INTRODUCTION
Liver regeneration after 70% partial hepatectomy in an adult rat involves initiation of proliferation of the remaining parenchymal cells and is a useful model for studying signaling molecules and other factors involved in cell proliferation. Cell proliferation begins very early during liver regeneration, peaking at 24 h, followed by proliferating biliary epithelium at 48 h, and kupffer cells and stellate cells at 72 h. The proliferation of sinusoidal endothelial cells was peaked at 96 h. Through its regenerative ability, the liver provides a model system for in vivo study of cell proliferation events following reentry into the cells cycle from the quiescent G0 phase. The damage caused by surgical resection or treatment with toxins results in a cascade of growth factors and cytokines to restore the liver mass to its original size[1-6]. The residual hepatic parenchymal cells and nonparenchymal cells can proliferate and differentiate through the action of some cytokines, hormones and growth factors. When liver regeneration is induced by partial hepatectomy or inflammation, the cell cycle can transit from G0 phase to G1 phase, enter into the preparing stage of division and proliferation, and then into S phase, G2 phase and M phase in turn. The late G1 phase contains a restriction point (R point), which has a selective division function and decides cell entry into S phase or reverse to G0 phase. The hepatic parenchymal and nonparenchymal cells can reconstitute the hepatic volume when they proliferate to some extent, and the liver regenerating response can be terminated by some factors[1,7,8], but the mechanisms of initiation and termination of liver regeneration have not been eventually clarified and need further study. Recently, a novel gene named GANKyrin was cloned and identified from human hepatocellular carcinoma[9]. The GANKyrin gene sequence is identical to one subunit of 26S proteasome named p28 which was firstly cloned from human cDNA library by comparing a subunit amino acid of purified bovine erythrocyte PA700 complex (also defined as 19S complex) with protein structure of human protein cDNA library databases. The product of GANKyrin or p28 gene (p28GANK protein) was an oncoprotein consisting of six conservative ankyrin repeats. The mRNA and protein level of p28GANK increased substantially in hepatocellular carcinomatous tissues, compared with levels in the respective non-cancerous portion of the resected livers, but the increase was not related to the grade or stage of the cancer. The finding that the increase occurred regardless of the staging or grading of cancer indicates that p28GANK may be involved in an early and essential step of liver carcinogenesis. However, the liver regeneration is involved in hepatocyte division, proliferation and termination. It is unclear whether p28GANK can participate in hepatocyte proliferation. This study was intended to disclose the biological function of p28GANK by establishing a liver regeneration rat model and determining the expression of p28GANK mRNA and protein levels.
MATERIALS AND METHODS
Experimental animal
One hundred and thirty two adult male Sprague-Dawley rats were obtained from the Experimental Animal Center of the Second Military Medical University, weighing from 200-250 g, and were randomly divided into sham operation (SO) group and partial hepatectomy (PH) group. Each group had eleven time points: 0, 2, 6, 12, 24, 30, 48, 72, 120, 168 and 240 h, six rats in each time point. The rats were housed in a room at 21 °C under a 12 -hour light/dark cycle and given tap water and commercial rat chow. The animals were acclimatized to the laboratory conditions for 1 wk prior to the experiments.
Establishment of liver regenerating model
The rats were undergone 70% PH under methoxyflurane anesthesia by resection of the anterior and left lateral lobes of the liver[10]. SO was undertaken only by laparotomy plus slight mobilization of the liver without resection. The rats were injected intraabdominally with 0.5-1 mL normal saline for fluid resuscitation after operation. Liver specimens were collected at 0, 2, 6, 12, 24, 30, 48, 72, 120, 168 and 240 h after PH or SO. The liver specimens obtained were snap frozen in liquid nitrogen. Total RNA was extracted using the guanidine isothiocyanate and phenol-chloroform method and aliquots of RNA samples were stored at -85 °C until use. The remaining liver was fixed in 4% formaldehyde polymerisatum, embedded in paraffin and then sectioned for histological examination and analysis of protein expression.
