Published online Jan 15, 2003. doi: 10.3748/wjg.v9.i1.9
Revised: May 23, 2002
Accepted: July 26, 2002
Published online: January 15, 2003
AIM: To characterize the gene expression profiles in different stages of carcinogenesis of esophageal epithelium.
METHODS: A microarray containing 588 cancer related genes was employed to study the gene expression profile at different stages of esophageal squamous cell carcinoma including basal cell hyperplasia, high-grade dysplasia, carcinoma in situ, early and late cancer. Principle component analysis was performed to search the genes which were important in carcinogenesis.
RESULTS: More than 100 genes were up or down regulated in esophageal epithelial cells during the stages of basal cell hyperplasia, high-grade dysplasia, carcinoma in situ, early and late cancer. Principle component analysis identified a set of genes which may play important roles in the tumor development. Comparison of expression profiles between these stages showed that some genes, such as P160ROCK, JNK2, were activated and may play an important role in early stages of carcinogenesis.
CONCLUSION: These findings provided an esophageal cancer-specific and stage-specific expression profiles, showing that complex alterations of gene expression underlie the development of malignant phenotype of esophageal cancer cells.
- Citation: Zhou J, Zhao LQ, Xiong MM, Wang XQ, Yang GR, Qiu ZL, Wu M, Liu ZH. Gene expression profiles at different stages of human esophageal squamous cell carcinoma. World J Gastroenterol 2003; 9(1): 9-15
- URL: https://www.wjgnet.com/1007-9327/full/v9/i1/9.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i1.9
Cancer development is a complex multi-step process, involving various genetic and epigenetic changes. Progress of phenotypes from normal to advanced carcinoma is controlled by a transcriptional hierarchy that coordinates the action of hundreds of genes. Conventional approaches investigating one or several candidate genes at a time can not show the whole story of carcinogenesis. The generation of vast amounts of DNA sequence information, coupled with advances in technologies developed for the experimental use of such information, allows the description of biological processes from a view of global genetic perspective. One such technology, DNA microarray, permits simultaneous monitoring of thousands of genes[1,2]. Fuller et al[3] and Sgroi et al[4] have used this new technique in analyzing gene expression profiles in human glioblastoma, human breast cancer and matched normal tissues. However, little is known about the exact expression changes in each stage of tumorigenesis, which will help us identify the exact series of events that leads to the initiation and progression of cancer development.
In this study, esophageal cancer was chosen as a model to analyze the changes of expression profiles in different stages of carcinogenesis. By using microarray membranes, expression of 588 known cellular genes were profiled in normal esophageal tissues, basal cell hyperplasia, high-grade dysplasia, carcinoma in situ, early and advanced cancer. Our results revealed some genes differently expressed in a certain stage, and some kept up or down regulated in all stages toward cancer.
Biopsy specimens were collected from the individuals who were more than 35 years old, underwent endoscopy examination in a screening for early cases of esophageal cancer in Henan province in North China, where has the highest incidence rate of this fatal cancer in the world. Agreements have been obtained from all individuals informed consents issued. In each case, 4 pieces of tissues with the size of 0.01 cm3 were separately removed from the mucosa, two of them were instantly frozen and kept in liquid nitrogen until use, and the other two underwent pathologic diagnosis according to the criteria of Riddell and associates. Specimens of normal esophagus, basal cell hyperplasia, high-grade dysplasia, carcinoma in situ, and early carcinoma were obtained in this study. Every tissue specimen was mostly composed of normal or abnormal epithelial cells.
Cancer and adjacent almost normal tissues with size of about 0.5 cm3 were collected from patients with primary esophageal cancer in Cancer Hospital of the Chinese Academy of Medical Sciences after informed consent was obtained. Fresh samples were dissected manually to remove mixed connective tissue and stored in liquid nitrogen. Pathological diagnosis showed that the cancers were from squamous cell at medium to high grade differentiation, the matched almost normal mucosa did not show any invasion of cancer cells.
