INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide, which ranks eighth in frequency among human cancer especially in Asia, Africa and South Europe, accounting for an estimated one million deaths annually. Men are afflicted at least twice as often as women. Although HCC ranks eighth in frequency among cancers worldwide, it is the sixth among men and eleventh among women[1]. It is one of the few human cancers for which an underlying etiology can be identified in most cases, and has a background of chronic inflammatory liver diseases caused by viral infection that induces cirrhosis[2]. HCC is unusual in patients with primary biliary cirrhosis but common when cirrhosis is secondary to chronic viral hepatitis[3,4]. However, it is not clear how this disorders result in HCC. Some tumor suppressor genes, such as RB and p53, may play a significant role in hepatocarcinogenesis[5,6]. Besides this, growth factor including transforming growth factor-α (TGF-α) have been implicated in the development of HCC[7]. It is early-stage metastasis that causes lower 5-year survival rate. However, these genetic changes do not precisely reflect the biological nature of cancer cells or the clinical characteristics of HCC patients. So, the molecular mechanism of metastasis of HCC is currently unknown. Cell adhesion and migration are fundamental properties of the metastasis. Changes in cell adhesion and migration are very important in the formation of tumors, and invasion and metastasis by neoplasms[8].
The integrin family of cell adhesion receptors plays a fundamental role in the processes involved in cell division, differentiation and movement. The extracellular domains of integrin alpha/beta heterodimers mediate cell-matrix and cell-cell contacts while their cytoplasmic tails associate with the cytoskeleton. Integrins are capable of transducing information in a bidirectional manner and the beta subunit is now recognized to play an important role in this process. Recent studies have led to the identification of a ligand-binding region on the beta subunit similar to that already characterized on some alpha subunits, and sequences in the cytoplasmic tails of the beta subunits that interact with cytoskeletal and signaling components. Adhesive events can also play a role in the progression of all four major classes of human disease--neoplastic, inflammatory, traumatic and infectious--and the specific nature of integrin adhesion mechanisms make them an attractive target for therapy[9].
Tumor development and progression involves a cascade of genetic alterations. Techniques frequently used to study gene expression alterations, such as RT-PCR, differential display PCR and Northern blot analysis, have their limitations: some need large amounts of RNA, others are time-consuming and can only study a small number of genes simultaneously. Hence, analysis of the expression profiles of a large number of genes in clinical HCC materials is an essential step toward clarifying the detailed mechanisms of metastasis and discovering target molecules for the development of novel therapeutic drugs.
The cDNA microarray technology, recently developed, enables investigators to study the gene expression profile and gene activation in thousands of genes and sequences[10-15]. This technique allows the large-scale comparison of multiple genes in a single hybridization and has been used successfully to explore the gene expression profiles in some kinds of carcinomas and other diseases[16-20].
In this study, we used cDNA expression microarray technology containing 17 intergrin genes related to cell adhesion and invasion that are differentially expressed in human HCC. This large body of information not only furthers an understanding of the mechanism of metastasis but also reveals novel features of intergrin genes and identifies potential candidates for cancer metastasis detection and HCC therapy.
MATERIALS AND METHODS
Tissues and specimen
Twenty-four pairs of primary HCC and corresponding noncancerous liver tissues without cirrhosis were obtained with informed consent from patients who underwent hepatectomy at the First Clinical College of Harbin Medical University and Cancer Hospital of Chinese Academy of Medical Science. Cancerous and noncancerous tissues were enucleated separately from the tumourous and nontumourous part of resected liver. The normal tissue block was from the distal incision tissue. Histopathological diagnosis and classification were performed by the same pathologist. The specimen were immediately frozen in liquid nitrogen.
RNA isolation and purification
Total RNA were extracted from frozen tissues in Trizol (Life Technologies Inc, Gaitherbur, MD) according to the manufacturer’s instructions. Normal liver tissues and HCC tissues were made in spices and homogenized in Trizol solution (1 mL/100 mg). Homogenates were incubated for 15 min on ice, and then a 1/5 volume of choloform was added to the homogenates. After vigorous agitation for 5 min, the inorganic phase was separated by centrifugation at 12000 × g for 20 min at 4 °C. RNA were then precipitated in the presence of 1 volume of isopropanol and centrifugated at 10000 × g for 15 min at 4 °C. RNA pellets were washed with 70% ice-cold ethanol and then dissolved in diethyl pyrocarbonate (DEPC) - treated H2O. Total RNA concentration and quantity was assessed by absorbency at 260 nm using an Nucleic Acid and Protein Analyzer (Beckman 470, USA).
cDNA microarray
Atlas human cancer cDNA expression array (7742-1) were purchased from Clonech Laboratories Inc (Palo Alto, USA). The membrane contained 10 ng of each gene-specific cDNA from 588 known genes and 9 housekeeping genes. Several plasmid and bacteriophage DNAs and blank spots are also included as negative and blank controls to confirm hybridization specificity. These genes analyzed in this study related to cell adhesion and motility, includeing laminin, collagen, fibronectin and intergrin. A complete list of the intergrin genes with the array positions and GeneBank accession number spotted on the array is available at Clontech’s web site (http://www.clontech.com).
cDNA synthesis, labeling and purification
Total RNA was reverse-transcribed into cDNA and labeled with α-32P dCTP using SuperscriptTM. Preamplification System for First Strand cDNA Synthesis (Life Technologies, Gaithersburg, MD). Before labeling, 5 μg total RNA of each sample was treated with 2 μL DNase I (10 units/μL, Boehringer Mannheim, Germany), 1 μL RNasin (40 units/μL, Promega, Madison, USA) at 37 °C for 15 min to remove contaminated DNA. For each labeling, the treated 5 μg total RNA was raised in volume to 6 μL with diethyl pyrocarbonate (DEPC)-treated H2O and first incubated with 4 μL oligo dT (0.5 μg/μL, Life Technologies, USA) at 70 °C for 10 min. Then, 6 μL 5 × first strand reaction buffer, 1 μL 0.1 MDTT (Life Technologies, Gaithersburg, MD), 1.5 μL dNTP mixture containing dATP, dGTP, and dTTP at 20 μM (Promega, Madison, USA), 10 μL α-32P dCTP (10 μCi/μL, NEN Life Science, Boston, MA) and 1.5 μL SuperScript II reverse transcriptase (200 units/μL, Life Technologies, USA) were added and incubated at 37 °C for 90 min. The labeled first strand cDNA probes were purified by Spin 200 column (Clontech, Palo Alto, CA) to remove the unincorporated nucleotides. After purification, labeled cDNAs were denatured in a boiling water bath for 3 min before use.
Membrane hybridization and exposure
Five milliliters ExpressHyb hybridization solution (Clontech, Palo Alto, CA) and 5 μg Cot-1 DNA (Life Technologies, USA) were added to the tube containing Atlas human cancer cDNA expression array, which was pre-hybridized at 68 °C for 2 hr. Then different incubated probes were added to different tubes and hybridization was performed at 68 °C for 18 hr in a rolling bottle. The membranes were washed twice at 50 °C in 2 × SSC, 0.1% SDS for 20 min, once at 68 °C in 1 × SSC, 0.5% SDS for 20 min, once at room temperature in 0.5 × SSC, 1% SDS for 20 min and 0.5 × SSC 10 min. Membrane were then exposed to X-ray films (Fuji Films, Tokyo, Japan) at -70 °C for 1-3 d.
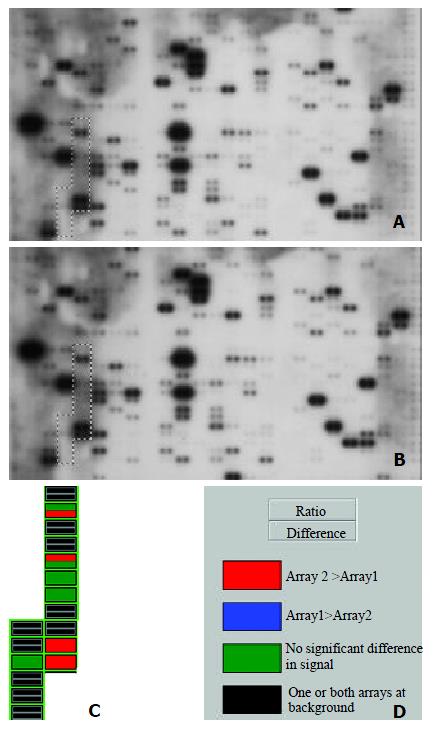
Image and Analysis
The images were scanned with Fluor-S MultiImager (Bio-Rad, Hercules, CA) and saved as TIFF format files. The TIFF images were imported into the AtlasImage analysis software Version 1.01 a (Clontech, Palo Alto, CA) and analyzed step by step with the guide. Housekeeping genes Ubiquitin and GAPDH were selected for normalization, because their expression was constant in cancer array hybridization system. Then the normalized intensity of each spot representing a unique gene expression level was acquired. Genes were considered to be up-regulated when the intensity ratio was ≥ 1.5 and the difference was ≥ 10000 between the expression of HCC tisssues and normal liver tissues.
