Published online Dec 15, 2003. doi: 10.3748/wjg.v9.i12.2801
Revised: August 16, 2003
Accepted: October 12, 2003
Published online: December 15, 2003
AIM: Little has been known about the pathogenesis of non-erosive reflux disease (NERD). Recent studies have implicated interleukin 8 (IL-8) in the development and progression of gastroesophgeal reflux disease (GERD). The purpose of this study was to determine IL-8 RNA expression levels in NERD patients with or without subtle mucosal changes.
METHODS: We studied 26 patients with NERD and 13 asymptomatic controls. Biopsy sample was taken from the esophagus 3 cm above the gastroesophageal junction and snap frozen for measurement of IL-8 mRNA levels by real-time quantitative polymerase chain reaction (PCR). We also examined mRNA expression of IL-8 receptors, CXCR-1 and -2 by reverse transcriptase PCR. The patients were endoscopically classified into grade M (mucosal color changes without visible mucosal break) and N (neither minimal involvement nor mucosal break) of the modified Los Angeles classification.
RESULTS: The relative IL-8 mRNA expression levels were significantly higher in esophageal mucosa of NERD patients than those of the controls. There was a significant difference in IL-8 mRNA levels between grade M and N. The CXCR-1 and -2 mRNAs were constitutively expressed in esophageal mucosa.
CONCLUSION: Our results suggest that high IL-8 levels in esophageal mucosa may be involved in the pathogenesis of NERD through interaction with its receptors. NERD seems to be composed of a heterogeneous population in terms of not only endoscopically minimal involvement but also immune and inflammatory processes.
- Citation: Kanazawa Y, Isomoto H, Wen CY, Wang AP, Saenko VA, Ohtsuru A, Takeshima F, Omagari K, Mizuta Y, Murata I, Yamashita S, Kohno S. Impact of endoscopically minimal involvement on IL-8 mRNA expression in esophageal mucosa of patients with non-erosive reflux disease. World J Gastroenterol 2003; 9(12): 2801-2804
- URL: https://www.wjgnet.com/1007-9327/full/v9/i12/2801.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i12.2801
Gastroesophageal reflux disease (GERD) is one of the most common chronic disorders in modern humans. In the United States, 44% of the adult populations reported experiencing heartburn at least once a month, 14% on a weekly basis, and 7% daily[1]. Esophageal erosions are the characteristic lesions of GERD seen on endoscopy[2]. A small number of GERD patients develop stricture, Barrett’s esophagus and adenocarcinoma of the esophagus. In fact, the majority of GERD patients have endoscopically normal-appearing esophageal mucosa; this group is termed non-erosive reflux disease (NERD) or endoscopy-negative reflux disease[2,3].
The Los Angeles (LA) classification is widely used for endoscopic assessment of GERD[4]. The slightest degree of esophagitis, i.e., grade A, is defined as one or more mucosal breaks confined to the mucosal folds, each no longer than 5 mm. Accordingly, this classification scheme ignores subtle mucosal damage in the absence of mucosal breaks. In this regard, Hoshihara et al[5,6] have proposed a modified LA system, in which grade O was subdivided into M and N, based on the concept of mucosal color changes. Thus, NERD patients can be classified into two subgroups (grade M and N) based on minimal esophageal involvement during endoscopy.
Recently, several studies have shown that mucosal immune and inflammatory responses, characterized by specific cytokine and chemokine profiles, may determine the diversity of esophageal phenotypes of GERD[7-9]. Of note, Fitzgerald et al[7] reported significantly higher expression levels of interleukin 8 (IL-8) messenger ribonucleic acid (mRNA) in patients with reflux esophagitis (RE), assessed by competitive reverse transcriptase polymerase chain reaction (RT-PCR), compared with subjects with non-inflamed or Barrett’s esophagus. Studies from our laboratories have also demonstrated high IL-8 protein levels in esophageal biopsy samples of patients with erosive esophagitis by enzyme linked immunosorbent assay (ELISA)[8]. Furthermore, we also showed significantly high mucosal IL-8 production, which paralleled the endoscopic severity of RE[8]. However, little has been known about the role of IL-8 in NERD.
The aim of the present study was to assess esophageal expression levels of IL-8 mRNA in NERD patients by quantitative real-time PCR procedure, with special reference to the difference between grade M and N subgroups of the modified LA scheme.
We studied 26 patients with NERD and endoscopically confirmed normal-appearing esophageal mucosa who visited the Outpatient Department between August 2002 and July 2003. They included 19 men and 7 women, aged between 28 and 80 years (mean, 62.0 years). The diagnosis of GERD was made with more than 6 points in the questionnaire for the diagnosis of reflux disease (QUEST) described by Carlsson et al[10]. None of these patients had been treated with non-steroidal anti-inflammatory drugs, proton pump inhibitors, histamine H2-receptor antagonists, anti-cholinergic agents or antibiotics within 4 weeks prior to the present study. Furthermore, patients with severe concomitant diseases, prior esophageal or gastric surgery, peptic ulcer diseases and comorbid conditions that might interfere with esophageal or gastric motility including diabetes mellitus, systemic sclerosis and neurological disorders were excluded. As a control group, we recruited 13 asymptomatic subjects with no hiatal hernia or any lesions in the esophagus, stomach and duodenum at endoscopy for a health check-up.
In each case, a biopsy specimen was obtained from the esophageal mucosa, 3 cm above the gastroesophageal junction[8], snap-frozen in an ethanol-dry ice mixture for quantitative analysis of IL-8 mRNA expression and stored at -80 °C until use.
NERD was endoscopically classified into grade M and N in accordance with the modified Los Angeles (LA) classification system proposed by Hoshihara et al[5,6]. The criteria were: grade M represents minimal changes (irregular redness or whiteness) without any mucosal breaks and grade N represents esophageal mucosa with neither the minimal changes nor mucosal injury. In addition, we also evaluated the presence of hiatal hernia by endoscopy[11].
Total RNA from the biopsy samples was extracted using a commercial kit according to the instructions provided by the supplier (Isogen, Nippon Gene Co., Toyama, Japan). One μg of total RNA was reversely transcribed into complementary DNA (cDNA) in a volume of 25 μl with MuLV reverse transcriptase and random hexamers (both from PE Applied Biosystems, Warrington, UK).
Real-time PCR measurement of IL-8 cDNA was performed in the ABI PRISM 7700 sequence detector (PE Applied Biosystems) with TaqMan assay. The primers and probe sequences for IL-8 were synthesized (PE Applied Biosystems) as described previously[12]: IL-8 forward primer, 5’-CTCTTGGCAGCCTTCCTGATT-3’, reverse primer, 5’-TATGCACTGACATCTAAGTTCTTTAGCA-3’ and probe, 5’-CTTGGCAAAACTGCACCTTCACACAGA-3’, labeled with the reporter dye 6-carboxyfluorescein at the 5’ end and quencher dye 6-carboxytetramethylrhodamine at the 3’ end. PCR was performed in a total volume of 50 μl of each amplification mixture containing 1 μl of each RT product, 25 μl of 2 × Universal Master Mix (PE Applied Biosystems), 200 nM IL-8 forward and reverse primers, 100 nM fluorogenic probe. Thermal cycling was initiated with at 50 °C for 2 min, followed by a first denaturation step at 95 °C for 10 min, and followed by 50 cycles of at 95 °C for 15 s and at 60 °C for 1 min.
The tubulin alpha 3 gene cDNA (internal control) was quantified in the same machinery using SYBR Green PCR Core reagents kit (PE Applied Biosystems). The primers used were: forward, 5’-AGATCATTGACCTCGTGTTGGA-3’ and reverse, 5’-ACCAGTTCCCCCACCAAAG-3’, which correspond to nucleotides 437-458 and 537-519, respectively (TUBA3, GenBank accession number 17986282). PCR was performed in a total volume of 25 μl of each amplification mixture containing 1 μl of each RT product, 3 μl of 25 mM MgCl2, 2.5 μl of 10 × SYBR Green buffer, 2 μl of dNTP Mix (5 mM adenosine, deoxycytosine and deoxyguanosine triphosphate and 2.5 mM deoxyuridine triphosphate), 0.625 U AmpliTaq Gold polymerase, 0.125 U AmpErase and 100 nM tubulin alpha 3 forward and reverse primers. Thermal cycling was initiated at 50 °C for 2 min, followed by a first denaturation step at 95 °C for 10 min, and continued with 40 cycles of at 95 °C for 15 s and at 59 °C for 1 min.
Each assay included a standard curve, a no-template control and cDNA samples in triplicate. The standard curve was generated by serial 5-fold dilutions of pooled cDNA obtained from gastric tissues that were found to contain high levels of mRNAs of both genes. Contents of the tubulin alpha 3 and IL-8 cDNAs were expressed in arbitrary units calculated according to the standard curve. The relative expression level of IL-8 was expressed as the ratio of IL-8/tubulin alpha 3 in arbitrary units[13].
Based on the technique described previously[14] with slight modification, the target sequence of CXCR-1 mRNA was amplified through 35 cycles, each consisting of denaturation at 94 °C for 30 sec, annealing at 53 °C for 30 sec and extension at 72 °C for 30 min, followed by a final extension at 72 °C for 5 min with specific primers (forward, 5’-CAGATCCACAGA-TGTGGGAT-3’ and reverse, 5’-TCCAGCCATTCACCTTG-GAG-3’) using an RT-PCR kit (Takara Shuzo Co., Otsu, Japan). Similarly, CXCR-2 mRNA expression was detected under the following conditions: amplification through 35 cycles, each consisting of denaturation at 94 °C for30 sec, annealing at 60 °C for 30 sec and extension at 72 °C for 30 min, followed by a final extension at 72 °C for 5 min with specific primers (forward, 5’-AGCTGCTCTTCTGGAGGTGT-3’ and reverse, 5’-TTAGAGAGTAGTGGAAGTGTGC-3’)[14]. A 10-µl aliquot of each PCR product was analyzed by electrophoresis on 2% agarose gel containing ethidium bromide, and the bands were examined under ultraviolet light for the presence of amplified DNA. Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene transcript was also amplified as described previously[15], and used as an internal control of the processed RNA for each preparation.
H pylori status was assessed by serology (anti-H pylori Immunoglobulin G antibody, HEL-p TEST, Amrad Co., Melbourne, Australia), rapid urease test (Helicocheck, Otsuka Pharmaceutical Co., Tokushima, Japan) and histopathology (hematoxylin-eosin and Giemsa staining) using additional biopsy specimens obtained during endoscopy from the antrum within 2 cm of the pyloric ring and the corpus along the greater curvature. Patients were considered positive for H pylori infection when at least two of these examinations yielded positive results. On the other hand, patients were defined as H pylori-negative if all the test results were negative[16].
All the samples were obtained with written informed consent of the patients prior to their inclusion in this study, in accordance with the Helsinki Declaration.
Statistical analyses were performed using Fisher’s exact, χ2, Student’s t, Mann-Whitney U, and Kruskal-Wallis tests, whenever appropriate. A P value of less than 0.05 was accepted as statistically significant. Data are expressed as mean ± standard deviation (SD).
According to the modified LA system, 14 patients were classified as grade M and 12 as grade N. There were no significant differences in age, gender, current tobacco use, alcohol intake, body mass index, the presence of hiatus hernia and H pylori status among the patients with grade M and N and the controls (Table 1). None had such complications as stricture, bleeding and columnar-lined esophagus. The overall incidence of H pylori infection in our series was 51.3%.
| Control group n = 13 | Nonerosive reflux disease group | ||
| Grade Man = 14 | Grade Nan = 12 | ||
| Mean age, yr, (range) | 61.6(39-80) | 58.7(28-75) | 62.5 (33-75) |
| Male/female | 8/5 | 11/3 | 8/4 |
| Smoker | 53.8%(7/13) | 28.6%(4/14) | 33.3%(4/12) |
| Alcohol drinker | 46.2%(6/13) | 57.1%(8/14) | 33.3%(4/12) |
| Hiatal hernia | 0%(0/13) | 35.7%(5/14) | 50.0%(6/12) |
| H pylori infection | 53.8%(7/13) | 57.1%(8/14) | 41.7%(5/12) |
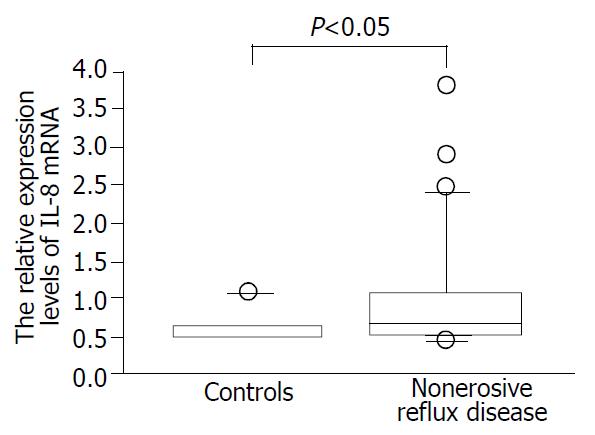
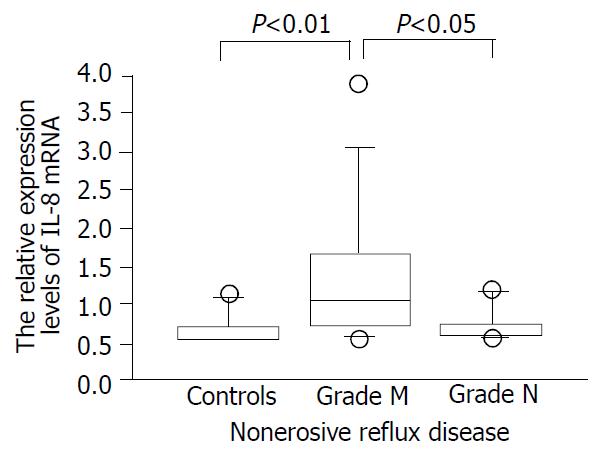
We confirmed that both RT-PCR procedures for IL-8 and tubulin alpha 3 yielded 87- and 101-base pair (bp) specific bands, respectively (data not shown). As a whole, NERD patients had significantly higher expression levels of IL-8 than the controls (Figure 1, P < 0.05). The expression levels of IL-8 in esophageal mucosa of grade M patients with NERD were significantly higher than those of grade N patients (P < 0.05, Figure 2). In addition, the expression levels of IL-8 were higher in grade M than control group (P < 0.01, Figure 2), but not significantly different between NERD-grade N and control group.
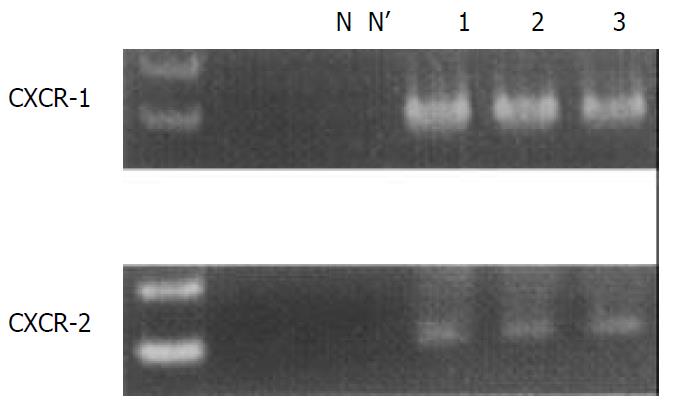
We identified the CXCR-1 and -2 gene-specific products as 257- and 1 154-bp bands, respectively, by RT-PCR (Figure 3). These mRNAs were constitutively expressed in each subject examined, irrespective of the presence of GERD-related symptoms or endoscopic grading. We confirmed that RT-PCR procedures for G3PDH housekeeping gene expression yielded 983 bp specific bands (data not shown).
Several lines of evidence indicated that a prototype of CXC chemokine, IL-8, played a crucial role in the development and progression of erosive esophagitis[7,8]. In the present study, we focused on the implication of this potent inflammatory mediator in NERD. We found significantly higher expression levels of IL-8 mRNA in esophageal mucosa of patients with NERD than those in asymptomatic controls, suggesting that IL-8 is implicated in the pathogenesis of this incipient form of GERD.
The striking finding of this study was that NERD patients classified as grade M subgroup based on the modified LA system had significantly higher expression levels of IL-8 mRNA compared to those of grade N. IL-8 mediated the recruitment of neutrophils into sites of inflammation[17]. In addition, this potent chemoattractant acted on neutrophils to respiratory burst and release of a variety of reactive oxygen species (ROS), leading to tissue damage[18]. In our previous work employing ELISA, we found a significant association between the presence of intraepithelial neutrophils and increased IL-8 levels in esophageal mucosa of patients with RE[13]. Although we did not perform histopathological evaluation in the current study, elevated mucosal IL-8 expression may be involved even in such subtle mucosal changes as seen in the grade M subcategory, probably through its action on neutrophils, thus triggering chemotaxis and generating harmful ROS. This result also highlighted the possibility that NERD patients could encompass diverse subpopulations in terms of immune and inflammatory reactions. Further studies on the implication of other members of chemokines and proinflammatory cytokines can shed light on our understanding of the mechanisms underlying this poorly studied disorder.
In the present study, we demonstrated constitutive mRNA expression of CXCR-1 and -2 in esophageal biopsy samples by RT-PCR procedure. To date, CXCR-1 and -2 are two distinct receptors for IL-8[19]. It is suggested that the increased IL-8 may facilitate trafficking of neutrophils into the mucosa affected by GERD process through the interaction with these receptors. Again, recent data from our laboratories showed a significant correlation between IL-8 protein levels and basal layer hyperplasia as well as papillary elongation in patients with RE[13]. IL-8 also exerted mitogenic actions directly or by binding to its receptors on epithelial cells[20,21]. Taken together, it is possible that IL-8, together with other cytokines as well as growth factors[14,22], could contribute to epithelial cell proliferation even in NERD, and could be eventually linked to carcinogenesis. Again, unlike CXCR-1, CXCR-2 is not specific for IL-8 and can bind to other chemokines such as growth related oncogene α, but it has 2- to 5-fold higher affinity for IL-8 than CXCR-1[19]. Further studies on the distribution of diverse IL-8 receptors and the receptor-mediated signaling pathway may help to elucidate the pathogenesis of GERD via IL-8 action.
In conclusion, our study demonstrated significantly enhanced expression of IL-8 mRNA level in NERD by real-time PCR technology. The interaction of IL-8 with CXCR-1 and -2 is likely to be involved in the pathogenesis of NERD. We also showed a significant difference in IL-8 mRNA levels between grade M and N subgroups of the modified LA classification, indicating the heterogeneity of NERD patients both immunologically and endoscopically.
Edited by Zhu LH
| 1. | Fass R, Ofman JJ. Gastroesophageal reflux disease--should we adopt a new conceptual framework. Am J Gastroenterol. 2002;97:1901-1909. [PubMed] |
| 2. | Frierson HF. Histology in the diagnosis of reflux esophagitis. Gastroenterol Clin North Am. 1990;19:631-644. [PubMed] |
| 3. | Quigley EM. Factors that influence therapeutic outcomes in symptomatic gastroesophageal reflux disease. Am J Gastroenterol. 2003;98:S24-S30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Armstrong D, Bennett JR, Blum AL, Dent J, De Dombal FT, Galmiche JP, Lundell L, Margulies M, Richter JE, Spechler SJ. The endoscopic assessment of esophagitis: a progress report on observer agreement. Gastroenterology. 1996;111:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 779] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 5. | Hoshihara Y, Hashimoto M. [Endoscopic classification of reflux esophagitis]. Nihon Rinsho. 2000;58:1808-1812. [PubMed] |
| 6. | Nishiyama Y, Koyama S, Andoh A, Moritani S, Kushima R, Fujiyama Y, Hattori T, Bamba T. Immunohistochemical analysis of cell cycle-regulating-protein (p21, p27, and Ki-67) expression in gastroesophageal reflux disease. J Gastroenterol. 2002;37:905-911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 7. | Fitzgerald RC, Onwuegbusi BA, Bajaj-Elliott M, Saeed IT, Burnham WR, Farthing MJ. Diversity in the oesophageal phenotypic response to gastro-oesophageal reflux: immunological determinants. Gut. 2002;50:451-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 211] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 8. | Isomoto H, Wang A, Mizuta Y, Akazawa Y, Ohba K, Omagari K, Miyazaki M, Murase K, Hayashi T, Inoue K. Elevated levels of chemokines in esophageal mucosa of patients with reflux esophagitis. Am J Gastroenterol. 2003;98:551-556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 96] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 9. | Fitzgerald RC, Abdalla S, Onwuegbusi BA, Sirieix P, Saeed IT, Burnham WR, Farthing MJ. Inflammatory gradient in Barrett's oesophagus: implications for disease complications. Gut. 2002;51:316-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 153] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 10. | Carlsson R, Dent J, Bolling-Sternevald E, Johnsson F, Junghard O, Lauritsen K, Riley S, Lundell L. The usefulness of a structured questionnaire in the assessment of symptomatic gastroesophageal reflux disease. Scand J Gastroenterol. 1998;33:1023-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 300] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 11. | Jones MP, Sloan SS, Rabine JC, Ebert CC, Huang CF, Kahrilas PJ. Hiatal hernia size is the dominant determinant of esophagitis presence and severity in gastroesophageal reflux disease. Am J Gastroenterol. 2001;96:1711-1717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 156] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 12. | Yuan A, Yang PC, Yu CJ, Chen WJ, Lin FY, Kuo SH, Luh KT. Interleukin-8 messenger ribonucleic acid expression correlates with tumor progression, tumor angiogenesis, patient survival, and timing of relapse in non-small-cell lung cancer. Am J Respir Crit Care Med. 2000;162:1957-1963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 124] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Rogounovitch TI, Saenko VA, Shimizu-Yoshida Y, Abrosimov AY, Lushnikov EF, Roumiantsev PO, Ohtsuru A, Namba H, Tsyb AF, Yamashita S. Large deletions in mitochondrial DNA in radiation-associated human thyroid tumors. Cancer Res. 2002;62:7031-7041. [PubMed] |
| 14. | Kondo S, Yoneta A, Yazawa H, Kamada A, Jimbow K. Downregulation of CXCR-2 but not CXCR-1 expression by human keratinocytes by UVB. J Cell Physiol. 2000;182:366-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | Ohnita K, Isomoto H, Mizuta Y, Maeda T, Haraguchi M, Miyazaki M, Murase K, Murata I, Tomonaga M, Kohno S. Helicobacter pylori infection in patients with gastric involvement by adult T-cell leukemia/lymphoma. Cancer. 2002;94:1507-1516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Isomoto H, Furusu H, Morikawa T, Mizuta Y, Nishiyama T, Omagari K, Murase K, Inoue K, Murata I, Kohno S. 5-day vs. 7-day triple therapy with rabeprazole, clarithromycin and amoxicillin for Helicobacter pylori eradication. Aliment Pharmacol Ther. 2000;14:1619-1623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Mukaida N. Interleukin-8: an expanding universe beyond neutrophil chemotaxis and activation. Int J Hematol. 2000;72:391-398. [PubMed] |
| 18. | Graça-Souza AV, Arruda MA, de Freitas MS, Barja-Fidalgo C, Oliveira PL. Neutrophil activation by heme: implications for inflammatory processes. Blood. 2002;99:4160-4165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 225] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 19. | Chuntharapai A, Kim KJ. Regulation of the expression of IL-8 receptor A/B by IL-8: possible functions of each receptor. J Immunol. 1995;155:2587-2594. [PubMed] |
| 20. | Metzner B, Hofmann C, Heinemann C, Zimpfer U, Schraufstätter I, Schöpf E, Norgauer J. Overexpression of CXC-chemokines and CXC-chemokine receptor type II constitute an autocrine growth mechanism in the epidermoid carcinoma cells KB and A431. Oncol Rep. 1999;6:1405-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Zachrisson K, Neopikhanov V, Samali A, Uribe A. Interleukin-1, interleukin-8, tumour necrosis factor alpha and interferon gamma stimulate DNA synthesis but have no effect on apoptosis in small-intestinal cell lines. Eur J Gastroenterol Hepatol. 2001;13:551-559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 22. | Sfakianakis A, Barr CE, Kreutzer DL. Localization of the chemokine interleukin-8 and interleukin-8 receptors in human gingiva and cultured gingival keratinocytes. J Periodontal Res. 2002;37:154-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |