INTRODUCTION
Aldose reductase (AR) is a NADPH-dependent enzyme that is involved in diabetic complications[1-17]. Some reports showed that AR was induced in rat hepatoma, suggesting that it may be essential for detoxifying harmful metabolites produced by fast growing cancer cells[18-27]. Partial sequence determination of a protein induced in rat hepatoma called Spot 17 showed that it was highly homologous to the rat AR[28,29]. Cao et al[22] found about 29% of liver cancers over-expressed AR. Moreover, they identified a novel human protein that was highly homologous to AR, then submitted it to GenBankTM(HARL U37100). This protein, known as ARL-1, consists of 316 amino acids, the same size as AR, and its amino acid sequence is 71% identical to that of AR. About 54% of hepatocellular carcinoma (HCC) over-express ARL-1 gene. These suggest ARL-1 might be related to liver cancers.
ARL-1 is a novel gene, its function and exact relationship with liver cancers are unclear. In order to clarify the role of ARL-1 in HCC, two recombinant proteins of ARL-(His)6 and ARL-GST were expressed, and highly specific polyclonal antibodies against ARL-1 protein were prepared.
MATERIALS AND METHODS
Restrictive endonuclease and T4 DNA polymerase enzymes were purchased from TaKaRa Biotech Corp. Glutathione sepharose 4B, CNBr-activated sepharose 4B and pGEX-4T-1(His)6C were obtained from Pharmacia Biotech. Rainbow markers and mid-range protein molecular weight markers were obtained from Amersham Pharmacia Biotech and Promega, respectively. PVDF membrane was from Schleicher & Schüll. T4 DNA ligase and GeneRulerTM 100 bp DNA ladder plus were products of MBI Fermentas. TALONR metal affinity resin was obtained from Clontech. Freund's adjuvant incomplete and Freund's adjuvant complete were purchased from Sigma. E.coli strains were from Stratagene. Horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody was from New England Biolabs Inc.
The following primers used in this study were made by HaoJia Corp: cggaattcatggccacgtttgtggagc, cgctcgagtcaatattctgcatcgaaggg according to GenBankTM(accession number HARLU37100).
RT-PCR to subclone cDNA of ARL-1
According to the reference[22], cDNA of ARL-1 was obtained by RT-PCR (data not shown).
Expression of ARL-1 in E. coli
ARL-1 polymerase chain reaction-amplified products were inserted into the E. coli expression vector pGEX-4T-1(His)6C, and their proteins would be translated in-frame from the vector's start codon. The plasmid DNA was then transformed into bacteria host BL21. Induction of expression of ARL-1 inserts and purification of the recombinant proteins named ARL-GST were done according to the manufacturer's manual. Briefly, bacteria growing to A600 = 0.4 were induced by 0.05 mM isopropyl-1-thio-β-D-galactopyranoside at 30 °C overnight. Cells were harvested and lysed by sonication. After centrifugation to remove debris, the supernatant was mixed with a 50% slurry of glutathione sepharose 4B, which bound to the glutathione S-transferase (GST) at the amino terminus of the recombinant protein. The slurry was then loaded onto a column and eluted with 5 bed volumes of elution buffer (10 mM reduced glutathione in 50 mM Tris-HCL, pH8.0). Fractions of 500 μL were collected, and samples from the column were analyzed in 15% SDS-polyacrylamide gel electrophoresis. Fractions containing the recombinant protein ARL-GST were stored at -20 °C.
We subcloned ARL-1 gene into another E. coli expression vector pQE-30 (Qiagen), expressed another recombinant protein named ARL-(His)6. According to the user manual, ARL-(His)6 was purified with TALONR metal affinity resin whose cobaltions bound to the 6-histidine residues at the amino terminus of the recombinant protein. Briefly, TALONR metal affinity resin was saturated with a bacterial polyhistidine-tagged GFPuv lysate in extraction buffer (pH8.0). The resin was then washed twice with 5 mL of wash buffer (pH7.0). At last, bound protein was eluted by washing three times with 1 mL elution buffer (pH7.0), a sample of each eluate was analyzed by electrophoresis on a 15% SDS-polyacrylamide gel to verify the purity of elution protein. The purified ARL-(His)6 was stored at -20 °C.
Preparation of polyclonal antibodies against ARL-1 protein
Domestic rabbits were injected with 0.4 mg recombinant protein ARL-(His)6 with Freund's complete adjuvant. Three weeks later, they were injected with 0.16 mg ARL-(His)6 with Freund's incomplete adjuvant, which was repeated four times at 2-week intervals. After a little serum was collected from rabbit ear and proved positive by double agar diffusion test, rabbit sera were taken by carotid intubation and stored in 0.1% sodium azide at -20 °C.