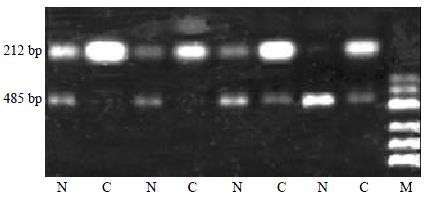
Semi-quantitative RT-PCR
To confirm the cDNA array results, semi-quantitative RT-PCR of 24 pairs of HCC tissues and normal liver tissues was performed for intergrin beta1 displaying expression alterations. Five micrograms of total RNA in each hybridization sample was used to synthesize the first strand cDNA with SuperScript Preamplification System For First Strand cDNA Synthesis kit (Life Technologies, USA). Then 1 μL product was used as the template to amplify specific fragments in a 25 μL reaction mixture under the following conditions: denaturation at 95 °C (3 min); 22 cycles of 94 °C (25 s), 60 °C (25 s) and 72 °C (45 s); then 72 °C extension (3 min). In each PCR reaction, primers for the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene were used as an internal control. GAPDH, which is considered a housekeeping gene, showed similar expression levels by cDNA array analysis for the control and tumor samples. The 5 μL RT-PCR reaction product was analyzed by electrophoresis on a 1.5% agarose gel. The electrophoresis images were scanned by Fluor-S MultiImager (Bio-Rad, USA) and the original intensity of every specific band was quantityed with the software Multi-Analyst (Bio-Rad, USA). The data were compared after being normalized by the intensity of GAPDH. After normalization, the adjusted intensities were calculated for the amplified gene products, and the ratios were calculated. The sequences of the PCR primer pairs of intergrin beta1 and GAPDH were designed using Primer3 Internet software program. (Whitehead Institute, Boston, USA). Their specificity was confirmed by a BLAST Internet software assisted search for a nonredundant necleotide sequence database (National Library of Medicine, Bathesda, MD, USA).
Primer sequences were as follows: GAPDH, forward primer 5'-ACCACAGTCCATGCCATCAC-3' and reverse primer 5'-TCCACCACCCTGTTGCTGTA-3' Intergrin beta1, forward primer 5'-GGAGTCAGGCAAATGCTCTC-3' and revese primer 5'-GCTAAGGCCACTTCTGCATC-3'
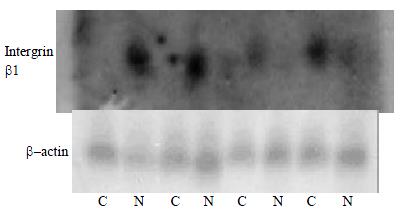
Northern blot
RNAs of HCC and normal liver tissues were seperated by electrophoresis in a 1.5% agrose gel containing 2.2 M formaldehyde in pairs then transferred onto a nylon membrane (Zeta-Probe, Bio-Rad, USA) by capillary action under 10 × SSPE. RNA was permanently attached to the membrane by UV illumination for 150 s (GS Gene Linker, Bio-Rad, USA). RNA intactness was estimated by comparing the intensities of the 28S and 18S ribosomal RNA bands. Hybridization was performed overnight in a rolling bottles containing 8 mL of hybridization buffer (5 × SSPE, 5 × Denhardt's solution, 0.5% SDS, 0.2 mg/mL heat-denatured salmon sperm DNA, and 50% formamide) and the hybridization probe that was obtained by PCR amplification. The primers were as follow: β-actin, forward primer 5'-CGTCTGGACCTGGCTGGCCGGGACC-3' and reverse primer 5'-CTAGAAGCATTTGCGGTGGACGATG-3' Intergrin beta1, forward primer 5'-GTGTGGCCCAAGACA GTTCT-3' and reverse primer 5'-GGTTACCCCACCCTCTGACT-3' α-32P-labeled cDNA probes were synthesized using Primer-a-Gene Random Labeling Kit (Promega, USA) and following the protocol.
The membranes were washed twice at room temperature in 2 × SSPE, 0.1% SDS for 10 min, once at 42 °C in 1 × SSPE, 0.1% SDS for 15 min and once at 50 °C in 0.5 × SSPE, 0.1% SDS for 20 min. Membrane were then exposed to X-ray films (Fuji Films, Tokyo, Japan) at -70 °C for 24-48 h.
DISCUSSION
In this study, we have investigated the gene expression profiles in human HCC and adjacent noncancerous liver tissues using Atlas human cancer array which contains 17 genes that were classified according to intergrin. cDNA array technology is used to examine simultaneously the expression of specific genes on a single hybridization. Although human genome project has generated a large-scale sequence data for a great number of genes, the biological functions of such genes remain to be deciphered. It is very important to define differential gene expression profiles of tumors and normal tissues before understanding the functional significance of specific gene products. Although expression analysis techniques such as differential display polymerase chain reaction (DD-PCR), northern blot, and serial analysis of gene expression (SAGE) and RT-PCR have been widely used in the past, these studies are time-consuming and can only be used to deal with a limited number of genes. Thus a systematic approach to examine large number of genes simultaneously is required. Microarray techniques have been developed in this condition[21].
Microarray technique was firstly reported in 1995 by Schena and Brown[10], which allows simultaneous parallel expression analysis of thousands of genes. There is considerable interest in the potential application of cDNA microarray analysis for gene expression profiles in human cancers[22]. Such information might be useful for tumor classification, elucidation of key factors in tumor metastasis, and identification of genes which might be useful for diagnostic purposes or as therapeutic targets[23-25]. The analytical principle of human cDNA expression array is based on reverse northern blot hybridization. DNA fragments representing human genes are immobilized in duplicate onto a nylon membrane. Each cDNA fragment is 200-500 bp long and is selected as a unique sequence. This sequence is without a poly (A) tail, repetitive elements or homologous sequence to avoid cross-hybridization and nonspecific binding of a cDNA probe. α-32P or α-33P labeled cDNA probes generated by RT-PCR of total or messenger RNA samples are then hybridized to a microarray membrane. The hybridization image can be obtained by autoradiography after a high-stringency wash. The final result will be exported by a specific software designed to analyze the membrane[26,27].
All intergrin genes identified were up-regulated in human HCC. Integrins are cell surface receptors that mediate the physical and functional interactions between a cell and its surrounding extracellular matrix (ECM). Expressed as heterodimers, the specific alpha or beta chains that constitute the integrin receptor determine the repertoire of ECM proteins to which a specific integrin may bind. While classically, the role ascribed to integrins has been that of anchoring cells to the ECM, the more contemporary spectrum of integrin function greatly exceeds that of mere cell adhesion. Recent reports have demonstrated that the interaction between the ECM and cell surface integrins leads to intracellular signaling events that affect cell migration, proliferation, and survival, which in the context of neoplastic cells, can translate directly into the malignant phenotype. Indeed, the role of specific integrins in tumorigenesis has been demonstrated in numerous cancer types. Integrins are a major family of cell adhesion molecules involved in cell-cell and cell-extracellular matrix interactions[28].
Each integrin is a heterodimeric glycoprotein composed of an alpha and a beta subunit. Intergrin beta 7 was mainly reported in hematology. The beta 7 chain of integrin forms heterodimers with the alpha 4 or alpha E chains. Mature B cells express alpha 4 beta 7, which is a receptor for vascular cell adhesion molecule-1 and fibronectin[29]. The alpha 4 beta 7 integrin acts as a gut homing receptor. High levels of beta 7 mRNA was restricted to intra-epithelial mucosa T-lymphocytes[30,31]. Beta 7 integrins have been implicated in the autoimmune process in rheumatoid arthritis[32]. It also involved in hematologic tumors that alpha 4 beta 7 ligand in the adhesion of in vivo activated multiple myeloma blood B cell adhesion to bone marrow fibroblasts. The adhesion properties distinguish them from normal B cells. Although the malignant status of these cells is as yet undefined, their adhesion properties implicate multiple myeloma blood B cells in migratory spread of the disease[33].
