Published online May 15, 2003. doi: 10.3748/wjg.v9.i5.888
Revised: December 23, 2002
Accepted: January 13, 2003
Published online: May 15, 2003
AIM: To understand the response of human REV3 gene to gastric cancer inducing carcinogen N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) and its role in human mutagenesis.
METHODS: The response of the human REV3 gene to MNNG was measured in human 293 cells and FL cells by RT-PCR. By using antisense technology, mutation analysis at HPRT locus (on which lesion-targeted mutation usually occurs) was conducted in human transgenic cell line FL-REV3- by 8-azaguanine screening, and mutation occurred on undamaged DNA template was detected by using a shuttle plasmid pZ189 as the probe in human transgenic cell lines 293-REV3- and FL-REV3-. The blockage effect of REV3 was measured by combination of reverse transcription-polymerase chain reaction to detect the expression of antisense REV3 RNA and Western blotting to detect the REV3 protein level.
RESULTS: The human REV3 gene was significantly activated by MNNG treatment, as indicated by the upregulation of REV3 gene expression at the transcriptional level in MNNG-treated human cells, with significant increase of REV3 expression level by 0.38 fold, 0.33 fold and 0.27 fold respectively at 6 h, 12 h and 24 h in MNNG-treated 293 cells (P < 0.05); and to 0.77 fold and 0.65 fold at 12 h and 24 h respectively in MNNG-treated FL cells (P < 0.05). In transgenic cell line (in which REV3 was blocked by antisense REV3 RNA), high level of antisense REV3 RNA was detected, with a decreased level of REV3 protein. MNNG treatment significantly increased the mutation frequencies on undamaged DNA template (untargeted mutation), and also at HPRT locus (lesion-targeted mutation). However, when REV3 gene was blocked by antisense REV3 RNA, the MNNG-induced mutation frequency on undamaged DNA templates was significantly decreased by 3.8 fold (P < 0.05) and 5.8 fold (P < 0.01) respectively both in MNNG-pretreated transgenic 293 cells and FL cells in which REV3 was blocked by antisense RNA, and almost recovered to their spontaneous mutation levels. The spontaneous HPRT mutation was disappeared in REV3-disrupted cells, and induced mutation frequency at HPRT locus significantly decreased from 8.66 × 10-6 in FL cells to 0.14 × 10-6 in transgenic cells as well (P < 0.01).
CONCLUSION: The expression of the human REV3 can be upregulated at the transcriptional level in response to MNNG. The human REV3 gene plays a role not only in lesion-targeted DNA mutagenesis, but also in mutagenesis on undamaged DNA templates that is called untargeted mutation.
-
Citation: Zhu F, Jin CX, Song T, Yang J, Guo L, Yu YN. Response of human
REV3 gene to gastric cancer inducing carcinogenN -methyl-N’ -nitro-N -nitrosoguanidine and its role in mutagenesis. World J Gastroenterol 2003; 9(5): 888-893 - URL: https://www.wjgnet.com/1007-9327/full/v9/i5/888.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i5.888
It has long been known that exposure to certain chemicals is associated with the development of specific human cancers, which is largely the outcome of interaction between environmental agents and genetic susceptibility. Examples include the associations between amine dyes and bladder cancer, benzene and leukemia, aflatoxin and hepatocellular carcinoma, and tobacco smoke and lung cancer[1-5]. Recent studies have also revealed that tobacco smoke significantly increases the risks for oral[6,7], esophageal[3-5,8], bladder[9-12], pancreas[11], gastric[13] and colorectal cancers[14]. In addition, men who have a history of chronic indigestion or gastroduodenal ulcer have substantially higher mortality rates associated with concurrent cigarette smoking[13].
