Published online Aug 15, 2002. doi: 10.3748/wjg.v8.i4.668
Revised: November 22, 2001
Accepted: November 29, 2001
Published online: August 15, 2002
AIM: The loss of heterozygosity (LOH) on tumor suppressor genes is believed to play a key role in carcinogenesis of colorectal cancer. In this study, we analyzed the LOH at 5 loci on the long arm of chromosome 22 in sporadic colorectal cancer to identify additional loci involved in colorectal tumorigenesis.
METHODS: Five polymorphic microsatellite markers were analyzed in 83 cases of colorectal and normal DNA by PCR. PCR products were eletrophoresed on an ABI 377 DNA sequencer; Genescan 3.1 and Genotype 2.1 software were used for LOH scanning and analysis. Comparison between LOH frequency and clinicopathological data were performed by χ² test. P < 0.05 was considered as statistically significant.
RESULTS: The average LOH frequency on chromosome 22q was 28.38%. The region between markers D22S280 and D22S274 (22q12.2-q13.33) exhibited relatively high LOH frequency. The two highest LOH loci with frequencies of 35.09% and 34.04% was identified on D22S280 (22q12.2-12.3) and D22S274 (22q13.32-13.33).8 cases showed LOH at all informative loci, suggesting that one chromosome 22q had been completely lost. On D22S274 locus, LOH frequency of rectal cancer was 50% (9/18), which was higher than that of proximal colon cancer (12%, 2/17) (P = 0.018). The frequency of distal colon cancer was 42% (5/12), also higher than that of proximal colon cancer. But there was no statistical significance. Putting both the tumors in distal colon and rectum together into consideration, the frequency, 47% (14/30), was higher than that of proximal colon cancer (P = 0.015), suggesting the mechanism of carcinogenisis was different in both groups.
CONCLUSIONS: This study provided evidence for the involvement of putative tumor suppressor genes related to the sporadic colorectal carcinoma on chromosome 22q. The tumor-suppressor-gene (s) might locate on the 22q12.2-12.3 and/or 22q13.32-13.33.
- Citation: Zhou CZ, Peng ZH, Zhang F, Qiu GQ, He L. Loss of heterozygosity on long arm of chromosome 22 in sporadic colorectal carcinoma. World J Gastroenterol 2002; 8(4): 668-673
- URL: https://www.wjgnet.com/1007-9327/full/v8/i4/668.htm
- DOI: https://dx.doi.org/10.3748/wjg.v8.i4.668
Colorectal cancer is one of the three leading causes of worldwide cancer mortality. The progression of the cancer is thought to result from an accumulation of genetic alteration at numerous loci controlling growth and proliferation. As a model for both multistep and multipathway carcinogenesis, colorectal neoplastic progression provides paradigms of both oncogenes and tumor suppressor genes[1,2]. The loss of heterozygosity (LOH) on tumor suppressor genes is believed to be one of the key steps to carcinogenesis of colorectal cancer[3]. The loss of one allele at a specific locus is caused by a deletion mutation or loss of a chromosome from a chromosome pair[4]. When this occurs at a tumor suppressor gene locus where one of the alleles is already abnormal, it can result in neoplastic transformation. In colorectal cancers, frequent allelic loss has been identified in chromosome 5q (30%), 8p (40%), 17p (75%-80%), 18q (80%), and 22q (20%-30%)[5,6]. Indeed, much has been published on tumor suppressor genes APC, p53, and DCC, which have been localized to chromosome 5q, 17p, and 18q, respectively. The LOH analysis became an effective way to find informative loci and then to find candidate tumor suppressor genes[7,8]. In this study we analyzed the LOH at 5 loci on chromosome 22 in sporadic colorectal cancer to identify additional loci involved in colorectal tumorigenesis.
This study was based on 83 consecutively collected tumors, including 40 males and 43 females, from unrelated patients with colorectal cancer, treated at the surgical department in Shanghai First people's hospital, China, between 1998 and 1999. The patients’s ages ranged from 31 to 84 years with a median of 66. All patients were confirmed by pathology, and were staged by Dukes criterion. Dukes stage A, B, C, D were 8, 21, 40, 14 cases respectively. Well-differentiated adenocarcinoma was 23 cases, moderate differentiated adenocarcinoma was 39, poorly differentiated adenocarcinoma was 6 and mucinous adenocarcinoma was 15. HNPCC patients were ruled out by Amsterdam criteria[9,10]. Each patient gave his or her informed consent for the use of his or her tissue in this study.