Integrin beta 8 subunit mRNA has been shown to be expressed at higher levels in the central nervous system than in other organs[34]. The present study demonstrates that αvβ8 is a growth-inhibitory molecule and provides the first evidence for an in vivo function of the divergent integrin subunit β8[35,36] and β8 integrin subunit is growth inhibitory in epithelial cells and that the diverge nt β8 cytoplasmic domain is sufficient to confer growth inhibition. It was not known why intergrin beta 8 were up-regulated in HCC.
Intergrin alpha 8 subunit always associates with beta1 subunit. Integrin beta-1 is essential for epithelial-mesenchymal interactions and can interact with a number of alpha subunits. The integrin alpha 8 beta 1 has been reported to bind to fibronectin, vitronectin, tenascin-C and osteopontin in cell adhesion or neurite outgrowth assays[37-39]. Integrin alpha8 beta1 may help regulate kidney development and other morphogenetic processes[40]. Mice with a mutation in the alpha8 gene do not express the integrin alpha8 beta1 and exhibit profound deficits in kidney morphogenesis[39]. Alpha 8 beta 1 mediates neurite outgrowth of embryonic sensory and motor neurons on tenascin-C extracellular matrix protein[41]. The integrin family consists of a series of related alpha beta heterodimers involved in a variety of cell-matrix and cell-cell adhesion functions.
Integrin beta1 is essential for epithelial-mesenchymal interactions and can interact with a number of alpha subunits[42]. Cell adhesion to fibronectin can be mediated by the interaction of an integrin (alpha 5 beta 1) with the Arg-Gly-Asp-Ser (RGDS)-containing cell adhesion region of fibronectin. The beta 1 subunit plays an important role in binding and assembly of exogenous fibronectin, perhaps by participation in the organization, regeneration, or cycling of the assembly site rather than by a direct interaction with fibronectin[43]. Alpha 5 beta 1 and alpha 2 beta 1 integrins play an important role in transducing mechanical stimuli into intracellular signals. The integrin alpha 4 beta 1 (also known as very late antigen-4, VLA4) interacts with the immunoglobulin superfamily member vascular cell adhesion molecule-1 (VCAM-1), and with an alternatively spliced form of fibronectin[44]. The integrin alpha 3 beta 1 is a multiligand extracellular matrix receptor (VLA-3) found in many cell types. It may function as a receptor for fibronectin, laminin, collagen and also for ladsin, epiligrin and entactin[45,46].
To investigate the role of integrins in HCC invasion, we analyzed the relationship between the expression and activity of beta1 integrins. Some studies showed that different types of HCC cells showed various levels of constitutive activity of beta1 integrins as assessed by the TS2/16 requirement in cell adhesion. Remarkably, as a result of in vitro chemoinvasion assay, the levels of constitutive activity of beta1 integrins correlated with the invasive ability of HCC cells[47]. A new human HCC cell line with a highly metastatic potential was established from subcutaneous xenograft of a metastatic model of human HCC in nude mice (LCI-D20) by means of alternating cell culture in vitro and growth in nude mice, which has a high intergrin[48]. But there was still another result showed integrins beta 1 down-regulated in poorly differentiated HCC, whereas relatively high activity in metastatic tumors and the presence of all integrins in cirrhotic liver[49]. Most results showed neoplastic progression of HCCs may be correlated with an aberrant expression of adhesion molecules[50]. Our results showed magnificent up-regulation in HCC compared with adjacent normal liver tissues.
Members of the beta 1 subfamily of integrins contribute to cell adhesion, cytoskeletal organization and signal transduction processes. In some transformed cell lines and tumors, a correlation has been established between the level of expression of the beta 1 and neoplastic behavior. In other instances, normal and neoplastic tissues differ in beta 1 integrin expression or sub-cellular distribution. The level of expression of beta 1 integrins in tumor cells may affect tumor growth properties in several ways, including: (a) effects on anchorage dependence of growth; (b) direction of signaling processes; (c) organization of the extracellular matrix and presentation of matrix bound growth factors; (d) effects on the functions of host defense cells. Thus the interplay between integrin expression and tumor behavior is complex and might be viewed as a series of interactive feedback loops rather than in terms of a straightforward cause and effect relationship[51].
In conclusion, our study demonstrated that cDNA array is a powerful tool to explore gene expression profiles in cancer. The intergrin genes described in this study should therefore provide valuable resources not only for basic research, such as molecular mechanism of metastasis, progression and prognosis, but also for clinical application, such as development of new diagnostic markers and identification of therapeutic intervention in human HCC.