Tobacco smoke consists of many chemicals. One important substance found in tobacco smoke is chemical carcinogen N-methyl-N’-nitro-N-nitrosoguanidine (MNNG), a direct acting carcinogen, that targets the cellular DNA and induces severe genotoxic stress to the cell that can result in various DNA damages[15]. Epidemiologic studies have suggested an etiological role for N-nitroso compounds from dietary sources in the development of gastric and colorectal cancer in humans[16,17], and animal experiments have shown that MNNG induces gastric cancer[18-21] and colorectal cancer[22,23]. Obviously, the link between DNA damages and MNNG induced cancers is closely related to mutagenesis. To ensure normal growth control and accuracy in DNA replication, cells have developed a variety of responses to stress, such as DNA repair, cell cycle checkpoints, DNA damage avoidance, or in extreme cases, apoptosis[24]. In addition, cells have also evolved a sophisticated lesion bypass system (also called translation synthesis, or TLS) to repair the damaged DNA, resulting in DNA damage lesion-targeted mutation. However, mutation can also occur on undamaged DNA template, which is designated untargeted mutation (UTM), which has been described in SOS-induced mutagenesis in E. coli[25]. It has been known that untargeted and targeted mutations caused by SOS response in E. coli both are resulted from the inhibition of DNA polymerase functions that normally maintain fidelity and the involvement of DNA polymerases with low fidelity, which include DNA pol IV (dinB), pol V (UmuD’2C) and other factors[26-30]. In eukaryote, it has been found that up to 40% of cycl-91 revertants induced by ultraviolet (UV) is untargeted using mating experiments with excision deficient strains of Saccharomyces cerevisiae[31], and that stress response induced by DNA damaging agents (8-methoxy-psoralen or UV) leads to specific and delayed UTM in mouse T-lymphoma cells[32]. Previous studies in our laboratory also shown that low concentration MNNG induces UTM in mammalian cells[33]. Currently, it has been known that specialized DNA polymerases are responsible for DNA damage lesion-targeted mutation in eukaryote. However, it is not clear which factor can be activated and involved in UTM on undamaged DNA templates.
The human REV3 gene, encoding the catalytic subunit REV3 of human pol ζ, has been received intensive attention in recent years[34]. REV3 gene is thought to be the major component of error-prone TLS pathway[34,35], although a number of other polymerases might also be involved in this process[36]. It is responsible for most of spontaneous and UV-induced mutation in yeast and humans, as well as somatic hypermutation in humans[34,35,37-43,44-47]. The expression of REV3 appears to be elevated at the transcriptional level in some tumor cell lines[48]. However, the response of REV3 gene to gastric cancer inducing carcinogen MNNG and its role in MNNG-induced mutagenesis are still not clear. In order to understand the relationship between the human REV3 gene and the etiology of gastric cancer and colorectal cancer in humans, the response of REV3 to MNNG and its role in MNNG-induced mutagenesis, including both lesion-targeted and untargeted mutation, were explored.
Human 293 cells were grown in DMEM (Dulbecco’s Modified Eagle Medium, Gibco) containing 10% fetal bovine serum (Gibco), 200 units/ml penicillin, 100 μg/ml streptomycin and 200 μg/ml kanamycin. Human FL cells were grown in MEM (Minimum Essential Medium, Gibco), containing 10% newborn calf serum (Gibco), 200 units/ml penicillin, 100 μg/ml streptomycin and 200 μg/ml kanamycin. Transgenic cell line 293-REV3-[49] and FL-REV3- (unpublished data) were established in this laboratory by transfecting 293 cells and FL cells with pM-RS- plasmid[50] that can express anti REV3 RNA when induced by dexamethasone (dex). 293-M and FL-M cell line were established by transfecting 293 cells and FL cells with the control vector pMAM neo-amp- alone. These transgenic cell lines were grown in MEM containing 200 mg/ml of G418 (geneticin, Gibco). For MNNG treatment, cells were exposed to 0.2 μM of MNNG (Sigma, dimethyl sulfoxide (DMSO) as solvent) in serum-free DMEM (for 293 cells) or MEM (for FL cells) for 2.5 h, and then MNNG was removed and replaced with fresh medium. DMSO treated cells were used as control.
The response of the human REV3 gene to MNNG was measured at the transcriptional level by using reverse transcription-polymerase chain reaction (RT-PCR) with ARF1 (encoding ADP-ribosylation factor 1) as the internal control. RNA from 2 × 106 293 or FL cells was extracted at different time point using TRIzol agent (Gibco) after 0.2 μM MNNG treatment, followed by the first-strand cDNAs synthesis with 3 μg of RNA using M-MuLV reverse transcriptase (MBI fermentas) and random hexamer primer. After exponential phase selection, PCR was performed with the appropriate cycles: 5 min pre-denaturation at 95 °C, 30 sec denaturation at 94 °C, 30 sec annealing at 59 °C, 1 min extension at 72 °C, and an additional 10 min extension at 72 °C. PCR primers: REV3, 5’-TGTCCAAGGCACCATATCTC-3’ (sense), 5’-TGCTACACGTGGTACTACTG-3’ (antisense); ARF1, 5’-GAACATCTTCGCCAACCTCTTC-3’ (sense), 5’-ACAGCCAGTCCAGTCCTTCATA-3’ (antisense). The sizes of the expected products are 635 bp for REV3 and 515 bp for ARF1. Ratios of ODREV3./ODARF1 representing REV3 transcript level were calculated.
The antisense blocking effect on REV3 function was analyzed by detecting the expression of antisense REV3 fragment with RT-PCR and the REV3 protein level with Western blotting. RNA was extracted from transgenic cells, which could express antisense REV3 fragment after 10 μM dex treatment for 3 days. 0.1 μg RNA from 1 μg RNA sample digested by 1unit DNaseI (Gibco) was reverse transcribed using the REV3 specific sense primer (5’-AAGGCCAGCATACAAGAC-3’). For the positive control (with no dex treatment), a random hexamer primer was used as the reverse transcription. Each cDNAs sample was amplified with the specific primers: 5’-GCCAAGGAATACAGAGGAAGT-3’ (sense), 5’-CCAGCTGAAGACATCAATACC-3’ (antisense). The PCR cycling parameter is as following: 5 min pre-denaturation at 94 °C, 30 cycles of 30 sec denaturation at 94 °C, 30 sec annealing at 59 °C, and 1 min extension at 72 °C. Amplifications were completed by an additional 8 min extension at 72 °C. For Western blotting, the nuclear protein were extracted from the cell strains as described before[24]. Each nuclear extract (30 μg) was used for Western blotting, and the Ku70 protein was used as the loading control.
2 × 105 cells of the FL, FL-M or FL-REV3- were seeded in 100 ml culture flasks, respectively. After 1 day incubation, the media were replaced with HAT medium (Gibco) for 24 h and HT medium (Gibco) for the next 48 h to remove the pre-existed HPRT- cells in the population, then the cells were induced with 10 μM dex for another 48 h. After treatment with 0.2 μM MNNG or DMSO for 2.5 h, the medium was removed and replaced with a fresh medium containing 10 μM dex for an additional 24 h incubation. Cells reaching approximately 80% confluent were subcultured three or four times, with a consistent density at 106 cells/flask. Then 200 cells were transferred to a 9-cm plate (5 plates total) for 15 days to count the relative cloning efficiency. In the meantime, 2 × 105 cells were seeded in 100 ml culture flask (5 flasks total). After 24 h, the medium was replaced with fresh one containing 5 μg/ml 8-azaguanine (Gibco). Cells were then maintained for 30 days, with the medium changed every 3 days. After washing with 0.9% NaCl, the clones were fixed with ethanol: acetic acid (3:1), stained with 1% methyllene blue, and the number counted. The mutation frequency was calculated as following:
Mutation frequency = (number of mutant clones/106 cells) × (1/relative cloning efficiency).
Statistical analysis was performed according to the method described by Kastenbaum and Bowman[52].
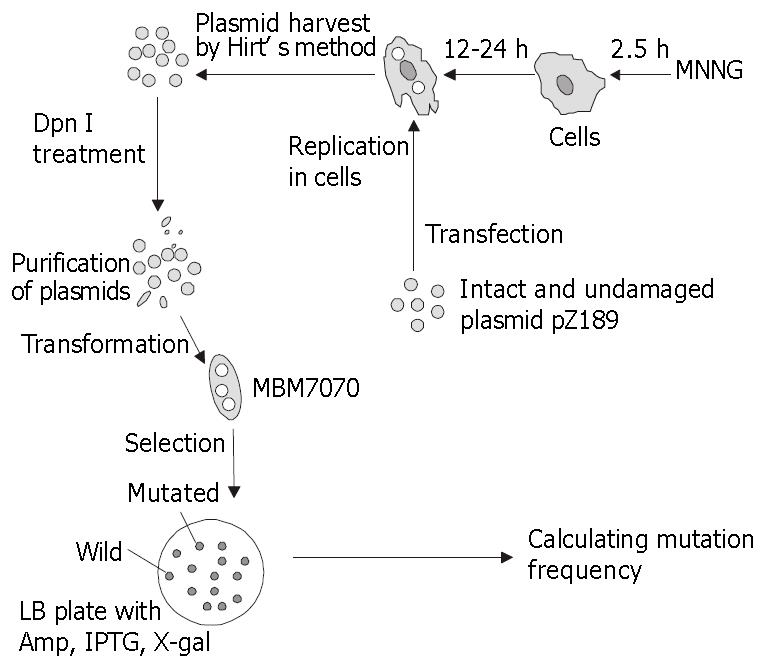
The detection of untargeted mutation was performed as described (Figure 1)[33].
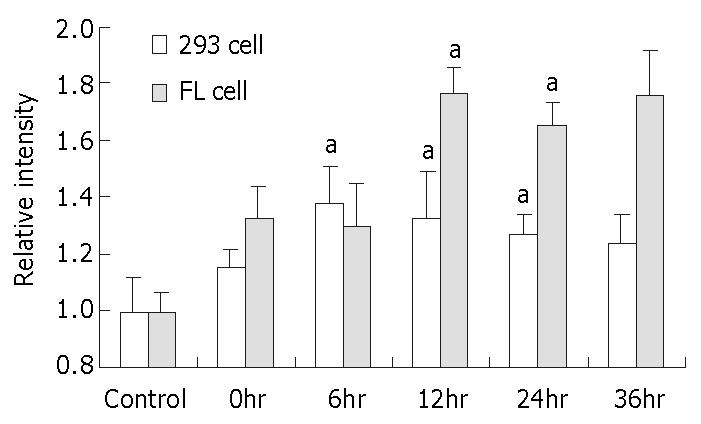
It was found that PCR with 31 cycles for 293 cells and 28 cycles for FL cells ensured the exponential amplification of REV3 and ARF1 within the same tube (data not shown). The expression of REV3 was upregulated at the transcriptional level in both 293 cells and FL cells after MNNG treatment. In MNNG-treated 293 cells, the level of REV3 expression was significantly increased by 0.38 fold at 6 h, 0.33 fold at 12 h and 0.27 fold at 24 h, when compared with the control (P < 0.05, Figure 2). Similarly, the transcriptional level of REV3 was also significantly increased by 0.77 fold at 12 h and 0.65 fold at 24 h in MNNG-treated FL cells, when compared with the control (all P < 0.05, Figure 2). The data suggest that the human mutator REV3 gene was activated by low concentration MNNG treatment and could be regulated at the transcriptional level.
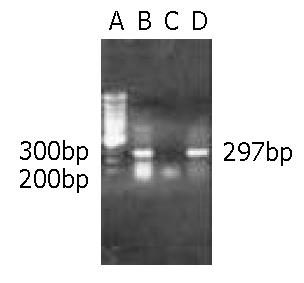
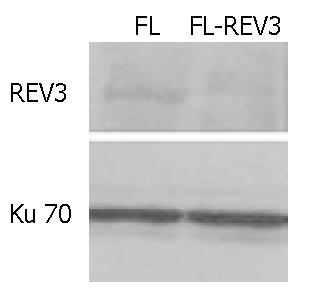
An expected high level of 297 bp antisense RNA to C-terminal of REV3 was detected in transgenic cells by RT-PCR (Figure 3). In addition, the results of Western blotting showed that the REV3 protein level was obviously reduced in transgenic cells (Figure 4). Therefore, it was indicated that the function of REV3 protein was partially blocked by antisense REV3 fragment in transgenic cells.
HPRT locus is traditionally used as a genetic marker for genome instability. Normally the spontaneous mutation frequency at HPRT locus was quite low. In the present study, we observed that the spontaneous mutation frequency was 2.87 × 10-6 in FL cells, and 4 × 10-6 in FL-M cells. Interestingly, in FL-REV3-cells, no spontaneous mutants were observed. This observation led to the speculation that REV3 may be involved in the process of spontaneous mutagenesis.
Previous studies found that MNNG could induce HPRT mutation in human cells[53]. It would be of interest to know if MNNG had the same effect on FL and the derived FL-M and FL-REV3- cells. As shown in Table 1, we observed that MNNG treatment significantly elevated the mutation frequency from 2.87 × 10-6 to 8.66 × 10-6 at HPRT locus in FL cells. Similarly, the mutation frequency was also increased in FL-M cells by MNNG treatment from 4 × 10-6 to 18.75 × 10-6. On the other hand, the induced mutation frequency was only 0.14 × 10-6 cells in FL-REV3- cells, which was significantly lower than that of the spontaneous mutation frequency in FL cells (Table 1).
Intact and undamaged shuttle plasmid pZ189 DNA was introduced into MNNG pretreated human cells. Progeny plasmids were harvested 48 h after transfection, and used to transform MBM7070. White and light blue colonies were picked and the frequency of SupF tRNA mutants was scored. As shown in Table 2, the spontaneous mutation frequencies were at comparable level between each cell lines. Untargeted mutation on undamaged DNA templates was increasingly induced in MNNG-pretreated 293, 293-M, FL and FL-M cells, with the mutation frequencies occurred in these cells being 4.5-5.8-fold higher than those in control groups. However, the untargeted mutation frequencies significantly decreased by 3.8 fold (from 7.37 × 10-4 to 1.52 × 10-4, P < 0.05) and 5.8 fold (from 27.4 × 10-4 to 4.0 × 10-4, P < 0.01) respectively in MNNG-pretreated transgenic 293 cells and FL cells in which REV3 was blocked by antisense RNA, and the mutation frequencies were almost similar to their spontaneous mutation levels.
| Cell line | DMSO | MNNG | ||||
| Number of transformant | Number of mutant | Mutation frequency(10-4) | Number of transformant | Number of mutant | Mutation frequency(10-4) | |
| 293 | 7954 | 1 | 1.26 | 12205 | 9 | 7.37ab |
| 293-M | 15358 | 2 | 1.3 | 12040 | 7 | 5.81ab |
| 293-REV3- | 39236 | 9 | 2.29 | 19758 | 3 | 1.52 |
| FL | 13495 | 7 | 5.2 | 13854 | 38 | 27.4cd |
| FL-M | 13272 | 7 | 5.3 | 10310 | 28 | 27.2cd |
| FL-REV3- | 10967 | 3 | 2.7 | 12609 | 5 | 4 |
The interconnections between environment and human health have been increasingly recognized. With the increasing cases of environmental cancer in the world range, especially in developing countries, investigation on the potential biomarkers for environmental risk assay or new targets for gene therapy is an emergent task to prevent and control the carcinogenesis. In China, the incidence of gastric cardia cancer has greatly increased in the past 2-3 decades, and dietary habits might be one of the risk factors for the cardia carcinogenesis among Chinese population[54]. Recently, it was found that COX-2 may contribute to progression of tumor in human gastric adenocarcinoma[55]. However, it has become clear that the induction of carcinogenesis is a complex multi-step process involving a series of genetic and epigenetic changes. For example, the induction of colon cancer requires alterations in at least three tumor-suppressor genes (MCC, DCC, and p53) and activation of the oncogene K-ras[56-58]. The genetic changes mainly occur in initiation, malignant conversion and progression stages in the development of malignant tumors[59]. DNA damaging agents can induce lesions in DNA template, causing the block on DNA replication fork. However, it also leads to the activation of several TLS DNA polymerases, especially the activation of pol ζ, to restart the replication process by replacing the normal replication polymerases and finally result in lesion-targeted mutation[40,42,60]. On the other hand, UV-light or chemical carcinogen can induce the UTM on undamaged DNA templates[31-33].
It was interesting to find that human REV3 gene, which encodes the catalytic subunit of TLS polymerase ζ, was activated by the carcinogen MNNG that can induce gastric and colorectal cancer. Our computational analysis indicated that transcriptional factor binding sites for CREB, AP-1 and NF-κB were found in the promoter region of REV3 (data not shown). Previous studies in our laboratory have shown that MNNG treatment activates CREB[61], AP-1 and NF-κB (unpublished data) in mammalian cells as early epigenetic events, which indicates that REV3 could be activated by MNNG via the activation of specific transcriptional factors in advance.
Mutation at HPRT locus can be used as an indicator to reflect the degree of genome instability[62]. It has been recognized that HPRT mutants are generated directly by DNA damage[62,63], i.e., the mutation spectrum belongs to lesion-targeted mutation. In human fibroblasts, the number of UV-induced HPRT mutants is significantly increased, whereas, the mutation is remarkably depressed in the human cells that express high levels of REV3 antisense RNA[47]. In this study, our data showed that the spontaneous mutation of HPRT locus in human cells was dependent on the function of REV3, since mutation at HPRT locus was eliminated in cells expressing antisense REV3 (Table 1). On the other hand, REV3 gene was also involved in MNNG-induced HPRT mutation, like in UV-induced mutation[47], as the antisense block of REV3 function significantly decreased the MNNG-induced mutation frequency. It is also possible that other factors might be involved in MNNG-induced HRPT mutagenesis, for example, the function of human REV1 gene is required for mutagenesis at HPRT locus induced by UV light[64].
Interestingly, our data further indicated that human REV3 gene also played a role in mutation genesis occurred on undamaged DNA templates. Unlike the role of REV3 in lesion-targeted mutation, the spontaneous mutagenesis in SupF tRNA gene in pZ189 replicated in human cells was REV3-independent, i.e., the antisense block of REV3 has no effect on the spontaneous mutations (Table 2). It was suggested that most of the spontaneous mutation occurring in such an experimental system are due to the deletion damage induced by the shear force during transfection. Different mechanisms are involved in repairing the base damage and deletion damage, in the later case no evidence was presented of REV3 dependent. In this study, however, we proved that MNNG-induced mutation on undamaged DNA templates was REV3-depedent (Table 2). To date, we still do not know whether there are other factors involved in untargeted mutation in addition to the human REV3 gene. Taken together, these data strongly suggest that human REV3 gene is capable of inducing mammalian genome instability, and this mutator gene could be a potential target for gastric and colorectal cancer prevention and gene therapy.
Edited by Xia HHX
| 1. | Stenius U, Högberg J. Re: Yang, J. and Duerksen-Hughes, P. (1998) A new approach to identifying genotoxic carcinogens: p53 induction as an indicator of genotoxic damage. Carcinogenesis, 19, 1117-1125. Carcinogenesis. 1999;20:181-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 66] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 2. | Li Y, Su JJ, Qin LL, Yang C, Luo D, Ban KC, Kensler T, Roebuck B. Chemopreventive effect of oltipraz on AFB(1)-induced hepatocarcinogenesis in tree shrew model. World J Gastroenterol. 2000;6:647-650. [PubMed] |
| 3. | Mortality trends for selected smoking-related cancers and breast cancer--United States, 1950-1990. MMWR Morb Mortal Wkly Rep. 1993;42:857, 863-866. [PubMed] |
| 4. | Bollschweiler E, Hölscher AH. [Carcinoma of the esophagus--actual epidemiology in Germany]. Onkologie. 2001;24:180-184. [PubMed] |
| 5. | Bonnin-Scaon S, Lafon P, Chasseigne G, Mullet E, Sorum PC. Learning the relationship between smoking, drinking alcohol and the risk of esophageal cancer. Health Educ Res. 2002;17:415-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 6. | Balaram P, Sridhar H, Rajkumar T, Vaccarella S, Herrero R, Nandakumar A, Ravichandran K, Ramdas K, Sankaranarayanan R, Gajalakshmi V. Oral cancer in southern India: the influence of smoking, drinking, paan-chewing and oral hygiene. Int J Cancer. 2002;98:440-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 210] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 7. | Bartal M. Health effects of tobacco use and exposure. Monaldi Arch Chest Dis. 2001;56:545-554. [PubMed] |
| 8. | Wang AH, Sun CS, Li LS, Huang JY, Chen QS. Relationship of tobacco smoking CYP1A1 GSTM1 gene polymorphism and esophageal cancer in Xi'an. World J Gastroenterol. 2002;8:49-53. [PubMed] |
| 9. | Badawi AF, Habib SL, Mohammed MA, Abadi AA, Michael MS. Influence of cigarette smoking on prostaglandin synthesis and cyclooxygenase-2 gene expression in human urinary bladder cancer. Cancer Invest. 2002;20:651-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 10. | Bernardini S, Adessi GL, Chezy E, Billerey C, Carbillet JP, Bittard H. Influence of cigarette smoking on P53 gene mutations in bladder carcinomas. Anticancer Res. 2001;21:3001-3004. [PubMed] |
| 11. | Borràs J, Borràs JM, Galceran J, Sánchez V, Moreno V, González JR. Trends in smoking-related cancer incidence in Tarragona, Spain, 1980-1996. Cancer Causes Control. 2001;12:903-908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Castelao JE, Yuan JM, Skipper PL, Tannenbaum SR, Gago-Dominguez M, Crowder JS, Ross RK, Yu MC. Gender- and smoking-related bladder cancer risk. J Natl Cancer Inst. 2001;93:538-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 188] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 13. | Chao A, Thun MJ, Henley SJ, Jacobs EJ, McCullough ML, Calle EE. Cigarette smoking, use of other tobacco products and stomach cancer mortality in US adults: The Cancer Prevention Study II. Int J Cancer. 2002;101:380-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 97] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Casimiro C. [Etiopathogenic factors in colorectal cancer. Nutritional and life-style aspects. 2]. Nutr Hosp. 2002;17:128-138. [PubMed] |
| 15. | Yang J, Duerksen-Hughes PJ. Activation of a p53-independent, sphingolipid-mediated cytolytic pathway in p53-negative mouse fibroblast cells treated with N-methyl-N-nitro-N-nitrosoguanidine. J Biol Chem. 2001;276:27129-27135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Palli D, Saieva C, Coppi C, Del Giudice G, Magagnotti C, Nesi G, Orsi F, Airoldi L. O6-alkylguanines, dietary N-nitroso compounds, and their precursors in gastric cancer. Nutr Cancer. 2001;39:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 17. | Knekt P, Järvinen R, Dich J, Hakulinen T. Risk of colorectal and other gastro-intestinal cancers after exposure to nitrate, nitrite and N-nitroso compounds: a follow-up study. Int J Cancer. 1999;80:852-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 18. | Sherenesheva NI, Mashkovtsev IuV. [Electron microscopy study of experimental stomach cancer]. Eksp Onkol. 1985;7:29-35. [PubMed] |
| 19. | Sasako M. [The effect of Nd: YAG laser irradiation on gastric cancer in rats induced by N-methyl-N'-nitro-N-nitrosoguanidine as a model of endoscopic laser treatment for early gastric cancers]. Nihon Geka Gakkai Zasshi. 1985;86:443-454. [PubMed] |
| 20. | Newberne PM, Charnley G, Adams K, Cantor M, Suphakarn V, Roth D, Schrager TF. Gastric carcinogenesis: a model for the identification of risk factors. Cancer Lett. 1987;38:149-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 21. | Yamashita S, Wakazono K, Sugimura T, Ushijima T. Profiling and selection of genes differentially expressed in the pylorus of rat strains with different proliferative responses and stomach cancer susceptibility. Carcinogenesis. 2002;23:923-928. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Amberger H. Different autochthonous models of colorectal cancer in the rat. J Cancer Res Clin Oncol. 1986;111:157-159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Narayan S, Jaiswal AS. Activation of adenomatous polyposis coli (APC) gene expression by the DNA-alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine requires p53. J Biol Chem. 1997;272:30619-30622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 24. | Li Z, Xiao W, McCormick JJ, Maher VM. Identification of a protein essential for a major pathway used by human cells to avoid UV- induced DNA damage. Proc Natl Acad Sci USA. 2002;99:4459-4464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 25. | Maenhaut-Michel G. Mechanism of SOS-induced targeted and untargeted mutagenesis in E. coli. Biochimie. 1985;67:365-369. [PubMed] |
| 26. | Pham P, Bertram JG, O'Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 91] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 27. | Otterlei M, Kavli B, Standal R, Skjelbred C, Bharati S, Krokan HE. Repair of chromosomal abasic sites in vivo involves at least three different repair pathways. EMBO J. 2000;19:5542-5551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 58] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci USA. 1997;94:13792-13797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 269] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 29. | Tang M, Bruck I, Eritja R, Turner J, Frank EG, Woodgate R, O'Donnell M, Goodman MF. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD'2C mutagenic complex and RecA protein. Proc Natl Acad Sci USA. 1998;95:9755-9760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 157] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 30. | Tang M, Pham P, Shen X, Taylor JS, O'Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 347] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 31. | Lawrence CW, Christensen RB. The mechanism of untargeted mutagenesis in UV-irradiated yeast. Mol Gen Genet. 1982;186:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 32. | Boesen JJ, Stuivenberg S, Thyssens CH, Panneman H, Darroudi F, Lohman PH, Simons JW. Stress response induced by DNA damage leads to specific, delayed and untargeted mutations. Mol Gen Genet. 1992;234:217-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 33. | Zhang X, Yu Y, Chen X. Evidence for nontargeted mutagenesis in a monkey kidney cell line and analysis of its sequence specificity using a shuttle-vector plasmid. Mutat Res. 1994;323:105-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 34. | Lawrence CW, Maher VM. Mutagenesis in eukaryotes dependent on DNA polymerase zeta and Rev1p. Philos Trans R Soc Lond B Biol Sci. 2001;356:41-46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Quah SK, von Borstel RC, Hastings PJ. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980;96:819-839. [PubMed] |
| 36. | Rechkoblit O, Zhang Y, Guo D, Wang Z, Amin S, Krzeminsky J, Louneva N, Geacintov NE. trans-Lesion synthesis past bulky benzo[a]pyrene diol epoxide N2-dG and N6-dA lesions catalyzed by DNA bypass polymerases. J Biol Chem. 2002;277:30488-30494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 167] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989;171:5659-5667. [PubMed] |
| 38. | Roche H, Gietz RD, Kunz BA. Specificity of the yeast rev3 delta antimutator and REV3 dependency of the mutator resulting from a defect (rad1 delta) in nucleotide excision repair. Genetics. 1994;137:637-646. [PubMed] |
| 39. | Lawrence CW, Hinkle DC. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surv. 1996;28:21-31. [PubMed] |
| 40. | Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase zeta. Science. 1996;272:1646-1649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 506] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 41. | Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017-1024. [PubMed] |
| 42. | Baynton K, Bresson-Roy A, Fuchs RP. Analysis of damage tolerance pathways in Saccharomyces cerevisiae: a requirement for Rev3 DNA polymerase in translesion synthesis. Mol Cell Biol. 1998;18:960-966. [PubMed] |
| 43. | Xiao W, Fontanie T, Bawa S, Kohalmi L. REV3 is required for spontaneous but not methylation damage-induced mutagenesis of Saccharomyces cerevisiae cells lacking O6-methylguanine DNA methyltransferase. Mutat Res. 1999;431:155-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 44. | Diaz M, Velez J, Singh M, Cerny J, Flajnik MF. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol. 1999;11:825-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 88] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 45. | Diaz M, Verkoczy LK, Flajnik MF, Klinman NR. Decreased frequency of somatic hypermutation and impaired affinity maturation but intact germinal center formation in mice expressing antisense RNA to DNA polymerase zeta. J Immunol. 2001;167:327-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 120] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 46. | Zan H, Komori A, Li Z, Cerutti A, Schaffer A, Flajnik MF, Diaz M, Casali P. The translesion DNA polymerase zeta plays a major role in Ig and bcl-6 somatic hypermutation. Immunity. 2001;14:643-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 167] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 47. | Gibbs PE, McGregor WG, Maher VM, Nisson P, Lawrence CW. A human homolog of the Saccharomyces cerevisiae REV3 gene, which encodes the catalytic subunit of DNA polymerase zeta. Proc Natl Acad Sci USA. 1998;95:6876-6880. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 252] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 48. | Xiao W, Lechler T, Chow BL, Fontanie T, Agustus M, Carter KC, Wei YF. Identification, chromosomal mapping and tissue-specific expression of hREV3 encoding a putative human DNA polymerase zeta. Carcinogenesis. 1998;19:945-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 49. | Xu F, Yu Y, Song TN. Establishment of cell lines whose hREV3 gene expression was inhibited by transfection of antisense RNA expression plasmids and their biological characteristics. Zhongguo Bingli Shengli Zazhi. 2000;16:293-297. |
| 50. | Xu F, Yu YN, Wu WW. Construction of eukaryotic expression plasmid expressing antisense fregment of hREV3. Zhongguo Bingli Shengli Zazhi. 1999;703-706. |
| 51. | Qian Y, Yu Y, Cheng X, Luo J, Xie H, Shen B. Molecular events after antisense inhibition of hMSH2 in a HeLa cell line. Mutat Res. 1998;418:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 52. | Kastenbaum MA, Bowman KO. Tables for determining the statistical significance of mutation frequencies. Mutat Res. 1970;9:527-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 779] [Cited by in RCA: 709] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 53. | Wang R. [Studies on human cervical carcinoma cell line. II. Establishment of HGPRT- cell line CC-80IAR2 and its biological characteristics]. Zhongguo Yixue Kexueyuan Xuebao. 1992;14:461-464. [PubMed] |
| 54. | Cai L, Zheng ZL, Zhang ZF. Risk factors for the gastric cardia cancer: a case-control study in Fujian Province. World J Gastroenterol. 2003;9:214-218. [PubMed] |
| 55. | Xue YW, Zhang QF, Zhu ZB, Wang Q, Fu SB. Expression of cyclooxygenase-2 and clinicopathologic features in human gastric adenocarcinoma. World J Gastroenterol. 2003;9:250-253. [PubMed] |
| 56. | Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R. Allelotype of colorectal carcinomas. Science. 1989;244:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 898] [Cited by in RCA: 879] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 57. | Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1202] [Cited by in RCA: 1191] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 58. | Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1217] [Cited by in RCA: 1129] [Article Influence: 35.3] [Reference Citation Analysis (0)] |
| 59. | Bast Robert C, Kufe Donald W, Pollock Raphael E, Weichselbaum Ralph R, Holland James F, Frei E, editors . Chemical carcinogenesis: Multi-stage carcinogenesis. Cancer Medicine 5th ed. Canada: BC Decker Inc. 2000;. |
| 60. | Kunz BA, Straffon AF, Vonarx EJ. DNA damage-induced mutation: tolerance via translesion synthesis. Mutat Res. 2000;451:169-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 53] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 61. | Wang G, Yu Y, Chen X, Xie H. Low concentration N-methyl-N'-nitro-N-nitrosoguanidine activates DNA polymerase-beta expression via cyclic-AMP-protein kinase A-cAMP response element binding protein pathway. Mutat Res. 2001;478:177-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 62. | Wilson VL, Wade KR, Yin X, Albertini RJ. Temporal delineation of sequential HPRT mutations arising in vivo in a T-cell clone with a mutator phenotype. Mutat Res. 2001;473:181-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 63. | Leonhardt EA, Trinh M, Chu K, Dewey WC. Evidence that most radiation-induced HPRT mutants are generated directly by the initial radiation exposure. Mutat Res. 1999;426:23-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 64. | Gibbs PE, Wang XD, Li Z, McManus TP, McGregor WG, Lawrence CW, Maher VM. The function of the human homolog of Saccharomyces cerevisiae REV1 is required for mutagenesis induced by UV light. Proc Natl Acad Sci USA. 2000;97:4186-4191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 140] [Article Influence: 5.6] [Reference Citation Analysis (0)] |