DNA Extraction The cancerous and adjacent normal tissues were fresh frozen within 30 min after removed. These tissues were then cut into cubes of approximately 2 mm3 and immediately frozen in liquid nitrogen. DNA was extracted using standard methods with proteinase K digestion and phenol/chloroform purification.
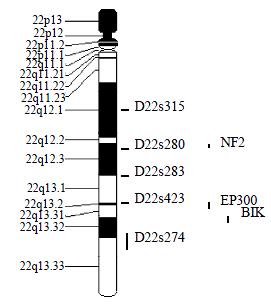
Microsatellite Markers and PCR Five fluorescence-labeled primers for polymorphic microsatellite markers (PE Applied Biosystems Foster city CA, USA), at a density of approximately one marker every 8 cm (Figure 1), was used to amplify matched pairs of normal and tumor DNAs for LOH analysis.
Polymorphic microsatellite markers were analyzed in each patient's tumor and normal DNAs by PCR (GeneAmp PCR System 9700, PE Applied Biosystems Foster city CA, USA). PCR conditions[11] were as follows: 5 μL total volume with approximately 1.4 ng of DNA as a template with 10 × standard buffer, 0.3 μL Mg2+, 0.8 μL deoxynueleotide triphosphetes, 0.3 unit of Hot-start taq polymerase and 0.06 mL of each oligonucleotide primer, with the forward primer fluorescence labeled with HEX, FAM or NED. Cycling conditions consisted of 3 stages: an initial denaturation at 96 °C for 12 min in Stage I; 14 cycles each at 94 °C for 20 s, 63-56 °C for 1 min (0.5 °C decreased per cycle), 72 °C for 1 min in Stage II: 35 cycles each at 94 °C for 20 sec, 56 °C for 1 min, 72 °C for 1 min in stage III.
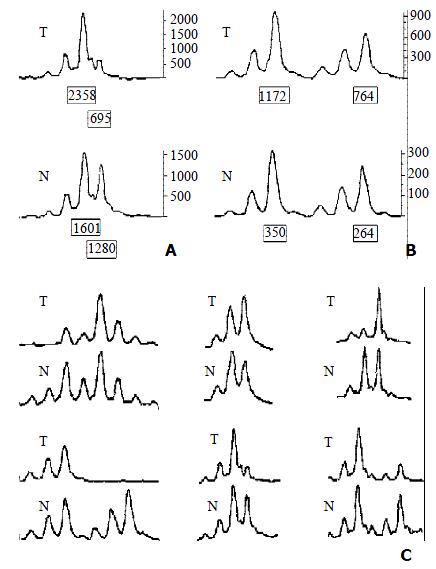
LOH Analysis A portion of each PCR product (0.5 μL) was combined with 0.1 μL of Genescan 500 size standard (PE Applied Biosystems Foster city CA, USA) and 0.9 μL of formamide loading buffer. After denaturation at 96 °C for 5 min, products were eletrophoresed on a 5% polyacrylamide gels on an ABI 377 DNA sequencer (PE Applied Biosystems Foster city CA, USA) for 3 h. Genotype 2.1 software displayed individual gel lanes as electopherograms with a given size, height, and area for each detected fluorescent peak. Stringent criteria were used to score the samples. Alleles were defined as the two highest peaks within the expected size range. A ratio of T1:T2/N1:N2 of less than 0.67 or greater than 1.50 was scored as a loss of heterozygosity (Figure 2). Most amplification of normal DNA produced two PCR products indicating heterozygosity. A single fragment amplified from normal DNA (homozygote) and those PCR reactions in which fragments were not clearly amplified were scored as not informative. The LOH frequency of a locus was equal to the ratio of the number between allelic loss and informative cases. The average LOH frequency of chromosome 22 long arm was the average value of each locus LOH frequency.
Comparison between LOH and clinicopathological data were performed by χ² test. P < 0.05 was considered as statistically significant.
The average LOH frequency at chromosome 22 q was 28.38%. The region between markers D22S280 and D22S274 (22q12.2-q13.33) exhibited relatively high LOH frequency, the two highest LOH loci with frequencies of 35.09% and 34.04% was identified on D22S280 (22q12.2-12.3) and D22S274 (22q13.32-13.33). Of these 83 cases, 8 cases had behaved LOH in all informative loci, suggesting that one chromosome 22q had been completely lost (Table 1, Table 2).
| Locus | Location | LOH case | Normal case | LOH rate (%) | Informative rate (%) |
| D22S315 | 22q12.1 | 12 | 56 | 17.65 | 81.93 |
| D22S280 | 22q12.2-12.3 | 20 | 37 | 35.09 | 68.67 |
| D22S283 | 22q12.3-13.1 | 17 | 43 | 28.33 | 72.29 |
| D22S423 | 22q13.2 | 15 | 41 | 26.79 | 67.47 |
| D22S274 | 22q13.32-13.33 | 16 | 31 | 34.04 | 56.63 |
| No | Gender | Age | Location | Gross Pattern | Size (cm) | Differentiation | Dukes stage |
| 125 | Female | 52 | Sigmoid Colon | Ulcerative | 5.5 × 4 | Moderately | A |
| 128 | Male | 70 | Descending Colon | Ulcerative | 4 × 4.5 | Moderately | C |
| 134 | Female | 70 | Ascending Colon | Massive | 5 × 5.5 | Moderately | C |
| 137 | Female | 76 | Sigmoid Colon | Ulcerative | 6 × 6 | Moderately | C |
| 138 | Female | 66 | Rectum Colon | Ulcerative | 3 × 3 | Well | A |
| 210 | Female | 41 | Ascending Colon | Massive | 5 × 4 | Well | A |
| 220 | Female | 79 | Ascending Colon | Massive | 7 × 4 | Well | B |
| 223 | Male | 63 | Rectum | Encroaching | 6 × 6.5 | Moderately | D |
On D22S274 locus, LOH frequency of rectal cancer was 50% (9/18), which was higher than that of proximal colon cancer (12%, 2/17) (P = 0.018). The frequency of distal colon cancer was 42% (5/12), which was also higher than the frequency of proximal colon cancer. But there was no statistical significance. Putting both the tumors in distal colon and rectum together into consideration, the frequency, 47% (14/30), was higher than that of proximal colon cancer (P = 0.015). There was no association between LOH of each marker on chromosome 22q and other clinicopathological data (patient sex, age, tumor size, growth pattern or Dukes stage). It indicated that LOH of 22q was a common phenomenon in sporadic colorectal cancer (Table 3).
| D22S315 | D22S280 | D22S283 | D22S423 | D22S274 | |||||||
| N | L | N | L | N | L | N | L | N | L | ||
| Gender | Male | 28 | 6 | 19 | 7 | 21 | 7 | 22 | 5 | 13 | 8 |
| Female | 28 | 6 | 18 | 13 | 22 | 10 | 19 | 10 | 18 | 8 | |
| Age | > 60 | 41 | 9 | 24 | 19 | 31 | 15 | 30 | 11 | 22 | 13 |
| ≤ 60 | 15 | 3 | 13 | 1 | 12 | 2 | 11 | 4 | 9 | 3 | |
| Location | Proximal Colon | 21 | 4 | 13 | 7 | 18 | 5 | 14 | 6 | 15 | 2 |
| Distal Colon | 13 | 4 | 12 | 4 | 11 | 5 | 12 | 6 | 7 | 5b | |
| Rectum | 22 | 4 | 12 | 9 | 14 | 7 | 15 | 3 | 9 | 9a | |
| Gross Pattern | Massive | 23 | 4 | 16 | 8 | 19 | 7 | 16 | 6 | 11 | 5 |
| Ulcerative | 21 | 7 | 15 | 9 | 14 | 7 | 18 | 7 | 14 | 8 | |
| Encroaching | 12 | 1 | 6 | 3 | 10 | 3 | 7 | 2 | 6 | 2 | |
| Size | ≥ 5 (cm) | 25 | 7 | 14 | 11 | 20 | 8 | 18 | 10 | 17 | 5 |
| < 5 (cm) | 31 | 5 | 23 | 9 | 23 | 9 | 23 | 5 | 14 | 11 | |
| LN Metastasis | LN (+) | 36 | 9 | 26 | 13 | 31 | 11 | 26 | 11 | 24 | 7 |
| LN (-) | 20 | 3 | 11 | 7 | 12 | 6 | 15 | 4 | 7 | 9 | |
| Differentiation | Well | 15 | 3 | 8 | 6 | 13 | 3 | 11 | 5 | 7 | 5 |
| Moderately | 28 | 3 | 17 | 10 | 19 | 10 | 24 | 5 | 15 | 7 | |
| Poorly | 3 | 3 | 4 | 2 | 3 | 1 | 1 | 2 | 3 | 3 | |
| Mucinous | 10 | 3 | 8 | 2 | 8 | 3 | 5 | 3 | 6 | 1 | |
| Dukes stage | A | 3 | 3 | 1 | 3 | 3 | 2 | 3 | 2 | 2 | 4 |
| B | 17 | 1 | 10 | 4 | 9 | 4 | 12 | 2 | 5 | 5 | |
| C | 26 | 4 | 18 | 11 | 22 | 9 | 19 | 7 | 16 | 5 | |
| D | 10 | 4 | 8 | 2 | 9 | 2 | 7 | 4 | 8 | 2 | |
During tumorigenesis, loss of the wild-type allele (inherited from the non-mutation-carrying parents) is frequently observed at the appropriate locus. To date, loss of heterozygosity (LOH) on tumor suppressor genes plays a key role in colorectal cancer transformation[3]. And LOH analysis of sporadic colorectal cancer can promote the discovery of unknown tumor suppressor genes[7,8]. In this study, LOH scanning was carried out in 83 sporadic colorectal cancer samples with 5 highly polymorphic markers and analyzed by Genotyper software, that is, by the ratio of the fluorescence intensity of allele, with an effort to identifying additional loci involved in colorectal tumorigenesis.
In this study, the average LOH frequency of chromosome 22q is 28.38%, which is consistent with previous observations[5,6]. D22S280 (22q12.2-12.3) and D22S274 (22q13.32-13.33) exhibited highest LOH frequency, indicating that colon cancer related tumor suppressor gene (s) located in this region and perhaps near D22S280 or/and D22S274. The previous study showed that 22q13.1-13.3 behaved high LOH frequency in sporadic colorectal cancer[12,13]. This study is consistent with the finding, and also showed that 22q12.2-12.3 existed obvious LOH phenomenon, which was similar to the pancreatic adenocarcinomas[14].
By database referring, there are three candidate tumor-suppressor genes related to colon cancer, NF2 (22q12.2)[15], EP300 (22q13)[16], NBK/BIK (22q13.3)[17] on 22q12.2-13.33. NF2 gene was confirmed to be a tumor-suppressor-gene in neurofibromatosis type 2 syndrome[18-21]. And NF2 gene inactivation was also reported in NF2-assoicated tumor and some sporadic cancer[22-28]. NF2 gene encodes a 587-amino acid protein with striking similarity to several members of the ERM family of proteins proposed to link cytoskeletal components with proteins in the cell membrane, including moesin, ezrin, and radixin. Because of the resemblance to these 3 proteins, Trofatter et al[29] called the NF2 gene product merlin. Stokowski et al[30] found that 80% of the merlin mutants significantly altered cell adhesion by causing cells to detach from the substratum. They stated that such changes in cell adhesion might be an initial step in the pathogenesis of NF2. And some scholars also studied the relationship between NF2 gene and sporadic colorectal cancer and found that NF2 gene was probably involved in some colorectal tumors, but was not the critical chromosome 22q tumor suppressor gene involved in colon tumorigenesis[31,32]. The results of this study suggested that there might be colon cancer related candidate tumor-suppressor-gene (s) on 22q12.2 and NF2 was the only known tumor-suppressor-gene in this region. So it was needed to evaluate the effect of NF2 gene on colorectal carcinogenisis, and the new tumor suppressor gene involved in colon tumorigenesis can not be excluded absolutely. There were 2 putative tumor-suppressor genes on 22q13.2-13.31, EP300 and NBK/BIK[33]. P300 is the number of the retinoblastoma protein family. Stein et al[34] supposed that p300 acted as a tumor suppressor firstly. Recently, Hasan et al[35] proposed the p300 might participate in chromatin remodeling at DNA lesion sites to facilitate proliferating cell nuclear antigen (PCNA) function in DNA repair synthesis. Muraoka et al[36] raised the possibility that inactivation of EP300 gene was involovled in the genesis or progression of colorectal cancer. And Gayther et al[37] described EP300 mutations that predicted a truncated protein in 6 (3%) of 193 epithelial cancers analyzed and provided the first evidence that it behaved as a classic tumor suppressorgene. But EP300 mutation was rare in colorectal cancer tissue. So Castells et al[12] presumed that NBK/BIK gene, a proapoptotic BCL-2 family member[38-41], acted as a candidate gene in that region. However, SSCP sequencing analysis excluded mutations of this gene. The results of this study showed that the LOH frequency was also high on 22q13, especial on 22q13.32-13.33, suggesting that colorectal cancer associated candidate tumor-suppressor genes are likely to locate on chromosome 22q13.
Yana et al[13] indicated that loss of heterozygosity correlated with Dukes staging. Iino et al[42] suggested that allelic loss on 22q was significantly associated with the presence of lymph node metastasis. However, Castells et al[12] did not support their opinion. This result also agreed with Castells’s study and suggested that there was no association between LOH of each marker on chromosome 22q and Dukes staging. However, we found on D22S274 locus, LOH frequency of rectal cancer was higher than that of proximal colon cancer. And the frequency of the tumors in distal colon and rectum was also higher than that of the tumors in proximal colon cancer. Now it was admitted that the mechanism of carcinogenisis in distal colon was different from that in proximal colon[43-45]. And the mechanism in rectal cancer was also different from that in the proximal colon[46]. Distal colonic cancer displayed a higher frequency of 17p and 18q allelic loss, p53 accumulation[47], c-myc expression and aneuploidy[48]. Right-sided tumors are more often diploid[48] and of the microsatellite instability (MSI) phenotype. Rectal cancers showed significantly more expression of p53 than that in proximal colon cancer[46], which was similar with distal colonic cancer. This study showed the D22S274 LOH was more frequent in distal colon and rectal cancer than in proximal colon ones, which proved the mechanism of carcinogenisis in distal colon and rectum was not completely same as that in the proximal colon.
Allelic loss on chromosome 22q is present not only in colorectal cancer but also in carcinomas of the ovary (55%)[49-52], breast (40%)[53-55], pancreatic endocrine (30%)[27], oral cavity (40%)[56], stomach[57], liver[58], lung[59], head and neck[60], and insulinoma[61]. After microsatellite DNA analysis, several attempts were made to identify a region of deletion and eventually the tumor suppressor genes responsible for these neoplasms. Allelic deletions were restricted to D22S274 (22q13) marker in oral squamous cell carcinoma[56]. Handel-Fernandez et al[14] found that LOH region presented between marker D22S444 and D22S922 (22q13.2-q13.3), indicating the locations of tumor suppressor genes that may contribute to the devolopment of pancreatic cancer. Considering these results, it is tempting to hypothesize that the same putative tumor suppressor genes might be involved in these different neoplastic processes. Further LOH scanning with high-density microsatellite markers in the region and the study of the relationship between these genes and the cacinogenesis of sporadic colorectal cancer may provide much more genetic information and find the potential tumor suppressor genes.
Edited by Zhang JZ
| 1. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8087] [Cited by in RCA: 8006] [Article Influence: 228.7] [Reference Citation Analysis (1)] |
| 2. | Hardy RG, Meltzer SJ, Jankowski JA. ABC of colorectal cancer. Molecular basis for risk factors. BMJ. 2000;321:886-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 3. | Kataoka M, Okabayashi T, Johira H, Nakatani S, Nakashima A, Takeda A, Nishizaki M, Orita K, Tanaka N. Aberration of p53 and DCC in gastric and colorectal cancer. Oncol Rep. 2000;7:99-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 4. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643-649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2881] [Cited by in RCA: 2818] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 5. | Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R. Allelotype of colorectal carcinomas. Science. 1989;244:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 898] [Cited by in RCA: 879] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 6. | Weber TK, Conroy J, Keitz B, Rodriguez-Bigas M, Petrelli NJ, Stoler DL, Anderson GR, Shows TB, Nowak NJ. Genome-wide allelotyping indicates increased loss of heterozygosity on 9p and 14q in early age of onset colorectal cancer. Cytogenet Cell Genet. 1999;86:142-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 7. | Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1354] [Cited by in RCA: 1422] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 8. | Kinzler KW, Nilbert MC, Vogelstein B, Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hamilton SR, Hedge P, Markham A. Identification of a gene located at chromosome 5q21 that is mutated in colorectal cancers. Science. 1991;251:1366-1370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 512] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 9. | Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC). Dis Colon Rectum. 1991;34:424-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1357] [Cited by in RCA: 1277] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 10. | Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453-1456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1765] [Cited by in RCA: 1691] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 11. | Peng Z, Ling Y, Bai S. [Loss of heterozygosity on chromosome 3 in sporadic colorectal carcinoma]. Zhonghua Yixue Zazhi. 2001;81:336-339. [PubMed] |
| 12. | Castells A, Ino Y, Louis DN, Ramesh V, Gusella JF, Rustgi AK. Mapping of a target region of allelic loss to a 0.5-cM interval on chromosome 22q13 in human colorectal cancer. Gastroenterology. 1999;117:831-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 13. | Yana I, Kurahashi H, Nakamori S, Kameyama M, Nakamura T, Takami M, Mori T, Takai S, Nishisho I. Frequent loss of heterozygosity at telomeric loci on 22q in sporadic colorectal cancers. Int J Cancer. 1995;60:174-177. [PubMed] |
| 14. | Handel-Fernandez ME, Nassiri M, Arana M, Perez MM, Fresno M, Nadji M, Vincek V. Mapping of genetic deletions on the long arm of chromosome 22 in human pancreatic adenocarcinomas. Anticancer Res. 2000;20:4451-4456. [PubMed] |
| 15. | Arai E, Ikeuchi T, Karasawa S, Tamura A, Yamamoto K, Kida M, Ichimura K, Yuasa Y, Tonomura A. Constitutional translocation t (4; 22) (q12; q12.2) associated with neurofibromatosis type 2. Am J Med Genet. 1992;44:163-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 859] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 17. | Verma S, Budarf ML, Emanuel BS, Chinnadurai G. Structural analysis of the human pro-apoptotic gene Bik: chromosomal localization, genomic organization and localization of promoter sequences. Gene. 2000;254:157-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 18. | Seizinger BR, Rouleau G, Ozelius LJ, Lane AH, St George-Hyslop P, Huson S, Gusella JF, Martuza RL. Common pathogenetic mechanism for three tumor types in bilateral acoustic neurofibromatosis. Science. 1987;236:317-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 187] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 19. | Evans DG, Trueman L, Wallace A, Collins S, Strachan T. Genotype/phenotype correlations in type 2 neurofibromatosis (NF2): evidence for more severe disease associated with truncating mutations. J Med Genet. 1998;35:450-455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 134] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Legoix P, Legrand MF, Ollagnon E, Lenoir G, Thomas G, Zucman-Rossi J. Characterisation of 16 polymorphic markers in the NF2 gene: application to hemizygosity detection. Hum Mutat. 1999;13:290-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 21. | Bruder CE, Hirvelä C, Tapia-Paez I, Fransson I, Segraves R, Hamilton G, Zhang XX, Evans DG, Wallace AJ, Baser ME. High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray-CGH. Hum Mol Genet. 2001;10:271-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 105] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Kluwe L, Mautner VF. Mosaicism in sporadic neurofibromatosis 2 patients. Hum Mol Genet. 1998;7:2051-2055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 60] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Kluwe L, Friedrich RE, Hagel C, Lindenau M, Mautner VF. Mutations and allelic loss of the NF2 gene in neurofibromatosis 2-associated skin tumors. J Invest Dermatol. 2000;114:1017-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Fukasawa T, Chong JM, Sakurai S, Koshiishi N, Ikeno R, Tanaka A, Matsumoto Y, Hayashi Y, Koike M, Fukayama M. Allelic loss of 14q and 22q, NF2 mutation, and genetic instability occur independently of c-kit mutation in gastrointestinal stromal tumor. Jpn J Cancer Res. 2000;91:1241-1249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Gutmann DH, Hirbe AC, Haipek CA. Functional analysis of neurofibromatosis 2 (NF2) missense mutations. Hum Mol Genet. 2001;10:1519-1529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Cheng JQ, Lee WC, Klein MA, Cheng GZ, Jhanwar SC, Testa JR. Frequent mutations of NF2 and allelic loss from chromosome band 22q12 in malignant mesothelioma: evidence for a two-hit mechanism of NF2 inactivation. Genes Chromosomes Cancer. 1999;24:238-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 27. | Schulze KM, Hanemann CO, Müller HW, Hanenberg H. Transduction of wild-type merlin into human schwannoma cells decreases schwannoma cell growth and induces apoptosis. Hum Mol Genet. 2002;11:69-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Quezado MM, Middleton LP, Bryant B, Lane K, Weiss SW, Merino MJ. Allelic loss on chromosome 22q in epithelioid sarcomas. Hum Pathol. 1998;29:604-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 29. | Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 891] [Cited by in RCA: 858] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 30. | Stokowski RP, Cox DR. Functional analysis of the neurofibromatosis type 2 protein by means of disease-causing point mutations. Am J Hum Genet. 2000;66:873-891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 31. | Arakawa H, Hayashi N, Nagase H, Ogawa M, Nakamura Y. Alternative splicing of the NF2 gene and its mutation analysis of breast and colorectal cancers. Hum Mol Genet. 1994;3:565-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 73] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 32. | Rustgi AK, Xu L, Pinney D, Sterner C, Beauchamp R, Schmidt S, Gusella JF, Ramesh V. Neurofibromatosis 2 gene in human colorectal cancer. Cancer Genet Cytogenet. 1995;84:24-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Zou Y, Peng H, Zhou B, Wen Y, Wang SC, Tsai EM, Hung MC. Systemic tumor suppression by the proapoptotic gene bik. Cancer Res. 2002;62:8-12. [PubMed] |
| 34. | Stein RW, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421-4427. [PubMed] |
| 35. | Hasan S, Hassa PO, Imhof R, Hottiger MO. Transcription coactivator p300 binds PCNA and may have a role in DNA repair synthesis. Nature. 2001;410:387-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 131] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong JM, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565-1569. [PubMed] |
| 37. | Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, Chin SF, Daigo Y, Russell P, Wilson A, Sowter HM. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24:300-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 434] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 38. | Boyd JM, Gallo GJ, Elangovan B, Houghton AB, Malstrom S, Avery BJ, Ebb RG, Subramanian T, Chittenden T, Lutz RJ. Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene. 1995;11:1921-1928. [PubMed] |
| 39. | Han J, Sabbatini P, White E. Induction of apoptosis by human Nbk/Bik, a BH3-containing protein that interacts with E1B 19K. Mol Cell Biol. 1996;16:5857-5864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 147] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 40. | Radetzki S, Köhne CH, von Haefen C, Gillissen B, Sturm I, Dörken B, Daniel PT. The apoptosis promoting Bcl-2 homologues Bak and Nbk/Bik overcome drug resistance in Mdr-1-negative and Mdr-1-overexpressing breast cancer cell lines. Oncogene. 2002;21:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 41. | Marshansky V, Wang X, Bertrand R, Luo H, Duguid W, Chinnadurai G, Kanaan N, Vu MD, Wu J. Proteasomes modulate balance among proapoptotic and antiapoptotic Bcl-2 family members and compromise functioning of the electron transport chain in leukemic cells. J Immunol. 