Human cancer cDNA expression array membrane was purchased from Clontech Laboratories Inc. (Palo Alto, CA), each membrane contains of 588 well characterized genes along with 9 housekeeping genes as internal control. During this study, membranes of the same lot were used to ensure the reproducibility.
The biopsy specimens of the same histological diagnosis were pooled and the total RNA of different kinds of samples was extracted with Micro RNA isolation kit (Stratagene, La Jolla, CA). Before labeling, 5 μg total RNA of each type was treated with 2 μl DNase I (10 unit/ml, Boehringer Mannheim, Mannheim, Germany), 1 μl RNasin (40 units/μl, Promega) at 37 °C for 15 min to remove contaminated DNA.
Following the recommendation of the manufacturer, 32P-labelled cDNA probes were generated by reverse transcription from 5 μg of RNA sample in the presence of [α-32P]dATP (3000 ci/mmol, Du Pont). Each cDNA probe was then hybridized to a membrane at 65 °C overnight. The membranes were washed twice in 2 × SSC, 0.5% SDS at 65 °C for 30 min, then twice in 0.2 × SSC, 0.5% SDS at 65 °C for 30 min, then exposed to X-ray film at -80 °C for 2-3 days.
Images of individual autoradiography for each stage of carcinogenesis were digitalized by Fluor-STM MultiImager System (Bio-Rad Laboratories, Inc.) The hybridization pattern of arrays was analyzed and compared using AtlasImageTM 1.01 software (Clontech). To normalize the relative gene expression, beta-actin and alpha tubulin were used as internal references whose expression level was found stable in different developmental stages in our study. These genes were preferred also because they are conventionally used as internal references for measuring gene expression levels in Northern blot, RNase protection and semi-quantitative RT-PCR.
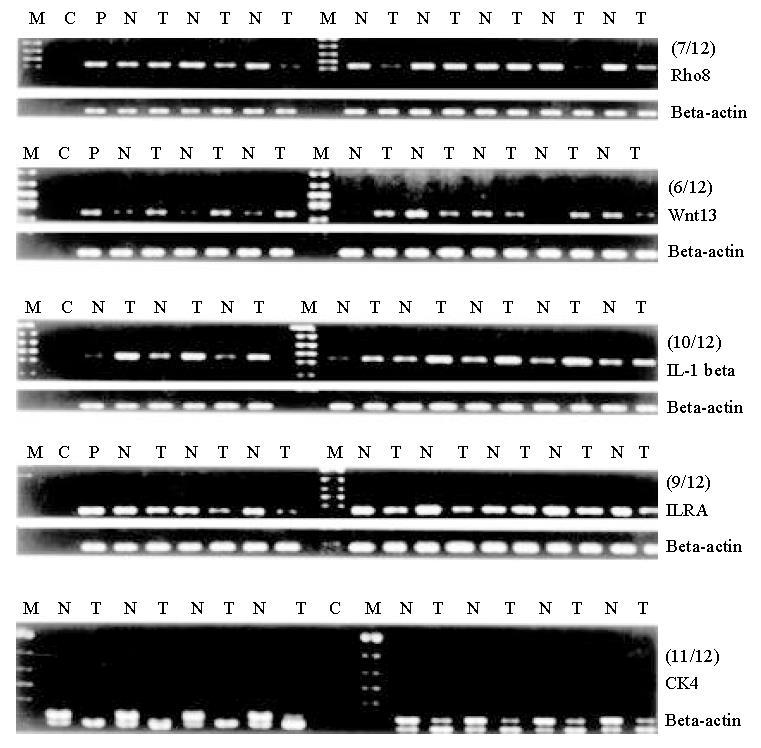
To validate the expression pattern identified on the expression arrays, 5 genes were randomly picked and semi-quantitative RT-PCR was performed to confirm their differential expression with cDNA template from esophageal cancer and adjacent almost normal tissues. First strand cDNA was synthesized from 5 μg RNase-free DNase treated total RNA using the SuperscriptTM Preamplification system for First Strand cDNA Synthesis (Life Technologies, Inc) as described by the manufacturer. An aliquot of 1/20 of the reverse transcribed product was used as template in the following PCR amplification. Reactions undergone in 25 μl total volume containing 1 × PCR buffer, 1.5 mmol/L MgCl2, 200 μmol/L dNTPs, 1.5 U Taq Polymerase (Life Technologies, Inc) and 40 μmol/L gene specific primers under the conditions: 94 °C 5 min, followed by 25-28 cycles each of 94 °C 20 s, 58 °C 30 s, 72 °C 1 min. Amplification of beta actin with same aliquot of cDNA template was used as internal reference to determine the relative gene expression.
