-4]. Many of these responses are mediated by the cytosolic aryl hydrocarbon receptor (AhR) and modulated by the interaction of DLCs: AhR complex with its DNA recognition sequence [the dioxin-responsive element (DRE)][5-9]. Generally, the combination of DLCs with cytoplasmic AhR is the initial step of DLCs-triggered cell signaling pathway. After that, two molecules of hsp90 dissociate from DLCs: AhR complex, and the latter enters the nucleus to form a new complex with AhR nuclear translocator protein (ARNT), which further binds to the DRE, leading to the transcriptional activation of adjacent genes and toxic effects[10,11]. Therefore, AhR and its related signal pathway have been usually taken as the useful targets to be investigated for the determination of DLCs in different situations. Till now, the DLCs-responsive receptor assays have been established in mammalian cell lines[12] and in mice[13].
However, not all of these assays have been considered to be satisfactory yet. For example, the ethoxyresorufin-O-deethylase (EROD) induced as a common and rapid response to DLCs under DREs control has been widely used to evaluate the relatively toxicological potency of a complex mixture containing DLCs[14-16] except that higher concentrations appear when the enzyme activity is inhibited. Some detecting methods are time-consuming, expensive and inadequate to be used for screening and diagnosing dioxin and dioxin-like compounds in large numbers of samples[17-19]. Even if the chemical-activated luciferase expression (CALUX) bioassay method based on the pGL2 vector[20] is to a certain extent lack of sensitivity and needs to be improved.
We therefore conducted the present study in an attempt to establish a more effective luciferase reporter cell line that responds to the alteration of DLCs concentrations and to evaluate its characteristics and applications in measurement of DLCs.
MATERIALS AND METHODS
Materials
Plasmids pHAV and pMcat were generously provided by Dr. James P. Whitlock Jr., Stanford University. Restriction endonuclease and other reagents used for molecular cloning were from Huamei Ltd. (China, Shanghai) or Promega Corporation. 2, 3, 7, 8-TCDD and 3', 4, 4'- polychlorinated biphenyl (PCB) were purchased from Accustanfards Inc. (New Haven, CT). Dimethylsulphoxide (DMSO) was from BIB Corporation. Luciferase assay reagents came from Promega Corporation.
Construction of inducible luciferase expression vector
Plasmid pHAV containing DREs was cleaved with HindIII. Following purification by agarose gel electrophoresis, the smallerHindIII fragment was further digested by BamHI and EcoRV to produce a 630-bp segment containing DREs, which was then ligased with BglII linkers and cleaved with BglII to create cohesive ends. To get the MMTV promoter, plasmid pMCat was cleaved with HindIII and the smaller HindIII fragment was then cleaved with BglII to produce cohesive 5' BglII and 3' HindIII termini. The two segments containing DREs and MMTV promoter were connected with T4 DNA ligase to form a 1020 bp fragment with cohesive 5' BglII and 3' HindIII termini, which was further subcloned into the BglII- HindIII site immediately upstream of the luciferase gene in the pGL3-promoter plasmid (Promega). As such, the luciferase's expression was under the control of DREs. The recombinant vector was identified structurally with restriction endonuclease analysis.
Inducible expression of recombinant vector
HepG2 cells were seeded in a 60 mm culture dish at a density of 3 × 105 cells in 5 mL of RPMI1640 supplemented with 10% heat-inactivated fetal calf serum at 37 °C with saturated humidity and a 5% (v/v) CO2 atmosphere[21]. After cultured for 24 h, the cells were transiently or stably transfected by 15 μg of recombinant vector with calcium phosphate mediated method[22]. For transient transfection, the cells were allowed to grow for 48 h, followed by adding 0.5% DMSO or 0.1-1 nmol/L TCDD dissolved in DMSO. After cultivated for another 24 h[22], cells were harvested to determine luciferase expression.
For the establishment of stable transfection, HepG2 cells were cotransfected with the selective plasmid PTK-Hyg and the recombinant vector simultaneously. Following 24 h of cultivation in nonselective medium, the transfected cells were transferred into a selective medium containing hygromycin and cultured for 4 wk when TCDD-induced luciferase expression was conducted. The clone with the largest ratio of inducible to constitutive expression of luciferase was selected for further study[23]. To identify the time course of inducible luciferase expression, the stably transfected cells (HepG2-Luc) were cultured with exposure to 0.5% DMSO or 1 nmol/L TCDD and the induced luciferase activities were determined at every 2 h up to 28 h post-exposure. For determination of the responsive period of luciferase induction, the HepG2-Luc cells were treated with the same kinds of inducers for 24 h and their luciferase activity was assayed, which was performed at every month up to 12 mo. The structure-inducibility and dose-effect relationships of different inducers that might represent their ability to bind AhR were also analyzed in HepG2-Luc cells with the indicated concentrations of 3, 3', 4, 4'-PCB, another member of DLCs, and TCDD.