Cloning of rat p28GANK cDNA
The rat p28GANK cDNA was cloned from rat placenta based on the GenBank (Rattus novegicus mRNA for p28GANK homologue: AB022014), sense (11 bp-30 bp, G1): 5’-GTGTGTCTAACCTAATGGTC-3’, anti-sense (643 bp-662 bp, G2): 5’-TTGAGTATTAACCCCAGGCC-3’. The segment length was 651 bp. The primers were synthesized by Sangon Company, Shanghai, P.R.China. Briefly, 5 µg of total RNA extracted from rat placenta served as template to synthesize the first strand cDNA. Polymerase chain reaction system was consisted of 1 × buffer, 1.5 mM MgCl2, 200 μM dNTPs, 0.5 μm of the oligonucleotide primers specific for rat p28GANK gene, 1 μL of the first strand cDNA synthesized as described above, and 1.25 units of Taq DNA polymerase. Amplification was undertaken by initial denaturation at 94 °C for 4 min, followed by 30 reaction cycles (for 50 s at 94 °C, for 50 s at 53 °C, and for 60 s at 72 °C) and a final cycle at 72 °C for 10 min. The PCR product with an expected size of 651 bp fractionated on a 2% agarose gel was purified with QIAQUICK gel extract kit (QIAGEN, Germany) and subcloned into PMD18 vector[11], and then sequenced and compared. The synthetic segment was verified the same sequence as the GenBank.
Northern blot analysis
Rat p28GANK cDNA released from PMD18 vector was labeled with α-32P-dCTP using Promega-gene labeling system (Promega, USA) according to the protocol of the manufacturer[12]. 90 μg of total RNAs was separated on a 1% formaldehyde/agarose gel by electropherosis, transferred to nitrocellulose and hybridized with α-32P-labeled p28GANK cDNA[13]. The hybridized nitrocellulose filters were exposed to X-ray film at -80 °C for 2 wk. The developed signals were assayed for p28GANK mRNA expression level after normalizion with 18S rRNA as internal standard using FujiFilm image gauge V3.3 analysis software (FujiFilm, Japan).
Immunohistochemistry
Immunohistochemical assay was performed as described by the manufacturer using SP kit (Dako Reagent, Denmark). Briefly, the rat liver sections of four-micrometer were deparaffnized, followed by blockage of endogenous peroxidase activity and restoration of the interested antigens. The sections were then subjected to incubation with purified primary polyclonal antibody (1:50 dilution, obtained from signal transduction laboratory, Shanghai, China) against p28GANK protein at 4 °C overnight and with horseradish peroxidase-biotinylated anti-rabbit IgG at 37 °C for 30 min. The sections were finally incubated with benzidine and counterstained with hematoxylin for examination. The experiment was performed on control sections with omission of the primary antibody in the same way. The expression of p28GANK was analyzed comprehensively by the staining extent and range referring to Fromowitz standard[14]. 5 visual fields were observed randomly, 100 cells were counted in each visual field. The average number of stained cells in 5 visual fields was regarded as the percent ratio of positively stained cells in each section. Positive range score: 0, 0%-5%; 1, 6%-25%; 2, 26%-50%; 3, 51%-75%; 4, > 75%. Positive extent score: 0, no staining; 1, light yellow; 2, brown; 3, dark brown. Judged by positive range score plus positive extent score: < 2, negative (-); 2-3, slight positive (+); 4-5, moderately positive (++); 6-7, strongly positive (+++).
Statistical analysis
The expression level of p28GANK mRNA detected by Northern Blot was presented as mean ± SD. ANOVA software (SPSS 8.0) was used to analyze the difference in mRNA level between the groups and Rank sum test for p28GANK protein assay in immunohistochemical study. P values less than 0.05 were considered as having significant difference.
RESULTS
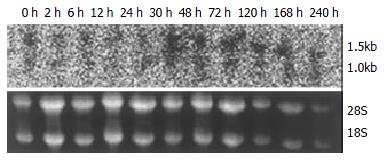
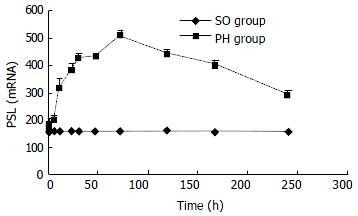
The expression of p28GANK mRNA in regenerating liver tissues possessed two transcripts, which were 1.5 kb and 1.0 kb respectively. There was a significant difference between SO and PH groups (P < 0.01). The expression of p28GANK mRNA increased 2 h after PH, the peak time was at 72 h (SO group: 163.83 ± 1.4720; PH group: 510.5 ± 17.0499, P < 0.01). There was a significant difference in the 1.5 kb transcript, which decreased gradually after 72 h (Figure 1, Figure 2).
Figure 1 Northern blot of p28GANK mRNA in rat regenerating liver tissues showed two transcripts of 1.
5 kb and 1.0 kb. Expression level of p28GANK mRNA increased 2 h after PH, and reached the peak level at 72 h. More significant variation was found for 1.5 kb transcript.
Figure 2 No significant difference of the expression of p28GANK mRNA was seen in the SO group.
In the PH group, the expres-sion increased in 1.5 kb transcript 2 h after PH. The peak time was at 72 h, but gradually decreased after 72 h.
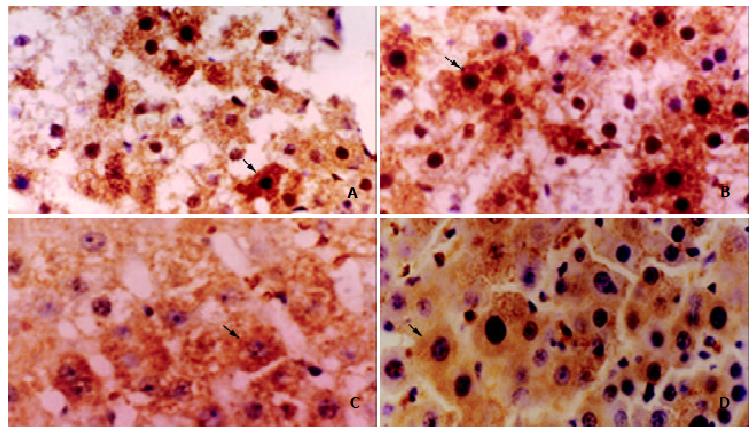
The protein expression of p28GANK was mainly in the cytoplasm of regenerating hepatocytes, and became increased near the central region 24 h after PH. The strongly positive expression was observed at 48 h (+++, vs the other time points, P < 0.05), which decreased 72 h after PH (Figure 3).
Figure 3 The expression of p28GANK protein in regenerating liver tissues was examined by immunohistochemistry.
A: Local brown expression of p28GANK in the cytoplasm and nucleus of regenerating hepatocyte near central region 24 h after PH. IHC × 400; B: Diffuse brown expression of p28GANK in the cytoplasm and nucleus of regenerating hepatocyte 48 h after PH. IHC × 400; C: Regional light yellow expression of p28GANK in cytoplasm of regenerating hepatocyte 72 h after PH. IHC × 400; D: Dispersed light yellow expression of p28GANK in cytoplasm of regenerating hepatocyte 120 h after PH. IHC × 400.
DISCUSSION
Hepatocytes proliferate firstly according to the cascade response induced by growth factors and cytokines after hepatectomy or toxin damage. The first peak of DNA synthesis in hepatocytes occurs at about 24 h, with a smaller peak between 36 and 48 h. The restoration of the original number of hepatocytes theoretically requires 1.66 proliferative cycles among the residual hepatocytes. Most of the hepatocytes (95% in the young and 75% in very old rats) in the residual lobes participate in one or two proliferative events. The hepatocytes begin to proliferate early, the peak time is at 24 h. Then biliary epithelial cells proliferate, the peak time is at 48 h. Then the Kupffer cells and stellate cells proliferate and the proliferative peak is at 72 h. The proliferative peak of sinusoidal endothelial cells is at 96 h. Thus these cell proliferations result in restoration of the original volume[3,15]. Hepatocyte proliferation starts in the periportal zone, and then proceeds to the pericentral zone at 36 to 48 h. The other cells of the liver enter into DNA synthesis about 24 h after the hepatocytes proliferation, with a peak of DNA synthesis at 48 h or later. The changes in the extracellular matrix after PH are undetectable until approximately 24 h post-PH. The hepatocytes begin to replicate in the periportal zones without corresponding increases in matrix within 3 d. The periportal areas consist of hepatocyte clusters of 10-14 cells without intervening sinusoids or matrix, as the wave of hepatocyte replication proceeds from periportal to pericentral regions of the lobule. The formation of cell clusters follows, the hepatocyte mitotic activity significantly decreases 4 d post-PH[16-20]. Liver regeneration is a complicated regulatory process, the volume and function of the liver restore after several periods. Many studies focus on the early factors associated with liver regeneration recently and on the important roles of these factors permitting the hepatocytes entering into cell division cycles. Activations of transcription factors (NF-κB and STAT3), immediately early protooncogenes (C-fos, C-myc, C-jun) and proinflammatory factors (TNF-α and IL-6) constitute the early responses to PH congenerically. The early changes play an important role in promoting the hepatocytes into cell cycles induced by hepatocytic reduction. Recruitment of early factors may be a part of ubiquitous response to stress protection and antiapoptosis of the liver, but it is unclear why the late downstream signal dissociates similar to the early response and certain time difference of cell proliferation process and atrophy or maintenance of liver size[21-24].
p28GANK is a novel gene cloned and identified from human hepatocellular carcinoma and is identical to one of the PA700 non-ATPase subunits, a 700 kDa multi-subunit regulatory complex of human 26S proteasome. p28GANK contains six conservative ankyrin repeats, which suggests it may contribute to 26S proteasome interaction with other proteins, and its effect in hepatocellular carcinoma may be related to ubiquitin-proteasome pathway which is often the target of cancer-related deregulation, and is involved in the processes such as oncogenic transformation, tumor progression, escape from immune surveillance and drug resistance[25-29]. The mRNA and protein levels of p28GANK was increased in cancerous tissues of hepatocellular carcinoma, and the increase was more substantial than that of non-cancerous tissues and was not related to the grading or staging of the tumors, indicating that p28GANK plays an early and crucial role in liver carcinogenesis. Moreover, the population doubling times in p28GANK-transfected mouse NIH/3T3 cells were 18-20 h, the control cells were 22-25 h. All cells transfected with p28GANK formed colonies in soft agar, whereas the control cell did not. In homozygous nude mice, inoculation of all positive clones overexpressing p28GANK produced tumors within 30 d, in contrast, none of nude mice inoculated with control clone developed tumors. These results demonstrated that p28GANK remarkably promoted cell proliferation and was an oncogene. p28GANK had the retinoblastoma (RB1)-binding motif LxCxE, resulted in an increased amount of the hyperphosphorylated form of RB1, and accelerated the degradation of RB1, activated the E2F transcription factors of nuclear partners of RB1 by increasing the phosphorylation of RB1, and induced the growth and oncogenity of anchoring independent cells. RB1 exerts its growth-inhibitory effects in part by binding to and inhibiting essential regulatory proteins, including members of the E2F family of transcription factors. E2F is selectively associated with hypophosphorylated RB1. Inactivation of RB1 by phosphorylation, mutation or binding to viral oncoprotein seems to release E2F from an inhibitory complex, enabling it to promote the transcription of genes necessary for progression into late G1 and S phases. p28GANK could bind to S6ATPase of the 26S proteasome and RB1, increase E2F activity and destabilize RB1. It was demonstrated that p28GANK was a cellular protooncogene whose functions were more similar to those of the viral oncoprotein E7 and related to protein degradation by 26S proteasome. Furthermore, ubiquitin-proteasome pathway played an important role in regulating cell growth and oncogenic transformation[9,30-34].
p28GANK, as a positive cell proliferation regulatory factor, could promote cell proliferating process. We found that the expression of p28GANK mRNA was not significantly different at indicated times in control liver tissues during liver regeneration, but the expression of p28GANK mRNA possessed two transcripts in regenerating liver tissues after 70% PH, about 1.5 kb and 1.0 kb. The mRNA level of p28GANK increased at 2 h, with a peak time at 72 h, but decreased gradually after 72 h, especially in the 1.5 kb transcript. The protein expression of p28GANK was mainly in the cytoplasm of the regenerating hepatocytes, strongly positive local expression of p28GANK in the cytoplasm and nucleus of regenerating hepatocyte near the central region at 24 h after 70% PH, strongly positive diffuse expression of p28GANK in the cytoplasm and nucleus of regenerating hepatocyte at 48 h, and the expression of p28GANK decreased gradually at 72 h. These demonstrated the mRNA and protein levels of p28GANK occurred a curved changes, because three mitotic replicative waves occurred from 18 to 72 h, the two peak times occurred at 24 h and 36-38 h. Moreover, the two peak times of DNA synthesis occurred at 24 h and 36-48 h. All these suggest that p28GANK might be involved in the whole process of hepatocyte proliferation and the expression of p28GANK restores to its basic level with the termination of liver regeneration. It is necessary to be studied further whether it is the effect of p28GANK on hepatocyte proliferation through S6ATPase-p28GANK-Rb-E2F1 pathway or through other pathways during liver regeneration and interaction with positive or negative cytokines, protooncogenes and anti-oncogenes associated with liver regeneration.