INTRODUCTION
High incidence of 50%-80% LOH (loss of hybridization) in gastric carcinoma cells reveals obvious chromosome fragments loss[1,2], e.g. 8p, 18q12.2, 21q22, 1p35-pter of regions of LOH were currently found[2-8]. These suggest the novel tumor suppressor genes in the regions of LOH are involved in gastric tumorigenicity[9]. But these novel tumor suppressor gene candidates have not been cloned. To reach the targets, we screened down-regulated genes in a suppression subtraction cDNA library established by counterpart normal gastric mucous membrane mixture mRNA (tester) subtracting gastric cancer cells mixture mRNA (driver) of five patients with gastric cancer based on the suppression subtractive hybridization (SSH) technique[10]. These down-regulated genes obtained from the library probably are tumor suppressor gene candidates. Up to now, down-regulated genes in gastric cancer cloned from gastric tissues have been seldom documented except CA11, LDOC1[11-13].
MATERIALS AND METHODS
RNA extraction and mRNA purification
RNA of gastric cancer tissues and counterpart normal gastric mucosa was respectively isolated with Trizol (Gibco) in five patients with gastric cancer from Xijing Hospital, Fourth Military Medical University. To ensure good total RNA quality 28S/18S ≥ 1.5, samples were immediately placed into liquid nitrogen after being removed intraoperatively, and trituration of the samples must be performed in liquid nitrogen. Then, mRNA was purified in Oligotex mRNA Kit (Qiagen). An equal mRNA mixture of gastric cancer tissues and counterpart normal gastric mucosa in five patients with gastric cancer contributed to driver group (gastric cancer tissues) and tester group (counterpart normal gastric mucosa) respectively. At last, reverse transcription products (the first stranded cDNAs) of mRNA mixture were electrophoresed to evaluate their size range and quality.
Suppression substraction library construction
First-strand was synthesized with 1 μL cDNA synthesis primer (10 μmol•l-1, Clontech) in a mixture containing 2 μg mRNA mixture, 20U AMV, 2 μL 5 × first-strand buffer in a final volume of 10 μL at 42 °C for 1.5 h. Then, second-strand synthesis was carried out in 10 μL first strand react volume, 4.0 μL 20 second-strand enzyme cocktail, 48.4 μL of sterile H2O, 1.6 μL dNTP Mix (10 mM•l-1), 16.0 μL 5 × second-strand buffer solution for 2 h at 16 °C. Polymeric reaction was performed at 16 °C for 0.5 h after 2 μL (6 units) of T4 DNA polymerase was added into the above reaction volume. The second-strand synthesis was terminated by adding 4 μL of 20 × EDTA/Glycogen Mix (Clontech). After phenol:chloroform: isoamyl alcohol (25:24:1) and chloroform: isoamyl alcohol (24:1) extraction twice respectively, and 4 M NH4OAc and 95% ethanol precipitation of the second stranded cDNAs, the pellet was dissolved in 50 mL of sterile H2O when precipitate was washed in 80% ethanol and residual ethanol was evaporated after the supertant was removed.
The second double-stranded cDNA was digested with 15U RsaI in a final volume of 50 μL at 37 °C for 1.5 h. Enzyme digestion was terminated by adding 2.5 μL 20 × EDTA/Glycogen Mix. After extraction and precipitation of digested second- strand cDNAs, the pellet was dissolved in 5.5 μL of sterile H2O when precipitate was washed in 80% ethanol and residual ethanol was evaporated after the supertant was removed.
One μL of digested first-strand cDNAs of normal gastric mucosa was diluted with 5 μL of sterile H2O. Each 2 μL of the cDNA was acted as tester1-1 and tester1-2 that were respectively mixed with adaptor1 and adaptor 2, and 1 μL T4 DNA ligase (400 kU•l-1) in a final volume of 10 μL, while mixture of 2 μL of each tester1-1 and tester1-2 was taken as control (Tester C). Ligation to adaptors completed at 16 °C overnight.
Then, 1.5 μL of tester1-1 with adaptor 1 and tester1-2 with adaptor 2 was respectively hybridized with 1.5 μL digested first-stranded driver cDNA of gastric cancer in 1.0 μL 4 × hybridization buffer solution at 68 °C for 10 h. Tester1-2 hybridization sample was drawn into the pipette tip. Afterwards, 1 μL denatured mixture from 1 μL digested second-stranded driver cDNA, 2μL H2O, 1 μL 4 × hybridization buffer solution at 98 °C was drawn into the pipette tip with a slight air space below the droplet of the above tester1-2 hybridization sample. Sequentially, the entire mixture of pipette tip was transferred to a tube containing the above tester1-1 hybridization sample overnight at 68 °C. After second hybridization, 200 μL dilution buffer was added into the tube. One μL of tester C was diluted with 1000 μL of sterile H2O. 1 μL of diluted tester C and the secondary hybridization sample were amplified with PCR primer 1 and 50 × advantage cDNA polymerase mix in a final volume of 25 μL respectively after adaptors were extended at 75 °C. 3 μL of primary PCR product was diluted with 27 mL of sterile H2O. 1 μL of diluted primary PCR products was again amplified with nested PCR primer 1, 2R and 50 × advantage cDNA polymerase mix in a final volume of 25 μL for 12 cycles.
Analyses of adaptor ligation efficiency and subtraction efficiency by PCR
One 1 μL of tester1-1 and tester1-2 with adaptors was diluted with 200 μL of sterile H2O respectively. 1 μL of diluted tester1-1 and tester1-2 was repeatedly amplified respectively using G3PDH 3'primer, PCR primer 1 as well as G3PDH 3'primer, G3PDH 5'primer after adaptors were extended at 75 °C. 1 μL of subtraction cDNA and secondary PCR product of tester C was diluted with 9 μL of sterile H2O. 1 μL of diluted subtraction cDNA and secondary PCR product of tester C was respectively amplified with G3PDH 3'primer, G3PDH 5'primer. 5 μL of PCR products collected at 18, 23, 28, and 33 cycles was electrophoresed on 2% agarose gel respectively.
Identification of suppression substraction library
Six μL of secondary PCR product of subtraction cDNA was ligated to T vectors in a mixture containing 2 μL T vectors, 1 μL T4DNA ligase in a final volume of 10 μL at 16 °C for 36 h. Then, 5 μL of ligated product was transformed into 100 μL of competent DH5a (stratgene) for electroporation. Competent DH5a transformed by ligated product was grown on LB medium plates. White colonies were placed into LB medium and shaken overnight at 37 °C. In a large scale, fragments inserted into library plasmids were identified by PCR amplification with SP6 and T7 primers after library plasmids were extracted. Each colony of plasmids with inserted fragments was inoculated twice on a LB medium plate (100 colonies per plate, and one pair of positive and negative controls per plate) and grown until colony diameter reached to 3 mm. At time, colonies were transfered onto a nylon filter (NEN), then the nylon filter was cross-linked by using an UV stratalinker (CL-1000, Upland). Each 200 ng mRNA of the normal mucosa and gastric carcinoma was reverseiy transcripted with 1 mg Oligo (dT)15 and super transcriptase II as a probe labeled with α-32PdCTP, hybridized respectively with filters. The filters were exposed to phosphore screen and analyzed.
RESULTS
Identification of mRNA quality
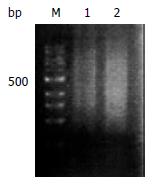
Good total RNA quality was confirmed by 28S/18S ≥ 1.5. Size range of reverse transcription product cDNAs was represented in a smear from 0.2-4 kb both in gastric cancer and normal mucosa (Figure 1).
Figure 1 Size range of mRNA on 2% agarose gel electrophorsis.
Normal gastric mucosa mRNA (lane1) and gastric carcinoma mRNA (lane 2). 100 bp size marker (lane M).
RsaI enzyme digestion
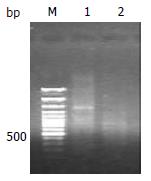
Size range of double-strand cDNA without digestion showed a normal size as expected. By comparison, RsaI enzyme digested double-strand cDNA on electrophoresis represented a smear from 0.2-2 kb caused by complete digestion (Figure 2).
Figure 2 Digested and undigested double strands cDNA products.
Undigested double strands cDNA products (lane 1) and RsaI digested double strands cDNA products (lane 2). 100bp size marker (lane M).
Detection of adaptor ligation efficiency and analyses of PCR products
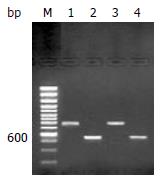
A 0.75 kb band of tester1-1 and tester1-2 PCR product accorded with the theoretic size as expected when they were amplified with G3PDH3' primer and PCR1 primer respectively. The 0.75 kb band intensity of tester1-1 and tester1-2 PCR product also was as same as the band of tester1-1 and tester1-2 PCR product amplified with G3PDH3’ and G3PDH5’ primers (Figure 3). Secondary PCR product of subtraction sample exhibited a smear from 0.2-2 kb with some distinct bands that were greatly different from that appeared in unsubtraction samples (Figure 4).
Figure 3 Detection of adaptors ligation efficiency.
Tester1-1 was amplified with G3PDH3' primer, PCR primer1 (lane 1) and tester1-1 with G3PDH3’, 5'primers (lane 2). Tester1-2 was amplified with G3PDH3'primer, PCR primer1 (lane 3) and tester1-2 with G3PDH3’, 5'primers (lane 4). 100 bp size marker (lane M).
Figure 4 Analyses of PCR products.
Secondary PCR products of subtraction samples (lane 1) and unsubtraction samples (lane 2). 100 bp size marker (lane M).
Analyses of subtraction efficiency by PCR
G3PDH persistently was expressed at 18, 23, 28 and 33 cycles of PCR and the bands exhibited greater and greater intensity with increasing cycle numbers in unsubtraction control group, but not expressed in subtraction group (Figure 5).
Figure 5 Identification of subtraction efficiency by PCR.
G3PDH PCR products of subtracted samples at 18, 23, 28 and 33 cycles (lanes1, 2, 3 and 4) respectively and G3PDH PCR products of unsubtracted samples respectively at 18, 23, 28 and 33 cycles (lanes 5, 6, 7 and 8). 100 bp size marker (lane M).
Identification of inserted fragments in plasmids of library by PCR and positive clone by reverse hybridization
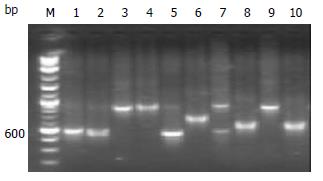
PCR products of library plasmids amplified with SP6 and T7 primers on a larger scale showed that each plasmid included one inserted fragment ranging from 300-700 bp (Figure 6). 86% of down-regulated genes between normal gastric mucosa and gastric carcinoma were confirmed by hybridization of a transferred filter with probes of reverse transcription product cDNAs.
Figure 6 Identification of inserted fragments in plasmids of library by PCR.
Inserted fragments in plasmids of library (lanes 1-10). 100 bp size marker (lane M).
DISCUSSION
Many genes involves in gastric tumorigenicity and tumor metastasis[14-24]. Occurrence and development of gastric carcinoma are closely associated with loss or lower expression of suppressor genes[25,26]. It contributes to a better understanding of the molecular mechanism of gastric tumorigenicity, and the expression profiles of down-regulated genes in gastric carcinoma, as well as cloning of novel genes , especially human stomach-specific gene. The novel genes usually express in lower abundance, and play an important role in cell differentiation and development. We have successfully established the cDNA suppression subtraction library to screen down-regulated genes in gastric carcinoma.
It is an important step to guarantee mRNA quality in constructing cDNA suppression subtraction library with a high subtraction efficiency because it is directly relatd to subtraction efficiency. Good mRNA quality depends on total RNA quality except for mRNA purification. To ensure good total RNA quality of 28S/18S ≥ 1.5, samples must be immediately placed into liquid nitrogen after removed intraoperatively, and trituration of samples must be performed also in liquid nitrogen. At last, reverse transcription products (the first stranded cDNAs) of mRNA were electrophoresed to evaluate its size and quality. The mRNA size range should accord with its theoretic value.
Each step for establishing cDNA suppression subtraction library was verified by corresponding methods provided by Clontech to ensure suppression subtraction efficiency. Many reports have shown that the suppression subtractive technique has successfully constructed a lots of cDNA suppression subtraction library with high efficiency, and cloned many novel genes[27-45]. Our experiments were carried out strictly according to the rule. Identifications of experimental results step by step revealed complete enzyme cutting, and enzyme digested size of double-strand cDNA accorded with theoretic size range, enough ligation of the digested fragments of double-strand cDNA and adaptors. It demonstrated that, in cDNA suppression subtraction library with high subtractive efficiency, G3PDH was persistently expressed at 18, 23, 28 and 33 cycles of PCR and the bands exhibited greater and greater intensity with increasing cycles in unsubtraction control group, but not expressed in subtraction group.
After establishment of the cDNA suppression subtraction library with a high efficiency, maximum cloning of novel down-regulated genes in gastric cancer depends on highly efficient plasmid transformation method and competent bacteria cells. Lower abundance of gene fragments will be likely cloned if commercially availiable high concentration competent cells are used (1 × 1012•l-1) for transformation with a high transformation rate (1-2 × 108/μg PUC19).Use of electroporation method can greatly enhance library plamids transformation rate by obtaining 108/μg PUC19. Additionally, it especially fits transformation of the small fragments produced in cDNA suppression subtraction library.
Several identified methods for cDNA suppression subtraction library were described below. The expression pattern of individual clones could be confirmed by Northern blot analysis. 10-20 clones were randomly picked from the subtracted library as probes on Northern blots. If less than two clones were confirmed as differentially expressed genes, the differential screening procedure should be performed to eliminate false positives. Dot or Southern blot analysis was performed. Secondary PCR products of the unsubtracted tester cDNA, the unsubtracted driver cDNA, and the subtracted cDNA were electrophoresed on a 1.5% agarose gel, transferred onto nylon filters and hybridized respectively with differential expressing genes as probes labeled with α-32PdCTP. But more background bands of unpredicted sizes often appeared. Nylon filters onto which the library colonies of bacteria were transferred and hybridized with reverse transcript product cDNA of gastric cancer tissues mRNA and normal gastric mucosa mRNA were used as probes labeled with α-32-32PdCTP respectively. This method has been extensively used. The disadvantage is that only a high abundance of mRNA can be detected. Another approach can bypass the problem of losing a low-abundance of sequences. By this method, the subtracted library was hybridized with forward- and reversely-subtracted cDNA probes. To make reversely-subtracted probes, subtractive hybridization was performed with the original tester cDNA as a driver and the driver cDNA as a tester. Plasmids colonies that are truly differentially expressed will hybridize only with the forward-subtracted probe. Plasmids colonies that hybridize with the reversely-subtracted probe may be considered as the background. This approach requires one additional step: before it can be used as probes, the forward- and reversely-subtracted probes must undergo restriction enzyme digestion to remove the adaptor sequences. Despite their small size, these adaptors can cause a very high background when the subtracted probes are hybridized to the subtracted cDNA library.