According to the manufacturer's manual, polyclonal antibodies against ARL-1 protein were purified with CNBr-activated sepharose 4B. Firstly, rabbit sera were filtered through a 0.22 μM filter and diluted one to four times with phosphate-buffered saline (PBS, pH7.4). Secondly, the gel was prepared with 1 mM HCL. The purified recombinant protein ARL-GST was then coupled with the prepared gel in coupling buffer (0.1 M NaHCO3, pH8.3, containing 0.5 M NaCL). Finally, the prepared rabbit sera were mixed with the sepharose coupled ARL-GST in a stopped vessel. The mixture was rotated end-over-end overnight at 4 °C and subsequently stood at room temperature for hours. The gel was washed with 1 × PBS (pH7.4), 2 M NaCL in 1 × PBS (pH7.4) and 1 × PBS (pH4.5) till the OD280 of washed solution was zero, respectively. At last, it was eluted with 0.1 M glycine (pH2.8). The polyclonal antibodies were collected in 0.05 mM Tris-base (pH9.5) and stored in 0.1% sodium azide at -20 °C.
Western blotting
All plasmids including pGEX-4T-1(His)6C-ARL, pGEX, pQE-30-ARL and pET-CTF that encoded protein ARL-GST, GST, ARL-(His)6 and CTF-(His)6, were transformed to BL21, respectively. After bacteria grew to A600 = 0.4, they were induced by 0.05 mM isopropyl-1-thio-β-D-galactopyranoside at 37 °C for 3 h. A 1/4 volume of SDS sample buffer containing Tris-HCl 0.33 mol•L-1, 10% SDS (w/v), 40% glycerol (v/v), and 0.4% bromophenol blue was added to cell lysates above and purified ARL-(His)6 and ARL-GST. After being boiled for 5-10 min, 10 mg protein was electrophoresed on 15% SDS-polyacrylamide gel. The protein was transferred to PVDF membrane, which was then blocked for 1 h at room temperature with 5% BSA in TBST (100 mmol/L Tris-HCl and 0.9% NaCl containing 0.05% Tween-20). The blots were incubated with the primary polyclonal antibodies against ARL-1 protein (1:2000 in TBST) and subsequently with HRP-labeled second antibody (1:5000 in TBST) at room temperature for 1 h, respectively. At last, immunoreactive signals were visualized by DAB in 0.03% H2O2.
RESULTS
Construction of pGEX-4T-1(His)6 C-ARL and pQE-30-ARL
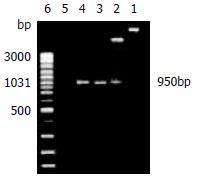
The recombinant plasmids were identified by restrictive endonuclease and/or polymerase chain reaction. All products of pGEX-4T-1(His)6C-ARL amplified by polymerase chain reaction and digested by EcoRI only or EcoR I and Xho I in combination, underwent 1.5% agarose electrophoresis. The expected 950 bp could be seen, which was ARL-1 gene (Figure 1). In the same way, pQE-30-ARL was identified by restrictive endonuclease. All fragments were expected including 4.5 kb fragment of pQE-30-ARL which was digested by Hind III alone, 0.7 kb and 3.8 kb fragments of pQE-30-ARL digested by BamHI only, and 3.4 kb, 0.7 kb and 0.3 kb fragments of pQE-30-ARL digested by BamHI and Hind III in combination (Figure 2).
Figure 1 Recombinant plasmid pGEX-4T-1(His)6C-ARL was identified by restrictive endonuclease and polymerase chain reaction (1.
5% agarose electrophoresis). Lane 1: The recombinant plasmid pGEX-4T-1(His)6C-ARL digested by EcoR I only, about 5.9 kb fragment could be seen; Lane 2: The recombinant plasmid pGEX-4T-1(His)6 C-ARL digested by EcoR I and Xho I, two fragments of 5.0 kb and 950 bp could be seen; Lane 3: pGEX-4T-1(His)6C-ARL being template, 950 bp was amplified by polymerase chain reaction; Lane 4: positive control of PCR; Lane 5: negative control of PCR; Lane 6: GeneRulerTM 100 bp DNA ladder plus.
Figure 2 Recombinant plasmid pQE-30-ARL was identified by restrictive endonuclease (1.
5% agarose electrophoresis). Lane 1: 3.4 kb and 0.7 kb and 0.3 kb fragments of pQE-30-ARL digested by BamHI and Hind III together; Lane 2: 0.7 kb and 3.8 kb fragments of pQE-30-ARL digested by BamHI only; Lane 3: 4.5 kb fragment of pQE-30-ARL digested by Hind III alone; Lane 4: GeneRulerTM 100 bp DNA ladder plus.
Expression of recombinant proteins ARL-GST and ARL-(His)6
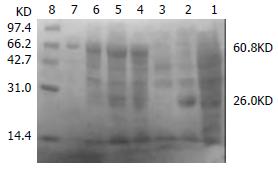
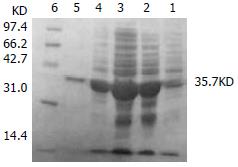
The transformed pGEX-4T-1(His)6C-ARL BL21 induced by 0.05 mM isopropyl-1-thio-β-D-galactopyranoside could express the protein with molecular weight of 60.8 KD, which was recombinant protein ARL-GST. Its expression level was 15.1% of total bacteria proteins. Under the same conditions, the transformed pQE-30-ARL BL21 expressed recombinant protein ARL-(His)6 with molecular weight of 35.7 KD. Among total bacteria proteins, ARL-(His)6 expression level was 27.7%. Two recombinant proteins were purified by non-denatured method because they were soluble, predominantly in supernatant. After purification, only one band could be seen, indicating that ARL-GST and ARL-(His)6 were highly purified (Figure 3 and Figure 4).
Figure 3 The transformed pGEX-4T-1(His)6C-ARL BL21 induced by 0.
05 mM isopropyl-1-thio-β-D-galactopyranoside. Lane 1: BL21 transformed with pGEX-4T-1(His)6C, no IPTG induced; Lane 2: BL21 transformed with pGEX-4T-1(His)6C, IPTG induced, GST (26KD) was expressed; Lane 3: BL21 transformed with pGEX-4T-1(His)6C-ARL, no IPTG induced; Lane 4: BL21 transformed with pGEX-4T-1(His)6C-ARL, IPTG induced, recombinant protein ARL-GST (about 60.8 KD) was expressed; Lane 5: supernatant; Lane 6: pellet; Lane 7: purified ARL-GST; Lane 8: mid-range protein molecular weight markers.
Figure 4 The transformed pQE-30-ARL BL21 induced by 0.
05 mM isopropyl-1-thio-β-D-galactopyranoside. Lane 1: BL21 transformed with pQE-30-ARL, no IPTG induced; Lane 2: BL21 transformed with pQE-30-ARL, IPTG induced, ARL-(His)6(35.7 KD) could express; Lane 3: supernatant; Lane 4: pellet; Lane 5: purified ARL-(His)6; Lane 6: mid-range protein molecular weight markers.
Preparation of polyclonal antibodies against ARL-1 protein
Purification of polyclonal antibodies against ARL-1 protein: The rabbit sera immunized with ARL-(His)6 were purified by CNBr-activated sepharose 4B coupled ARL-GST. After mixed with sera, washed with solution as described above, the gel was eluted with 0.1 M glycine (pH2.8), only one elution peak could be seen in the elution curve (Figure 5), suggesting that polyclonal antibodies against ARL-1 protein were highly purified.
Figure 5 The gel eluted with 0.
1 M glycine (pH2.8).
Specification of polyclonal antibodies against ARL-1 protein: Proteins of all bacteria described were tested by Western blotting using polyclonal antibodies against ARL-1. Figure 6 displays that positive signals could be seen in the two purified proteins and the bacteria transformed with pGEX-4T-1(His)6C-ARL and pQE-30-ARL individually.
Figure 6 Proteins of all bacteria described were tested by Western blotting using polyclonal antibodies against ARL-1.
Lane 1: purified ARL-GST; Lane 2: BL21 transformed with pGEX-4T-1(His)6C-ARL; Lane 3: BL21 transformed with pGEX-4T-1(His)6C; Lane 4: purified ARL-(His)6; Lane 5: BL21 transformed with pQE-30-ARL; Lane 6: BL21 transformed with pET-CTF; Lane 7: BL21; Lane 8: Rainbow markers.
DISCUSSION
The cDNA of ARL-1 gene was cloned and identified by Cao et al[22]. The amino acid sequence of this protein called ARL-1 was 71% identical to that of AR. It is more homologous to a group of AR-like proteins. Its amino acid sequence was 80%, 82%, 83%, and 94% identical to that of the mouse vas deferens protein (MVDP), the mouse fibroblast growth factor-regulated protein (FR-1), the Chinese hamster ovary (CHO) reductase, and Spot 17 in rat hepatoma, respectively.
ARL-1 is a new member of AKR1B group of aldoketo reductases including AR, MVDP, FR-1, CHO reductase, Spot 17, etc. The gene expression spectrum is not the same. MVDP gene was primarily expressed in the vas deferens and the adrenal gland[30-37]. AR, was different. In adults, AR message was most abundant in the prostate, testis, skeletal muscle, and the heart. Substantial amounts were found in the ovary, small intestines, colon, thymus, spleen, kidney, and the placenta. Lower levels were found in the brain, lung, leukocytes, and the pancreas. Of the four fetal tissues tested, AR mRNA was most abundant in the kidneys, a substantial amount was found in the brain, and a low level in the lung and liver[38,39]. FR-1 was expressed quite strongly in the small intestines, and it was expressed in testis, ovary, brain, heart, liver, kidney, and muscle, but FR-1 expression has not been screened in urinary bladder and jejunum[40-44]. CHO reductase mRNA was quite abundant in the urinary bladder and the jejunum, this gene was also expressed in the testis and the ovary. However, CHO reductase mRNA was not detectable in brain, heart, liver, kidney, and muscle[45-47]. ARL-1 was not expressed in many tissues. Its message was most abundant in the small intestines and colon, and a low level of its mRNA was found in the liver, thymus, prostate, testis, and skeletal muscle. No ARL-1 mRNA was detected in the four fetal tissues tested including the kidney, brain, lung and liver. Moreover, ARL-1 mRNA was not present in human vas deferens. Although many functions of all these AR-like genes are not known, different expression spectrums indicate that their physiological functions are quite different[22].
Not only 71% of ARL-1 amino acid sequence was identical to that of AR, but also all of the key amino acids being responsible for AR's enzymatic activities were conserved in this protein. They included Ser159, Asn160, Gln183, and Lys262 for cofactor NADPH binding, Tyr48 and His110 as potential hydrogen donors, and active site residues Lys77, Tyr209, and Cys298. The enzymatic activity of ARL-1 was similar to that of the well-characterized human AR. In terms of pH optimum, salt requirement, and kinetics in reducing some substrates, these two proteins were different. For example, the activity of ARL-1 was not affected by 0.3 M sulfate ion, which was the optimum concentration for stimulating AR activity[22]. In the present project, two recombinant ARL-1 proteins were expressed and highly purified, polyclonal antibodies against ARL-1 were prepared and characterized. Polyclonal antibodies prepared by us were only against ARL-1 protein, the yeast and fly tissues were detected by Western blotting using our antibodies, no signals were visualized (data not shown), suggesting that these polyclonal antibodies were highly specific. Other characters of ARL-1 protein should be studied, as well as the relation between ARL-1 and human diseases.
AR is thought to be involved in diabetic complications such as cataract, retinopathy, neuropathy, and nephropathy. It is not clear if ARL-1 also contributes to these diabetic complications. So two recombinant ARL-1 proteins and polyclonal antibodies against ARL-1 prepared in our research would help determine the relationship between ARL-1 and diabetes. AR and an AR-like protein called Spot 17 were induced in rat hepatomas, suggesting that these proteins may be needed to detoxify methylglyoxal or other metabolites generated by fast growing cancer cells. About 29% of individual human HCCs over-expressed AR and 54% of them over-expressed ARL-1. The deduced protein sequence of ARL-1 was 94% identical to that of Spot 17, indicating that it is most likely the human homologue of that protein[22]. In our studies, ARL-1 protein was detected in 94.4% (17/18) liver cancer tissues, but only in 82.4% (14/17) surrounding nontumorous tissues. Moreover, in all normal liver tissues (5 cases), ARL-1 protein was negative, ARL-1 expression in all detected liver cancer tissues was up-regulated (data not shown). All these studies suggest that ARL-1 is closely related to liver cancers.
In conclusion, two recombinant ARL-1 proteins and polyclonal antibodies against ARL-1 protein were prepared. As a basic research, these would help us identify the function of ARL-1, the exact relationship between ARL-1 and liver cancers. The life expectancy of HCC patients is hard to predict because of the high possibility of postoperative recurrence. Many factors, such as patients general conditions, macroscopic tumor morphology, as well as tumor histopathological features, have been proven of prognostic significance[48-58]. With the progress in ARL-1 studies, the early diagnosis rate and the therapy level of liver cancers might be elevated, which will improve the prognosis of patients with liver cancer in the future.