Gene specific primers: Rho8 5’-GGACACTTCGGGTTCTCCTTAC-3’, 5’-TGTGGCTCTCTGTGATTTGTTC-3’; IL-1 beta 5’-GCAGAAAGGGAACAGAAAGGTT-3’, 5’-AAGGAGGCACACCAGTCCAAAT-3’; Interleukin 1 receptor antagonist 5’-ACTCTCCTCCTCTTCCTGTTCC-3’, 5’-GCTTGTCCTGCTTTCTGTTCTC-3’; Cytokeratin 4 5’-GGGAACAAAAGCATCTCCAT-3’, 5’-ATCTCAGGGTCAATCTCCAC-3’; Wnt-13 5’-GCCAAAGTTAGATGGGACAAAG-3’, 5’-TTGAACAGGCAGCAAGTAAGC-3’.
A principle component analysis was used to explain the variance-covariance structure of the gene expressions in 6 stages of tumor development. The purpose of principle component analysis is data reduction, and interpretation through a few linear combinations of the original variables (gene expressions)[5].
Principle component analysis starts with k variables which represent the expression profiles of k genes. The t th element of the j variable corresponds to the expression level of the j th gene in the t stage of the tumor development. Principle component analysis intends to replace the k variables by p principle components where p is smaller than k, while minimizing loss of information. The principle component is a linear combination of the variables x1, x2, ..., xk and can be constructed by the eigenvector ei (i = 1, 2, .. k) of the covariance matrix V of the variables x1, x2, ..., xk, where the eigenvector ei of the matrix V is defined as any vector satisfying the equation V ei = li ei. and li is called the corresponding eigenvalue associated with the eigenvector ei. The elements of the eigenvector measure the importance (loading score) of the genes to the principle component and the eigenvalue measures the variance of the corresponding principle component explaining variations of expressions of a linear combination of genes represented by the principle component. The first principle component with the largest eigenvalue has the largest variance, the second principle component with the second largest eigenvalue has the second largest variance and so on. It can be shown that li is equal to the variance of the i th principle component.
Although k variables are required to reproduce the total system variability, often much of this variability can be accounted for by a small number of principle components with larger eigenvalues (variances) which explain large proportions of variations of gene expressions underlying some biological processes. Principle component analysis was used to identify a set of genes which may contribute to the tumor development.
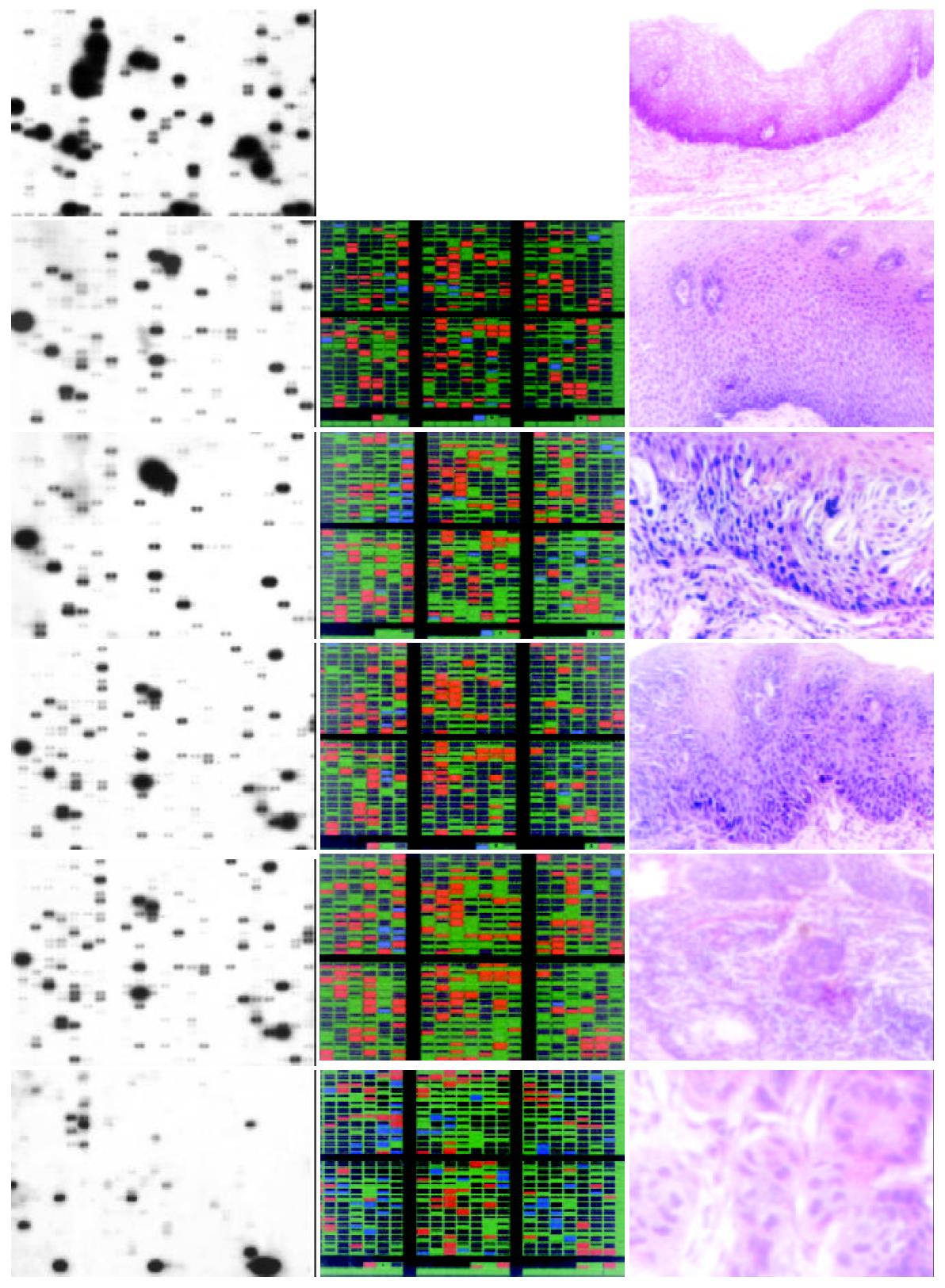
High quality total RNA was isolated from biopsy specimens. The RNA was reverse-transcribed into 32P-labeled cDNA probe and hybridized to AtlasTM Human Cancer cDNA expression array to profile the expression of 588 known genes. The hybridization patterns were imaged by the Fluor-STM MultiImage System and analyzed by AtlasImageTM 1.01 software. Genes with adjusted intensity difference > 20000 or ratio > 2 between two stages were supposed to be regulated differentially. Figure 1 showed the cDNA array images along with color charts indicating up-regulated genes with red, down-regulated ones with blue, and non-changed with green.
There were more than 100 genes up or down regulated substantially in every abnormal stage when compared to human normal mucosa. In stage of basal cell hyperplasia II, there were 122 genes up regulated and 17 down regulated; in stage of high grade dysplasia, there were 134 genes up regulated and 33 down regulated; in stage of squamous cell carcinoma in situ, there were 114 genes up regulated and 10 down regulated; in stage of early cancer, there were 169 genes up regulated and 13 down regulated; In stage of late cancer, there were 77 genes up regulated and 60 down regulated. As shown in the color charts of Figure 1 the types as well as expression level of genes are different among each stage.
To further investigate the reliability of our array data, we randomly picked 5 differentially expressed genes and measured the expression of the genes in the paired cancer and adjacent almost normal tissues using RT-PCR. Figure 2 showed that the different expression pattern of each of the five genes as determined by RT-PCR were similar to those observed with cDNA array in more than half of 12 pairs of matched tissues (squamous esophageal cancer and matched adjacent almost normal tissue), confirming the reliability of our array data. Our observed correlation between the cDNA array and RT-PCR are consistent with that observed by others[6,7].
Principle component analysis showed that there were 5 principle components in the 6 stages of cancer development. The three principle components with eigenvalues 233.52, 121.59 and 101.34 respectively, accounted 92% of total variations of gene expressions in 6 stages. Table 1 and Table 2 showed a set of genes with largest loading score (negative or positive) in the first three principle components in the ascending order. The loading scores are highly correlated with the expression level of genes. In Table 1 and Table 2 we also listed the ratio of the gene expressions in the corresponding stages of the tumor development. From Table 1 and Table 2 we can see that the first principle component roughly describes the changes of the gene expression level in the first and last stage of tumor development. Nine genes in the upper of the table were up-regulated in the stages of basal cell hyperplasia and early cancer, meanwhile seven genes in the bottom of the table were down-regulated in these stages. Genes GAF, CAPR, CASP8, BMP5, MIF and MYC may play important roles in the esophagus tumorigenesis. GAF (Glia-activating factor) is a novel heparin-binding growth factor purified from the culture supernatant of a human glioma cell line with growth-stimulating effect on glial cells in vitro. GAF domains are ubiquitous motifs present in cyclic GMP (cGMP)-regulated cyclic nucleotide phosphodiesterases and form a new class of cyclic nucleotide receptors distinct from the regulatory domains of cyclic nucleotide-regulated protein kinases and ion channels[8], the role of GAF in tumor development has not been characterized yet; CAPR is cadherin-associated protein related, also named as alpha 2 catenin, the abnormal expression of adhesion molecules (E-cadherin, alpha-catenin) has been reported as markers of high malignant potential in esophageal cancer[9], and it was suggested that alpha catenin has prognostic significance in colorectal and prostate cancer[10]; The human CASP8 gene, whose product is also well known as caspase 8, encodes an interleukin-1beta converting enzyme (ICE)-related cysteine protease that is activated by the engagement of several different death receptors. Caspase 8 is a cysteine protease regulated in both a death-receptor-dependent and -independent manner during apoptosis, it is well characterized in apoptosis and tumor development[11]; BMP5 (Bone morphogenetic protein-5) is a signaling molecule which have the ability to induce ecotopic bone when placed under the skin of an animal[12]; MIF (macrophage migration inhibitory factor) is involved in tumorigenesis via promotion of angiogenesis[13], it is an ubiquitary cytokine whose expression has been investigated in tumors, showing a correlation between tumor aggressiveness and production of this protein by neoplastic cells. MIF is known to function as a cytokine, hormone, and glucocorticoid-induced immunoregulator, it is likely that MIF may function as a novel growth factor that stimulates incessant growth and invasion of melanoma concomitant with neovascularization; Myc is a well known oncogene which was involved in most of tumorigenesis, and obviously it plays an important role in esophageal cancer development.
| Component 1 | Component 2 | ||||
| Gene | Ratio ELCA/LaCa | Ratio BCH/N | Gene | Ratio BCH/N | Ratio DYS/CAIN |
| GAF | 131 | 16 | PCTK1 | 122 | 8 |
| CAPR | 582 | 25 | DAXX | 5 | 95 |
| CASP8 | 743 | 37 | P70 | 7 | 62 |
| MMP16 | 143 | 17 | CCNH | 174 | 87 |
| DCC Precursor | 316 | 12 | CD153 antigen | 70 | 31 |
| Tenascin-R | 742 | 21 | TLAA | 9 | 39 |
| ZAP70 | 157 | 83 | XPG | 90 | 26 |
| IL-13 | 110 | 22 | Apoptosis Inhibitor Survivin | 3 | 107 |
| TRAIL receptor | 66 | 82 | Bcl-1 oncogene | 122 | 19 |
| LaCa/ELCA | N/BCH | N/BCH | CAIN/DYS | ||
| BMP5 | 284 | 170 | MERLIN | 3 | 411 |
| MIF | 79 | 72 | LERK-8 | 31 | 334 |
| N-myc | 14 | 110 | DSG2; HDGC | 355 | 395 |
| RPSA | 19 | 68 | MMP11 | 2 | 271 |
| RBL2 | 33 | 18 | CDK2 | 13 | 51 |
| RAD52 | 27 | 9 | CLK1 | 60 | 798 |
| RBQ1 | 15 | 55 | MDMX | 3 | 225 |
| Gene | Ratio N/BCH | Ratio DYS/BCH |
| Bcl-2L8 | 803 | 227 |
| SRC1 | 333 | 141 |
| Muscle cadherin precursor | 299 | 111 |
| Neurogenic locus notch protein | 488 | 104 |
| CASP3 | 232 | 92 |
| BCL2A1; GRS protein | 133 | 665 |
| Type 1 cytoskeletal 10 protein | 302 | 370 |
| TRAF5 | 83 | 650 |
| Transcription factor E2F5 | 615 | 30 |
| MAP kinase p38 | 147 | 86 |
| BCH/N | BCH/DYS | |
| KRT7 | 2 | 105 |
| GRB-IR | 5 | 96 |
| GAS1 | 4 | 210 |
| BMP8 | 21 | 21 |
| DVL | 12 | 187 |
| CAM-PDE1B | 18 | 18 |
| Apotosis regulator bcl-2 | 60 | 59 |
The second component involves three stages of the tumor development: basel cell hyperplasia, dysplasia and carcinoma in situ. Table 1 and Table 2 listed nine genes which were up-regulated and eight genes which were down-regulated in the stages of basel cell hyperplasia and dysplasia. The third principle component reflects the changes of the gene expression level in the transition from normal to basel cell hyperplasia and from basel cell hyperplasia to dysplasia. The table listed genes may play an important role in these transitions.
According to their specificity to stages, differentially expressed genes fall into two main categories. The first category contains genes that are up or down regulated in the same way in all 4 abnormal stages. 77 genes belong to this category, including G1/S-specific cyclin C, proliferating cyclic nuclear antigen (PCNA), E2F dimerization partner 2, Wnt5A, retinoic acid receptors (epsilon, gamma 1, beta), early growth response protein 1 (hEGR1), etc. Part of these genes are listed in Table 3. Most of which are genes related to cell proliferation or differentiation. These expression profiles are accordant with the generality of characteristic phenotype of all of the 4 abnormal stages, i.e. decrease in extent of differentiation and increase in rate of proliferation.
| Position | Gene Name |
| Up Regulated Genes | |
| A2k | G1/S-specific cyclin C |
| A4i | ERK5 |
| A4j | ERK6 |
| A5b | ERK activator kinase 1 |
| A5l | transcription factor DP2 |
| B3g | caspase-8 precursor |
| B3i | caspase-9 precursor |
| B3j | caspase-10 precursor (CASP10) |
| C1a | DNA-dependent protein kinase |
| C2c | DNA-repair protein XRCC1 |
| C2j | RAD1 |
| C4b | Wnt-5A |
| C4k | DVL1 |
| C6m | CCK4 |
| C7k | retinoic acid receptor epsilon |
| C7l | retinoic acid receptor gamma 1 |
| C7m | retinoic acid receptor beta |
| D4b | integrin alpha 8 |
| D4j | integrin beta 7 precursor |
| D4k | integrin beta 8 precursor |
| D5g | ezrin; cytovillin 2; villin 2 |
| D5j | CD56 antigen |
| D7f | placenta growth factor 1 |
| E1f | MMP9 |
| E2a | MMP18 |
| F3j | early growth response protein 1 |
| F4l | IL-6 |
| F5e | IL-13 |
| F5k | interferon gamma precursor |
| F5l | leukocyte interferon-inducible peptide |
| Down regulated Genes | |
| C6c | insulin-like growth factor binding protein 2 (IGFBP2P2) |
| F4d | interleukin-1 receptor antagonist protein precursor (IL-1RA; IRAP) |
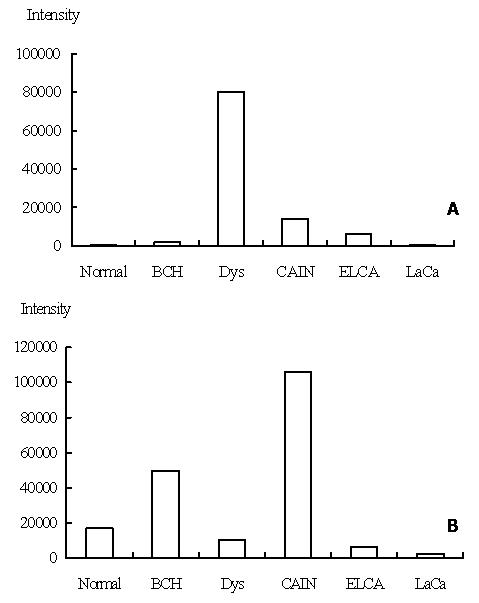
The second category contains genes that were up or down regulated in particular stages, some of them seemed to be activated in early stages of carcinogenesis, p160ROCK is one of such genes. Figure 3 showed the expression level of p160ROCK in 5 stages. P160 ROCK peaked its expression level in the stage of high grade dysplasia, down-regulated rapidly after this stage, recovered its expression level in early and late cancer, which revealed that this gene was transcriptionally activated at pre-cancerous stage.
p160ROCK belongs to a family of Rho-associated serine/threonine kinase isozymes and has been identified as a new class of Rho effectors. Its main function is participating the reorganization of the cytoskeletal, and plays an important role in signal transduction from Rho to cytoskeletal[14] and also get involved in cell motility and morphological changes[15]. In human hepatocellular carcinoma cells, dominant active p160ROCK transfectants showed increased motility, and dominant negative p160ROCK transfectants showed reduced motility under stimulation. Furthermore, implantation of dominant negative p160ROCK transfectants resulted in a reduced metastatic rate in vivo compared with the parent cells or a control transfectant[16]. In this study, p160ROCK was found transcriptionally activated at the stage of high grade dysplasia, a pre-malignant stage, which imply that activation of the gene may be one of the early events during progress of esophageal carcinogenesis and may be one of the events responsible for morphological changes and increase in cell motility in early stages of carcinogenesis.
JNK2 is another representative gene which peaked its expression level in the stage of carcinoma in situ. Its expression levels in the 5 stages were shown in Figure 3. JNK2, c-Jun N-terminal kinase 2, also named as protein kinase mitogen-activated 9, is a proline-directed serine/threonine kinase activated by dual phosphorylation on threonine and tyrosine residues in response to a wide array of extracellular stimuli. Multiple research showed that JNK2 plays a critical role in coordinating the cellular response to stress and has been implicated in regulating cell growth and transformation, antisense JNK2 induced growth inhibition which correlated with significant apoptosis[17]. It was shown that JNK2 plays an important role in cell transformation and carcinogenesis. In this study, JNK2 was found activated at stage of carcinoma in situ, and down regulated in early and late cancer, which showed that this may also be an early events of carcinogenesis and one of the forces pushing the cell from pre-malignancy to malignancy.
Tumorigenesis is a complex and multistage process with many genes involved in. As a step toward understanding the complicated changes between normal and malignant cells, this report focused on gene expression profile variations among normal and abnormal esophageal epithelium tissues. Analyzing alterations of gene expression profiles in different stages of neoplasia is necessary for establishing the preventive, diagnostic, therapeutic, and prognostic potential of each related gene. To illustrate the mechanisms controlling malignant changes at molecular level may provide a further understanding of tumorigenesis, as well as new approaches in early detection and treatment of esophageal cancer. Furthermore, since it is impossible to get tissue samples of different stages from one patient, we got the biopsy specimens from different patients and pooled the samples with same pathologic diagnosis together to avoid the individual differences, and semi-quantitative RT-PCR was performed to confirm the reliability of the data. Moreover, we observed that many genes expressed abnormally in this complicated process and those changes mainly involved in cell proliferation, apoptosis, DNA repair, growth factors and cytokines. These genes and their expression alteration constitute a molecular atlas that is stage-specific and esophageal cancer-specific, and possibly being an important supplement to the traditional morphological diagnosis.
Principle component analysis identified a set of genes which plays important role in different stages of tumor development. Although many genes were related to tumorigenesis and development of esophageal cancer, further investigation is still needed to elucidate which gene (s) is the most critical one (s) to this complex process.
Edited by Xu JY
| 1. | DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1283] [Cited by in RCA: 1150] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 2. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5103] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 3. | Fuller GN, Rhee CH, Hess KR, Caskey LS, Wang R, Bruner JM, Yung WK, Zhang W. Reactivation of insulin-like growth factor binding protein 2 expression in glioblastoma multiforme: a revelation by parallel gene expression profiling. Cancer Res. 1999;59:4228-4232. [PubMed] |
| 4. | Sgroi DC, Teng S, Robinson G, LeVangie R, Hudson JR, Elkahloun AG. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59:5656-5661. [PubMed] |
| 5. | Johoson RA, Wichern DW; Applied multivariate statistical analysis, Prentice Hall, Inc., New York. 1982;. |
| 6. | Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Boguski MS. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1467] [Cited by in RCA: 1313] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 7. | Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 507] [Cited by in RCA: 492] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 8. | Ho YS, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288-5299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 244] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 9. | Krishnadath KK, Tilanus HW, van Blankenstein M, Hop WC, Kremers ED, Dinjens WN, Bosman FT. Reduced expression of the cadherin-catenin complex in oesophageal adenocarcinoma correlates with poor prognosis. J Pathol. 1997;182:331-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 10. | Aaltomaa S, Lipponen P, Ala-Opas M, Eskelinen M, Kosma VM. Alpha-catenin expression has prognostic value in local and locally advanced prostate cancer. Br J Cancer. 1999;80:477-482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 594] [Cited by in RCA: 558] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 12. | DiLeone RJ, Marcus GA, Johnson MD, Kingsley DM. Efficient studies of long-distance BMP5 gene regulation using bacterial artificial chromosomes. Proc Natl Acad Sci USA. 2000;97:1612-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 13. | Shimizu T, Abe R, Nakamura H, Ohkawara A, Suzuki M, Nishihira J. High expression of macrophage migration inhibitory factor in human melanoma cells and its role in tumor cell growth and angiogenesis. Biochem Biophys Res Commun. 1999;264:751-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 156] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 14. | Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895-898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1179] [Cited by in RCA: 1249] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 15. | Hirose M, Ishizaki T, Watanabe N, Uehata M, Kranenburg O, Moolenaar WH, Matsumura F, Maekawa M, Bito H, Narumiya S. Molecular dissection of the Rho-associated protein kinase (p160ROCK)-regulated neurite remodeling in neuroblastoma N1E-115 cells. J Cell Biol. 1998;141:1625-1636. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 374] [Cited by in RCA: 405] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 16. | Genda T, Sakamoto M, Ichida T, Asakura H, Kojiro M, Narumiya S, Hirohashi S. Cell motility mediated by rho and Rho-associated protein kinase plays a critical role in intrahepatic metastasis of human hepatocellular carcinoma. Hepatology. 1999;30:1027-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 119] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Potapova O, Gorospe M, Dougherty RH, Dean NM, Gaarde WA, Holbrook NJ. Inhibition of c-Jun N-terminal kinase 2 expression suppresses growth and induces apoptosis of human tumor cells in a p53-dependent manner. Mol Cell Biol. 2000;20:1713-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 105] [Article Influence: 4.2] [Reference Citation Analysis (0)] |