Luciferase assay was performed by the routine procedures. Briefly, after removal of the culture medium, the incubated cells were washed twice with phosphate buffered saline (PBS) and lysed using luciferase lysis reagent for 15 min at room temperature. Following centrifugation, 20 μL of supernatant was added into 100 μL luciferase assay reagent, the resulting bioluminescence was quantified immediately with Lumate LB 9570 luminometer over 3 s. The concentration of the sample protein was determined with Bio-Rad method[24]. The luciferase activity was finally expressed as relative light unit (RLU) per microgram protein[21,25].
Comparative study with EROD determination
EROD activity was employed as a variable to evaluate the effectiveness of HepG2-luc cells in response to TCDD and to compare with luciferase. HepG2 (HepG2-wt) and HepG2-luc cells were seeded in 6-well dishes at a density of 2 × 105 cells in 3 mL of medium and cultivated for 24 h, then they were exposed to the indicated concentrations of TCDD in DMSO and further incubated for 72 h. Following removal of the culture medium, the cells were washed twice with PBS and stored at -80 °C. EROD activity was assayed using fluorescent method. The concentrations of sample protein were determined with Bio-Rad method[24]. EROD activity was finally expressed as pmol resorufin production per microgram protein per minute[26,27].
Statistical analysis
Data values in the present study were presented as the mean for each independent determination of four replicates. The detection limit was expressed as the average value plus three times of standard deviations (SD). Coefficients of variation (CV) were calculated by SD/mean × 100%. Correlation coefficients (r) were obtained using least-squares linear regression analysis.
RESULTS
Identification of inducible expression of recombinant vector
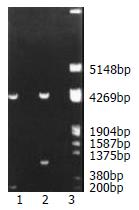
Structurally, segment analysis by restriction endonuclease digestion confirmed that the inserted fragment sequences containing DREs and MMTV promoter in the constructed plasmids were completely consistent with that of the theoretical calculations as shown in Figure 1.
Figure 1 Fragment mapping analysis of recombinant plasmid digested with BglII and HindIII in gel electrophoresis.
Lane 1: pGL3 plasmid was cleaved into two fragments, which showed about 5000 bp and 200 bp respectively. Lane 2: Recombinant plasmid was cut into a fragment of about 5000 bp and the one of about 1000 bp. Lane 3: DNA marker.
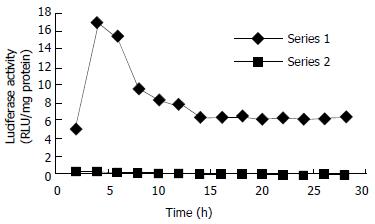
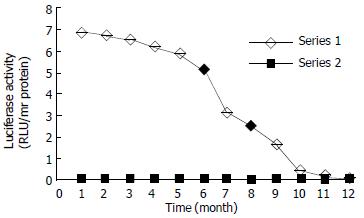
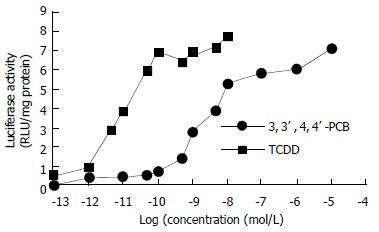
Inducible expression of luciferase by the recombinant plasmid was identified in transiently transfected HepG2 cells, which produced a significantly higher induction of luciferase activity (80-fold, data not shown) compared with that of their non-transfected counterparts when exposed to TCDD. Dynamically, TCDD-induced luciferase activity in HepG2-Luc cells peaked at 4 h and then decreased to a stable level at about 14 h after TCDD treatment (Figure 2). Responsiveness of HepG2-Luc cells to the TCDD induction was decreased with culture time and became undetectable at 10th month of HepG2-Luc cell formation (Figure 3), which indicats that the recombinant plasmid could not integrate stably with HepG2 chromosome. The structure-inducibility relationship of different DLCs inducers was demonstrated by the fact that the luciferase activity induced by 3, 3', 4, 4'-PCB was much less than that induced by TCDD and a dose-dependent increase of luciferase activity was evoked by both of the DLCs inducers as shown in Figure 4.
Figure 2 The time course of luciferase activities induced by TCDD and DMSO in stably transfected cells.
The TCDD-induced luciferase activity in HepG2-Luc cells peaked at about 4 h, and then decreased to a stable level at about 14 h after TCDD treatment.
Figure 3 Responsive period of HepG2-Luc cells to TCDD induction.
The responsiveness of HepG2-Luc cells to TCDD induction was decreased with culture time and became undetectable at about 10th month.
Figure 4 Comparison of luciferase activity induced by TCDD and 3, 3', 4, 4'-PCB in HepG2-Luc cells.
The luciferase activity induced by indicated concentrations of 3, 3', 4, 4'-PCB was much less than that induced by TCDD.
Comparative study
Within concentrations from 3.5 × 10-12 to 5 × 10-9 mol/L, significant correlations between TCDD doses and EROD activities were observed in both HepG2-luc and HepG2-wt cells. The correlation between TCDD doses from 1.1 × 10-13 to 1 × 10-8 mol/L and luciferase activities was also found to be significant in HepG2-luc cells (r = 0.997, P < 0.001), but not in their HepG2-wt counterparts. For comparison of the enzyme responsiveness between cell lines to TCDD, the luciferase sensitivity and reproducibility in HepG2-luc cells were better than those of EROD in HepG2-wt cells, which were 1.1 × 10-13 mol/L and 3.5 × 10-12 mol/L, whose CV was 15%-30% and 22%-38% (data not shown), respectively.
DISCUSSION
Few environmental compounds have generated much interest within the scientific community and in the lay public as polychlorinated dibenzo-p-dioxins (PCDDs) and polychlorinated dibenzofurans (PCDFs). Their ubiquitous presence in the environment and the risk of accidental exposure have raised concern over a possible threat of PCDDs or PCDFs to human health. The most extensively studied potent isomer is TCDD or dioxin, which is known to induce a wide range of toxic and biochemical responses in laboratory animals and humans[8,11]. Therefore, monitoring the levels of DLCs in environmental pollutants is important for assessing and maintaining the safety of food and the health of environment. Exploring new biodetectors such as bioassays, biomarkers, enzyme immunoassays (EIAs), or other bioanalytical tools has been a continuously growing area in recent years[15]; in which, however, establishing luciferase reporter cell lines for bioassays has been considered as the rapid, sensitive and inexpensive methods for the screening and diagnosis of dioxin and dioxin-like compounds[20,28].
In the present study, we constructed a recombinant expression plasmid that contained the luciferase gene under TCDD-inducible control of several DREs and responded to DLCs with the induction of firefly luciferase, which has been identified by both transient and stable transfection of this vector into HepG2 cells. Our results indicate that this established HepG2-luc cell bioassay reporter system can respond sensitively to DLCs with the induction of luciferase in a time-, dose-, and AhR-dependent manner and harbors lots of better features. Firstly, it bears higher level of sensitivity and can detect TCDD within the linear range from 1.1 × 10-13 to 1 × 10-8 mol/L, which is 10-fold more sensitive than that of previously similar studies and 30-fold more sensitive than the EROD assay. This is partially because the vector plasmid employed in the study is pGL3, whose expression efficiency of luciferase is 10-100 times more than that of pGL2. In addition, no post- transcriptional procedures are needed to regulate the expression of prokaryotic luciferase, thus its linear relationship is better than that of endogenous gene. Secondly, its pronounced accuracy has been demonstrated by the significant correlation between inducible luciferase activities and TCDD doses (r = 0.997, P < 0.001), and by the satisfactory reproducibility of TCDD determination with HepG2-luc cells (CV = 15%-30%). Finally, the simplicity and facility of HepG2-luc cells in analytical methodology make it possible to perform a rapid screening and semi-quantitation of DLCs[29,30] and to deal with lots of samples in a short period of time.
Besides the detection of DLCs described above, HepG2-luc cells can also be applied to many other DLCs-related research fields. Since ARNT is required to exert biological effects by some TCDD and related ligands, interactions between AhR and hypoxia signaling pathways can be studied by using luciferase transfected cell line[31]. In accordance with the fact that the toxic factor of 3, 3', 4, 4'-PCB equivalent to TCDD is 0.01[32], our results showed that the ability of 3, 3', 4, 4'-PCB to induce luciferase expression was much less than that of TCDD, suggesting that the structure-inducibility relationship existing in some DLCs could be reflected by HepG2-luc cells, and the latter can be used alone or with chromatographer to evaluate the relative toxicity of DLCs[19,33,34]. In addition, this kind of recombinant cell lines can also be used for the detection and relative quantitation of AhR agonists/antagonists in complex mixtures of environmental and biological samples, for identification and characterization of novel AhR agonists, and for examination of species differences in DLCs responsiveness[12].
One limitation of the present study is the relatively short duration of responsiveness to DLCs by established HepG2-luc cells, although it is in accordance with that reported by most of the other studies. Up to the present, many established luciferase reporter cell lines such as MLE/BV, H4IIE, GPC16 and HGL1.1c3 responding to DLCs maintain their stable TCDD responsiveness for 6-12 mo, except for Hepa1 which maintains its sensitivity for over 3 years[27]. Therefore, our further study has been designed aiming at improving and maintaining the sensitivity of HepG2-luc cells to DLCs for a longer time.