2001;166:3130-3142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 94] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 42. | Iino H, Fukayama M, Maeda Y, Koike M, Mori T, Takahashi T, Kikuchi-Yanoshita R, Miyaki M, Mizuno S, Watanabe S. Molecular genetics for clinical management of colorectal carcinoma.17p, 18q, and 22q loss of heterozygosity and decreased DCC expression are correlated with the metastatic potential. Cancer. 1994;73:1324-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 43. | Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 505] [Cited by in RCA: 550] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 44. | Distler P, Holt PR. Are right- and left-sided colon neoplasms distinct tumors? Dig Dis. 1997;15:302-311. [PubMed] |
| 45. | Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 151] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 46. | Kapiteijn E, Liefers GJ, Los LC, Kranenbarg EK, Hermans J, Tollenaar RA, Moriya Y, van de Velde CJ, van Krieken JH. Mechanisms of oncogenesis in colon versus rectal cancer. J Pathol. 2001;195:171-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 47. | Soong R, Grieu F, Robbins P, Dix B, Chen D, Parsons R, House A, Iacopetta B. p53 alterations are associated with improved prognosis in distal colonic carcinomas. Clin Cancer Res. 1997;3:1405-1411. [PubMed] |
| 48. | Lanza G, Maestri I, Dubini A, Gafa R, Santini A, Ferretti S, Cavazzini L. p53 expression in colorectal cancer: relation to tumor type, DNA ploidy pattern and short-term survival. Am J Clin Pathol. 1996;105:604-612. [PubMed] |
| 49. | Bryan EJ, Watson RH, Davis M, Hitchcock A, Foulkes WD, Campbell IG. Localization of an ovarian cancer tumor suppressor gene to a 0.5-cM region between D22S284 and CYP2D, on chromosome 22q. Cancer Res. 1996;56:719-721. [PubMed] |
| 50. | Englefield P, Foulkes WD, Campbell IG. Loss of heterozygosity on chromosome 22 in ovarian carcinoma is distal to and is not accompanied by mutations in NF2 at 22q12. Br J Cancer. 1994;70:905-907. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 51. | Lin H, Pizer ES, Morin PJ. A frequent deletion polymorphism on chromosome 22q13 identified by representational difference analysis of ovarian cancer. Genomics. 2000;69:391-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 52. | Bryan EJ, Thomas NA, Palmer K, Dawson E, Englefield P, Campbell IG. Refinement of an ovarian cancer tumour suppressor gene locus on chromosome arm 22q and mutation analysis of CYP2D6, SREBP2 and NAGA. Int J Cancer. 2000;87:798-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 53. | Allione F, Eisinger F, Parc P, Noguchi T, Sobol H, Birnbaum D. Loss of heterozygosity at loci from chromosome arm 22Q in human sporadic breast carcinomas. Int J Cancer. 1998;75:181-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 54. | Iida A, Kurose K, Isobe R, Akiyama F, Sakamoto G, Yoshimoto M, Kasumi F, Nakamura Y, Emi M. Mapping of a new target region of allelic loss to a 2-cM interval at 22q13.1 in primary breast cancer. Genes Chromosomes Cancer. 1998;21:108-112. [PubMed] |
| 55. | Castells A, Gusella JF, Ramesh V, Rustgi AK. A region of deletion on chromosome 22q13 is common to human breast and colorectal cancers. Cancer Res. 2000;60:2836-2839. [PubMed] |
| 56. | Miyakawa A, Wang XL, Nakanishi H, Imai FL, Shiiba M, Miya T, Imai Y, Tanzawa H. Allelic loss on chromosome 22 in oral cancer: possibility of the existence of a tumor suppressor gene on 22q13. Int J Oncol. 1998;13:705-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 57. | Sud R, Wells D, Talbot IC, Delhanty JD. Genetic alterations in gastric cancers from British patients. Cancer Genet Cytogenet. 2001;126:111-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 58. | Takahashi K, Kudo J, Ishibashi H, Hirata Y, Niho Y. Frequent loss of heterozygosity on chromosome 22 in hepatocellular carcinoma. Hepatology. 1993;17:794-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 59. | Nishioka M, Kohno T, Takahashi M, Niki T, Yamada T, Sone S, Yokota J. Identification of a 428-kb homozygously deleted region disrupting the SEZ6L gene at 22q12.1 in a lung cancer cell line. Oncogene. 2000;19:6251-6260. [PubMed] |
| 60. | Poli-Frederico RC, Bergamo NA, Reis PP, Kowalski LP, Zielenska M, Squire JA, Rogatto SR. Chromosome 22q a frequent site of allele loss in head and neck carcinoma. Head Neck. 2000;22:585-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 61. | Wild A, Langer P, Ramaswamy A, Chaloupka B, Bartsch DK. A novel insulinoma tumor suppressor gene locus on chromosome 22q with potential prognostic implications. J Clin Endocrinol Metab. 2001;86:5782-5787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |